Abstract
TR-700 (torezolid), the active moiety of the novel oxazolidinone phosphate prodrug TR-701, is highly potent against gram-positive pathogens, including strains resistant to linezolid (LZD). Here we investigated the potential of Staphylococcus aureus strains ATCC 29213 (methicillin-susceptible S. aureus [MSSA]) and ATCC 33591 (methicillin-resistant S. aureus [MRSA]) to develop resistance to TR-700. The spontaneous frequencies of mutation of MSSA 29213 and MRSA 33591 resulting in reduced susceptibility to TR-700 at 2× the MIC were 1.1 × 10−10 and 1.9 × 10−10, respectively. These values are ∼16-fold lower than the corresponding LZD spontaneous mutation frequencies of both strains. Following 30 serial passages in the presence of TR-700, the MIC for MSSA 29213 remained constant (0.5 μg/ml) while increasing eightfold (0.25 to 2.0 μg/ml) for MRSA 33591. Serial passage of MSSA 29213 and MRSA 33591 in LZD resulted in 64- and 32-fold increases in LZD resistance (2 to 128 μg/ml and 1 to 32 μg/ml, respectively). Domain V 23S rRNA gene mutations (Escherichia coli numbering) found in TR-700-selected mutants included T2500A and a novel coupled T2571C/G2576T mutation, while LZD-selected mutants included G2447T, T2500A, and G2576T. We also identified mutations correlating with decreased susceptibility to TR-700 and LZD in the rplC and rplD genes, encoding the 50S ribosomal proteins L3 and L4, respectively. L3 mutations included Gly152Asp, Gly155Arg, Gly155Arg/Met169Leu, and ΔPhe127-His146. The only L4 mutation detected was Lys68Gln. TR-700 maintained a fourfold or greater potency advantage over LZD against all strains with ribosomal mutations. These data bring to light a variety of novel and less-characterized mutations associated with S. aureus resistance to oxazolidinones and demonstrate the low resistance potential of torezolid.
Staphylococcus aureus infections pose a serious health threat worldwide. Increasing antibiotic resistance and the prevalence of methicillin (meticillin)-resistant S. aureus (MRSA) in clinical settings have created a demand for novel therapeutic agents. Linezolid (LZD) has a broad spectrum of activity against a variety of gram-positive pathogens, including MRSA, and was the first oxazolidinone antibiotic to gain FDA approval (1). LZD acts through inhibition of protein synthesis via binding to the peptidyl transferase center (PTC) of the 50S ribosomal subunit (37, 65, 68). Despite in vitro studies demonstrating a low resistance potential for LZD (31, 79), soon after its approval in 2000, LZD-resistant (LZDr) MRSA (72) and LZDr, vancomycin (VAN)-resistant enterococci (22) emerged in the clinic. Although rare, resistance has most commonly occurred in patients undergoing long-term LZD therapy (10, 17, 22, 45, 72, 74). Three classes of oxazolidinone resistance mechanisms have been previously characterized: mutations in the domain V region of 23S rRNA genes (69), acquisition of the ribosomal methyltransferase gene cfr (43), and mutations in the rplD gene encoding the 50S ribosomal protein L4 (76).
A variety of 23S rRNA mutations conferring resistance to LZD in S. aureus have been identified, including C2192T (26), G2447T (69), T2500A (45), A2503G (39), T2504C (39), G2505A (50), G2766T (38), and G2576T (72) (Escherichia coli numbering), the most common mutation observed clinically. To date, T2500A and G2576T are the only 23S rRNA mutations that have been documented in clinical S. aureus isolates. 23S rRNA-mediated resistance occurs in a gene dose-dependent fashion, with higher copy numbers of 23S mutant alleles associated with increased resistance (5, 38, 66, 78). Despite evidence of fitness costs associated with some 23S rRNA mutations (5, 8, 44), highly LZDr 23S rRNA homozygous mutant strains of S. aureus (72), Staphylococcus epidermidis (71), and Enterococcus faecalis (60) have been recovered clinically.
A second mechanism of LZD resistance has been identified in staphylococci possessing cfr, a gene encoding a ribosomal methyltransferase (32, 70) which catalyzes methylation at position 8 of nucleotide A2503 in the 23S rRNA domain V region (21). Cfr methylation confers resistance to five classes of 50S ribosomal subunit-targeted antibiotics defined by the PhLOPSA phenotype, including phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (43). Recent outbreaks of LZDr staphylococci possessing the transposon- and plasmid-associated cfr methyltransferase (11, 16a, 32, 46, 47, 70) are particularly problematic due to the potential for rapid gene transfer between strains of both human and animal origins, driven by the use of antibiotics in any of the classes to which resistance is conferred.
The third and least common class of LZD resistance involves mutations in ribosomal protein L4, which have been associated with reduced LZD susceptibility in streptococci (19, 76). L4 interacts closely with the PTC, and mutations are thought to confer antibiotic resistance by perturbing 23S rRNA structure (23).
Despite the characterization of these three primary resistance classes, there have been a number of reports describing LZDr staphylococci, streptococci, enterococci, and mycobacteria of clinical (28, 29, 58) and laboratory (12, 25, 40, 57, 62) origins possessing “unknown” resistance mechanisms. A recent report documents mutations in an endogenous ribosomal methyltransferase (modifying G2445) and altered efflux via upregulation of ABC transporters linked to LZD resistance (19). Such mutations may account for the resistance phenotype of some of the undefined LZDr strains previously observed.
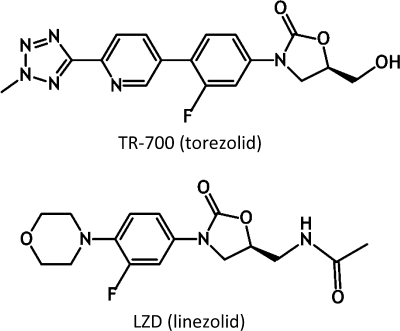
Increasing incidences of clinical resistance to LZD and the potential for rapid horizontal dissemination of cfr have contributed to the demand for expanded-spectrum oxazolidinones (4) such as TR-700 (torezolid), the active antibacterial moiety of the prodrug TR-701 (torezolid phosphate) (Fig. 1) (64). TR-700 demonstrates 4- to 16-fold greater potency than LZD against LZDs and LZDr strains (30, 38, 64). Notably, TR-700 has been shown to be 16-fold more active than LZD against S. aureus Cfr strains (64). Molecular modeling suggests that this is likely due to the smaller size of the hydroxymethyl group compared to the acetamide group of LZD, as well as additional C and D ring interactions with the PTC, allowing TR-700 to bind in the presence of the methylated A2503 base (64). Enhanced PTC binding properties are also thought to contribute to its potency advantage over LZD and may translate into lower frequencies of resistance (64). Preliminary reports suggested that resistance to TR-700 is very low (13, 30), although further work to characterize the frequency of resistance and resistance mechanisms was warranted.
FIG. 1.
TR-700 and LZD chemical structures.
In this study, we investigated the potential for two S. aureus strains to develop resistance to TR-700 and LZD through serial passage and analysis of spontaneous mutation frequencies. Underlying resistance mechanisms were elucidated by sequencing of genes encoding 23S rRNA and ribosomal proteins L3, L4, and L22. Mutations were analyzed structurally by using LZD-bound 50S ribosomal subunit crystallographic data.
(Portions of this work were presented at the 19th European Congress of Clinical Microbiology and Infectious Diseases [J. B. Locke, M. Hilgers, and K. J. Shaw, posters P1103 and P1104], Helsinki, Finland, 16 to 19 May 2009.)
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. aureus strains ATCC 29213 (methicillin-susceptible S. aureus [MSSA]) and ATCC 33591 (MRSA) were cultured aerobically at 37°C on cation-adjusted Mueller-Hinton II agar (MHA; Becton Dickinson, Franklin Lakes, NJ), on sheep blood agar (SBA; tryptic soy agar plus 5% sheep red blood cells; Remel, Lenexa, KS), or in MH broth (MHB). Enumeration of CFU was performed by serially diluting bacteria in phosphate-buffered saline (PBS) and plating on MHA.
Antimicrobial agents.
Solutions of TR-700 (torezolid; Trius Therapeutics, Inc., San Diego, CA), LZD (ChemPacific Corp., Baltimore, MD), VAN (Sigma-Aldrich Corp., St. Louis, MO), chloramphenicol (CHL; Sigma-Aldrich Corp., St. Louis, MO), and tiamulin (TIA; Wako Pure Chemical Industries, Ltd., Richmond, VA) were freshly prepared prior to use in MIC assays or selective medium.
MIC testing.
MIC assays were performed via broth microdilution in accordance with CLSI guidelines (14), and values were determined visually by detection with Alamar Blue (Invitrogen Corp., Carlsbad, CA) as previously described (2).
Amplification and sequencing of ribosomal genes.
Chromosomal DNA was isolated from S. aureus via lysostaphin (Sigma-Aldrich Corp., St. Louis, MO) digestion and concentration (DNA Clean and Concentrator-5 kit; Zymo Research, Orange, CA). PCR (Platinum PCR SuperMix High Fidelity; Invitrogen Corp., Carlsbad, CA) was used to amplify each of the six S. aureus rrn operons (including the 5S, 16S, and 23S rRNA genes) as previously described (45, 53, 55). The genes encoding PTC-associated ribosomal proteins L3 (rplC), L4 (rplD), and L22 (rplV) were amplified as a single amplicon (∼3.3 kb) with the flanking primer set rplCF/rplVR (Table 1). PCR was performed with cycling parameters of 94°C for 45 s, 54°C for 45 s, and 72°C for 6.5 min for 35 cycles for rrn operons. The extension time was modified to 3.5 min for the rplC-rplD-rplV amplicon. PCR products were cleaned and concentrated (DNA Clean and Concentrator-5 kit; Zymo Research, Orange, CA) and sequenced (Retrogen Inc., San Diego, CA) with primers flanking the 23S rRNA domain V region (VdomainF) (53) and primers for individual ribosomal protein genes (rplCF, rplDF, and rplVF) (Table 1).
TABLE 1.
Primers used to sequence S. aureus ribosomal genes
| Primer | Sequence (5′ to 3′) | Reference(s) |
|---|---|---|
| rrn1F | GCGGTGTTTTGAGAGATTATTTA | 53 |
| rrn1R | GCTTCATGATATACGCTTCCTTT | 53 |
| rrn2F | GCAGACGCACAGGACTTA | 53 |
| rrn2R | GATACCGTCTTACTGCTCTTCTC | 53 |
| rrn3F | AGGCCGGCAATATGTAAG | 53 |
| rrn3R | GTCGTCAAACGGCACTAATA | 53 |
| rrn4F | TGTGGACGGTGCATCTGTAG | 53 |
| rrn4R | ATCACCCGCTCCATAGATAAT | 53 |
| rrn5F | GCCGATAGCTCTACCACTG | 53 |
| rrn5R | AGGTGCGATGGCAAAACA | 53 |
| rrn6F | GAAAGGCGTAACGATTTGGG | 45, 55 |
| rrn6R | CGTTGACATATTGTCATTCAG | 45, 55 |
| VdomainF | GCGGTCGCCTCCTAAAAG | 53 |
| rplCF | ATGGGCTTAAACTTACCATC | This study |
| rplDF | AAAAGGTTTAGTAGAAATCAG | This study |
| rplVF | GTACATTCAAAGGACACGTTG | This study |
| rplVR | AATCACGGATAATACCAACACG | This study |
Spontaneous mutation frequency experiments.
Large-format assay plates (245 by 245 mm; Corning, Corning, NY) were prepared with 200 ml MHA containing TR-700 or LZD at 2× the MIC for the corresponding S. aureus strain. Mid-log-phase cultures (optical density at 600 nm, ∼0.8) were pelleted and resuspended in PBS to a concentration of ∼2 × 109 CFU/ml. From each culture, 5-ml aliquots were spread onto TR-700 and LZD MHA plates with sterile glass beads. Starting CFU were enumerated by triplicate plating of serial dilutions of the starting inocula in PBS. Plates were incubated at 37°C for 5 days. Putative mutant colonies with reduced susceptibility to TR-700 and LZD were confirmed by subculturing on MHA containing an equal amount of antibiotic prior to MIC and sequence analyses. Mutation frequency experiments were performed on 3 separate days with a total of 12 independent cultures for each strain. Spontaneous mutation frequencies were determined by dividing the number of resistant colonies on a given plate by the actual plated CFU. The mean and median frequencies were calculated. Mean mutation frequencies inherently include data from potential “jackpot” cultures where mutation events occur in an early growth phase and are subsequently overrepresented in the plated CFU. Additionally, mean frequencies exclude data from cultures producing no resistant colonies, for which actual values cannot be determined. Therefore, median mutation frequency values were used to calculate frequency ratios (LZD/TR-700) as a way to factor in these extreme data points without incurring their quantitative biases.
Serial passage experiments.
MHA antibiotic gradient plates (90 by 90 mm) were prepared as previously described (9). Starting plates for both strains had maximal concentrations of 4 μg/ml for TR-700 and 8 μg/ml for LZD. Overnight S. aureus cultures were diluted in MHB to an optical density at 600 nm of ∼0.6, and 100-μl aliquots (∼1 × 108 CFU) were spread onto TR-700 and LZD gradient plates with sterile glass beads. Following 48 h of incubation at 37°C, the leading edge of growth was sampled with a loop and subcultured into MHB without antibiotics. Overnight cultures were then diluted and plated as described for the initial passage, and additional aliquots were stored as −80°C glycerol stocks for MIC and sequence analyses. Gradient plate antibiotic concentrations were increased twofold once growth was observed at approximately half of the plate distance (∼45 mm).
Structural analysis of ribosomal mutations.
Proposed structural effects of the ribosomal mutations in this study were deduced by using the coordinates of the Deinococcus radiodurans and Haloarcula marismortui LZD-bound 50S ribosomal subunit crystal structures (Protein Data Bank accession codes 3DLL and 3CPW) (27, 73). Sequence alignments showed that the regions of the 50S ribosomal subunit discussed in this study are highly conserved, so the structural rationales proposed would be expected to hold for S. aureus and other species. Structural figures were generated with PyMOL (16) by using the D. radiodurans coordinates.
RESULTS
Spontaneous mutations in S. aureus conferring reduced susceptibility to TR-700 are less frequent than those conferring reduced susceptibility to LZD.
Large-format plating experiments were conducted to determine the frequency of spontaneous mutations conferring reduced susceptibility to TR-700 and LZD at 2× the MIC. The median spontaneous frequencies of MSSA 29213 and MRSA 33591 mutation to reduced susceptibility to TR-700 were 1.1 × 10−10 and 1.9 × 10−10, respectively (Table 2), in line with previous estimates in the range of 10−10 to 10−11 (13, 30). These values are approximately 16-fold lower than the corresponding median frequencies of spontaneous mutation of both strains to reduced LZD susceptibility. The mean spontaneous mutation frequencies for reduced TR-700 and LZD susceptibility were 2.8 × 10−10 and 2.0 × 10−9, respectively, for MSSA 29213 and 5.7 × 10−10 and 6.1 × 10−9, respectively, for MRSA 33591. Unlike the median values (Table 2), mean values are inherently biased toward “jackpot” cultures and do not factor in cultures producing no resistant colonies. Despite these limitations, the mean frequencies of mutation of MSSA 29213 and MRSA 33591 resulting in reduced susceptibility to TR-700 and LZD were significantly different (P < 0.001 and P < 0.05, respectively, two-tailed t test).
TABLE 2.
Frequencies of S. aureus spontaneous mutations resulting in decreased susceptibility to TR-700 and LZD
| Strain | Range (median) of mutation frequencies |
LZD/TR-700 ratio | |
|---|---|---|---|
| TR-700 | LZD | ||
| ATCC 29213 (MSSA) | 1.5 × 10−9 to <4.9 × 10−11 (1.1 × 10−10) | 4.1 × 10−9 to 2.9 × 10−10 (2.0 × 10−9) | 18 |
| ATCC 33591 (MRSA) | 3.6 × 10−9 to <4.1 × 10−11 (1.9 × 10−10) | 3.0 × 10−8 to <4.1 × 10−11 (3.0 × 10−9) | 16 |
Spontaneous TR-700- and LZD-selected mutants possess changes in 23S rRNA and ribosomal protein L3.
Sequence analysis was performed on a random subset of spontaneous mutants of both MSSA 29213 and MRSA 33591 that had been selected for decreased susceptibility to TR-700 and LZD. PCR products were amplified with appropriate primers (Table 1) to determine whether there were any changes in the domain V region of any of the six copies of the 23S rRNA genes or in rplC, rplD, and rplV genes encoding ribosomal proteins L3, L4, and L22, mutations in all of which have been implicated in resistance to various PTC-targeted antibiotics. MSSA 29213 mutants selected on TR-700 possessed T2500A 23S rRNA mutations, while mutants selected on LZD possessed either the G2447T or the T2500A mutation (Table 3), both previously described in S. aureus (45, 69).
TABLE 3.
Characteristics of S. aureus wild-type and isogenic mutant strains isolated from spontaneous mutation frequency and serial passage experiments
| Strain and antibiotic selection | Isolation experimenta | 23S rRNA mutation(s)b |
Ribosomal protein mutation(s)c |
MIC (μg/ml)d |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rRNA gene | Allele(s) | Proportion | rplC | L3 | rplD | L4 | TR-700 | LZD | TIA | CHL | VAN | ||
| ATCC 29213 (MSSA) | |||||||||||||
| Wild type | 0.5 | 2 | 1 | 8 | 1 | ||||||||
| TR-700 | Serial pass.e | 0.5 | 2 | 1 | 8 | 1 | |||||||
| LZD | Spont. mut. | G2447T | 4 | 1/6 | 0.5 | 4 | 1 | 8 | 1 | ||||
| LZD | Serial pass. | G2447T | 3, 4 | 2/6 | 1 | 8 | 2 | 8 | 1 | ||||
| LZD | Serial pass. | G2447T | 3, 4 | 2/5f | 2 | 16 | 4 | 8 | 1 | ||||
| LZD | Serial pass. | G2447T | 3, 4, 5 | 3/5f | 4 | 32 | 8 | 8 | 1 | ||||
| LZD | Serial pass. | G2447T | 3, 4, 5 | 3/5f | G455A | Gly152Asp | 8 | 128 | 32 | 8 | 1 | ||
| LZD | Serial pass. | G2447T | 1, 3, 4, 5 | 4/5f | G455A | Gly152Asp | 8 | 128 | 64 | 8 | 1 | ||
| TR-700, LZD | Spont. mut. | T2500A | 4 | 1/6 | 1 | 4 | 1 | 8 | 1 | ||||
| TR-700 | Spont. mut. | T2500A | 4, 5 | 2/6 | 2 | 8 | 2 | 16 | 1 | ||||
| LZD | Serial pass. | G463C | Gly155Arg | 1 | 4 | 4 | 8 | 1 | |||||
| TR-700, LZD | Spont. mut. | G463C/ A505T | Gly155Arg/ Met169Leu | 2 | 8 | 4 | 4 | 2 | |||||
| TR-700 | Spont. mut. | ΔA384-A443 | ΔPhe127-His146 | 2 | 8 | 4 | 8 | 2 | |||||
| ATCC 33591 (MRSA) | |||||||||||||
| Wild type | 0.25 | 1 | 0.5 | 32 | 1 | ||||||||
| LZD | Serial pass. | G2576T | 3 | 1/6 | 0.5 | 4 | 2 | 64 | 1 | ||||
| LZD | Serial pass. | G2576T | 3, 4 | 2/6 | 0.5 | 4 | 2 | 64 | 1 | ||||
| LZD | Serial pass. | G2576T | 3, 4, 5 | 3/6 | 1 | 16 | 2 | 64 | 1 | ||||
| LZD | Serial pass. | G2576T | 2, 3, 4, 5 | 4/6 | 2 | 32 | 4 | 128 | 1 | ||||
| TR-700 | Serial pass. | T2571C/G2576T | 4 | 1/6 | 0.5 | 2 | 0.5 | 32 | 1 | ||||
| TR-700 | Serial pass. | T2571C/G2576T | 4, 5 | 2/6 | 1 | 4 | 1 | 64 | 2 | ||||
| TR-700 | Serial pass. | T2571C/G2576T | 2, 4, 5 | 3/6 | 2 | 16 | 1 | 64 | 2 | ||||
| LZD | Serial pass. | A202C | Lys68Gln | 0.5 | 2 | 1 | 64 | 1 | |||||
| TR-700, LZD | Spont. mut. | G463C | Gly155Arg | 0.5 | 2 | 4 | 32 | 2 | |||||
| TR-700 | Spont. mut. | ΔA384-A443 | ΔPhe127-His146 | 1 | 4 | 4 | 32 | 2 | |||||
Representative strains derived from single colonies from serial passage (Serial pass.) and spontaneous mutation frequency (Spont. mut.) experiments are listed.
The genetic basis of 23S rRNA mutations (rRNA gene, E. coli numbering), mutant 23S rRNA allele copy numbers [Allele(s)], and the proportion of the total number of 23S rRNA alleles detected that were mutant alleles are given.
Ribosomal protein mutations (S. aureus numbering) are reported for the genes (rplC and rplD) and proteins (L3 and L4), respectively.
MICs of TR-700, LZD, TIA, CHL, and VAN were determined for both strains.
TR-700 serial passage no. 30 ATCC 29213 is included for reference, although no mutations or MIC increases were observed.
23S rRNA gene copy 6 (rrn6) was not detected by PCR in this strain.
S. aureus mutations in rplC were previously associated with decreased susceptibility to pleuromutilins, namely, TIA, but not to LZD (7, 20, 48). The MIC data in Table 3 indicate that decreased susceptibility to oxazolidinones is also associated with changes in rplC in both MSSA strain 29213 and MRSA strain 33591. MSSA 29213 mutants possessed two novel L3 mutations: Gly155Arg/Met169Leu and ΔPhe127-His146. Some of the MRSA 33591 mutants also possessed the novel ΔPhe127-His146 mutation, while the previously described Gly155Arg mutation (35) was identified in others (Table 3). Similar to previous studies (20), we observed greater decreased susceptibility in MSSA 29213 mutants possessing coupled L3 mutations versus those possessing single L3 mutations. The TR-700-selected ΔPhe127-His146 mutation involved a 60-bp deletion between two 6-bp repeats (TTTCCA) in rplC resulting in a 20-amino-acid deletion in L3. In mutants possessing rplC mutations, no mutations were identified in any of the 23S rRNA gene domain V regions, rplD, or rplV.
Serial passage of S. aureus in the presence of TR-700 results in smaller changes in oxazolidinone MICs than does serial passage with LZD.
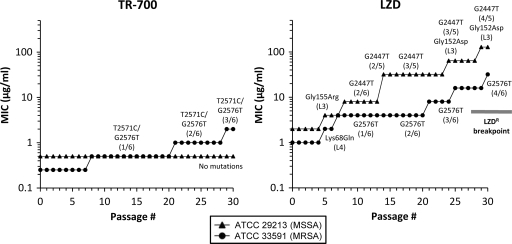
Serial passage on antibiotic gradient plates was used to access the potential of S. aureus to develop resistance upon prolonged exposure to TR-700 and LZD (Fig. 2). Gradient plates were utilized instead of liquid medium in order to allow for selection of mutants with intermediate MICs, rather than limiting the selection of mutants to those able to grow at or above a fixed drug concentration. Following 30 serial passages of MSSA 29213, the TR-700 MIC remained unchanged at 0.5 μg/ml, in sharp contrast to the parallel LZD serial passage experiment, where the MIC increased 64-fold, from 2 to 128 μg/ml (Fig. 2). For MRSA 33591, an eightfold increase was observed after 30 serial passages in TR-700 (0.25 to 2.0 μg/ml), whereas a 32-fold higher MIC (1 μg/ml to 32 μg/ml) was observed after passage on LZD (Fig. 2). These data are consistent with previous S. aureus LZD serial passage experiments which generated high levels of resistance (5, 52).
FIG. 2.
Serial passage of MSSA 29213 and MRSA 33591 in the presence of TR-700 and LZD. Total mutant population MICs are plotted for each serial passage of ATCC 29213 and ATCC 33591 with TR-700 and LZD. Underlying mutations in 23S rRNA genes and ribosomal proteins L3 and L4 are shown corresponding to the stepwise MICs at which they were identified by analysis of individual colonies. Fractional 23S rRNA values indicate the number of mutant alleles out of the total number of 23S alleles detected by PCR.
Serial passage-derived mutants have changes in genes encoding the 23S rRNA and ribosomal proteins L3 and L4.
Individual colonies from representative passages for each stepwise TR-700 and LZD MIC increase were analyzed for changes in the sequence of the domain V region of 23S rRNA genes, as well as the rplC, rplD, and rplV genes. After 30 serial passages in TR-700, the representative MSSA 29213 colonies tested showed no change in MIC and did not possess any mutations in any of the ribosomal genes sequenced (Fig. 2 and Table 3). In contrast, mutants of MSSA 29213 with decreased susceptibility to LZD became apparent after passage 5 in LZD, with the first increase in the MIC of LZD (2 to 4 μg/ml) associated with the L3 mutation Gly155Arg. Later serial passage MSSA 29213 mutants possessed G2447T 23S rRNA mutations or G2447T coupled with L3 Gly152Asp (Table 3). MRSA 33591 serial passage mutants included L4 Lys68Gln and G2576T for LZD and a novel T25761C/G2576T 23S rRNA double mutation for TR-700 (Table 3). No mutations were identified in the rplV gene encoding ribosomal protein L22 in either strain.
Increased copy number of mutated 23S alleles is associated with decreased susceptibility to TR-700, LZD, and TIA.
Previous studies have shown that an increased copy number of 23S rRNA mutations is correlated with higher levels of resistance to LZD in both S. aureus and enterococci (5, 38, 66, 78). Isogenic MSSA 29213 strains with G2447T mutations in one or two copies of the 23S rRNA demonstrate this trend for TR-700 and LZD, as well as TIA, but not for CHL or VAN (Table 3). Starting at passage 14, we were unable to amplify one of the 23S rRNA alleles (rrn6) in LZD-passaged MSSA 29213 colonies. The potential loss of a 23S rRNA gene correlated with twofold TR-700, LZD, and TIA MIC increases compared to the mutant from passage 8 with two of six 23S rRNA gene copies mutated to G2447T, presumably due to an increase in the proportion of mutant ribosomes in the cell (45). Additional twofold MIC increases resulted from another gene conversion event leading to three of five mutant copies of G2447T. A Gly152Asp L3 mutation appeared in passage 24 in conjunction with three of five mutant 23S rRNA gene copies of G2447T, resulting in further two- to fourfold MIC increases. Another G2447T gene conversion event in passage 29 resulted in four of five mutant alleles (in addition to the Gly152Asp L3 mutation) and was associated with marginal MIC increases. An independent Gly152Asp mutation was observed in laboratory-derived TIAr S. aureus associated with a twofold LZD MIC increase (4 to 8 μg/ml) (35), supporting its association here with oxazolidinone-pleuromutilin cross-resistance.
Similar trends were observed for mutants from the MRSA 33591 LZD serial passage experiment carrying G2576T, where an increasing mutant allele copy number from one of six to four of six was associated with decreasing susceptibility to TR-700, LZD, and TIA, reaching a maximum of 8- to 32-fold changes versus the susceptible isogenic wild-type MRSA 33591 strain. A fourfold increase in the MIC of CHL (32 to 128 μg/ml) was also observed for the four-of-six G2576T mutant (Table 3), a finding consistent with previous CHL susceptibility testing of G2576T S. aureus mutants (5). No cross-resistance between LZD and CHL was observed in MSSA 29213 G2447T mutants, consistent with previous observations of S. aureus G2447T mutants (51).
A novel T2571C/G2576T double mutant was identified after serial passage 7 of MRSA 33591 in TR-700. Despite examining several colonies from earlier passages, we were unable to identify either a single-copy G2576T or T2571C mutant from the TR-700 serial passage experiment, presumably due to marginal changes in the TR-700 MIC resulting in a low frequency in the population. However, the data in Fig. 2 support the hypothesis that continued selection on TR-700 resulted in gene conversion of this dual mutation such that mutants with higher T2571C/G2576T allele copy numbers were identified in serial passages 21 and 29. These mutants were associated with further MIC increases. The three-of-six T2571C/G2576T mutant demonstrated 8-fold and 16-fold increases in the MICs of TR-700 and LZD (2 μg/ml and 16 μg/ml, respectively). Changes in TIA and CHL resistance were less pronounced than those in TR-700 and LZD resistance but similar to what was observed for the G2576T one-, two-, and three-copy mutants.
In MSSA 29213, two types of spontaneous mutants containing the 23S rRNA mutation T2500A were identified: those which had a single T2500A mutation (one of six) and those which were mutated in two of six alleles. The single mutant demonstrated a twofold increase in the MICs of TR-700 and LZD, whereas the double mutant showed a fourfold increase in the MICs of TR-700 and LZD and a twofold increase in the MICs of TIA and CHL (Table 3). These data also suggest a gene dosage effect.
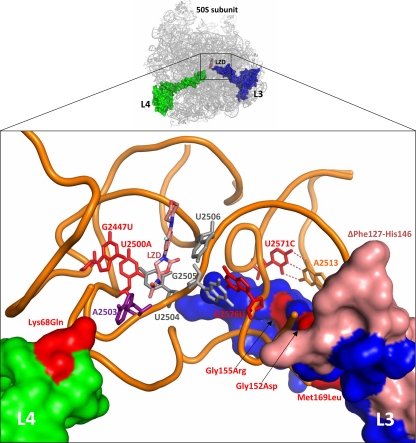
TR-700- and LZD-selected mutations occur in close proximity to the oxazolidinone binding site of the PTC.
Using the published coordinates of LZD bound to the H. marismortui and D. radiodurans 50S subunits (27, 73), we evaluated the effect of mutations in 23S rRNA domain V and ribosomal proteins L3 and L4 (Fig. 3). For the mutations that are novel, or for those not previously structurally rationalized, we propose hypotheses for their effects on oxazolidinone binding. Bases G2447 and U2500 both interact directly with the critical PTC base U2504, which is key to oxazolidinone binding. A detailed mechanistic review of G2447U and U2500A mutations has been previously reported (15).
FIG. 3.
TR-700- and LZD-selected ribosomal mutations cluster near the bases that compose the oxazolidinone binding site. Base/residue change mutations are red, deletion mutations are pink, LZD is salmon, and the 23S rRNA backbone is orange. The solvent-accessible surfaces of L3 and L4 are blue and green, respectively. A2503 (purple), the site of Cfr methylation, and bases 2504 to 2506 (gray), key bases lining the PTC, are shown for reference. In the D. radiodurans structure used to generate this image and Fig. 4, the U2571-A2513 base pair is a C · G base pair. The base pairing at this location is conserved across species, although the identity of the bases varies.
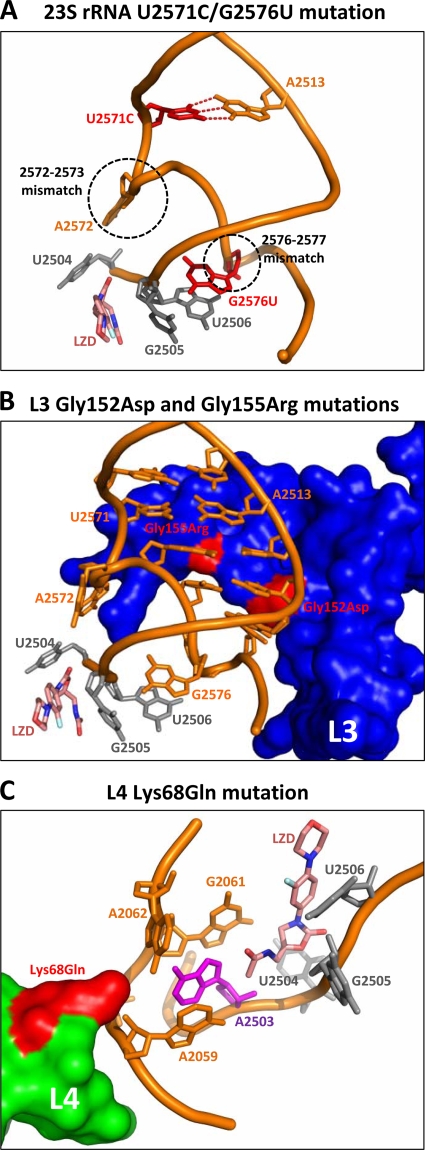
(i) 23S rRNA U2571C/G2576U mutation.
The 50S subunit structures reveal that bases 2571 and 2513 of the 23S rRNA form a base pair that immediately precedes a conserved two-base mismatch (bases 2572 to 2573). This brief interruption in the helical structure positions A2572 in contact with U2504, a residue forming a significant portion of the LZD binding site (Fig. 4A). The perturbation of U2504 by adjacent mutations has been reported to be a major mechanism of resistance to multiple classes of antibiotics that bind at the PTC (15). We propose that the loss of base pairing in the U2571C mutant leads to an expansion of the mismatched region, resulting in either A2572 impinging on U2504, causing a deformation of the LZD binding site, or a loss of interactions due to the movement of U2504 away from the PTC. Consistent with the latter hypothesis, a structural role for A2572 in preventing U2504 from tilting away from the PTC has been described (15, 54). The plausibility of both models is supported by the recent description of an A2572U mutation in Mycobacterium smegmatis that results in a twofold LZD MIC increase (42). Although selected in the presence of a pleuromutilin, this mutant shows modest cross-resistance to LZD and demonstrates that perturbations of A2572 can influence the conformation of U2504 such that oxazolidinone binding is diminished.
FIG. 4.
Putative structural consequences of key mutations. (A) U2571C creates an extended mismatch that may lead to A2572 impinging on U2504, a critical PTC base. G2576U directly influences the PTC through its interactions with bases 2505 and 2506. (B) Gly152Asp likely alters the conformation of G2576, which in turn affects bases 2505 and 2506, bases that line the PTC. Gly155Arg would disrupt the 2572-2573 mismatch, perturbing U2504 in a manner analogous to that of the A2572 mutant. (C) Lys68Gln results in the loss of charge compensation of the rRNA backbone such that the pocket surrounding A2503 is compromised. This may lead to the movement of A2503 toward the oxazolidinone binding site, with consequences similar to those of Cfr methylation.
Base G2576 interacts with U2506 and G2505, both of which form a critical portion of the PTC (27, 73). Thus, the combination of U2571C and G2576U may be additive (or even synergistic) by impacting multiple adjacent residues involved in oxazolidinone binding (Fig. 4A). This is consistent with the data in Table 3 that show a twofold difference in the MIC of TR-700 in the T2571C/G2576T versus the G2576T mutant when two or three copies of the mutant alleles are present. Together, these mutations would influence bases 2504 to 2506, which comprise the majority of contacts in the oxazolidinone binding site.
(ii) Ribosomal protein L3 mutations.
All of the L3 mutations identified in the present study cluster in a central extension of the L3 protein that approaches the PTC (Fig. 3). The Gly152Asp and Gly155Arg mutations have been previously identified in the context of pleuromutilin resistance (20, 35, 48). Given its proximity, Gly155Arg likely impacts the 2572-and-2573 conserved mismatch, disrupting U2504 in a manner similar to that described above for the U2571C mutation (Fig. 4B). The Gly152Asp mutation, packing beneath another conserved mismatch (bases 2576 and 2577), probably reduces oxazolidinone affinity by indirect perturbation of bases 2505 and 2506, analogous to the G2576T mutation (Fig. 4B). The ΔPhe127-His146 mutation is particularly noteworthy, given the extensive set of interactions this portion of the L3 protein has with the 23S rRNA, including portions of the H73, H90, and H94 helices. The effect of this mutation may be due to the loss of a few key interactions in the vicinity of the other observed mutations or a gross disruption of tertiary structure. The Met169Leu mutation (observed only in conjunction with Gly155Arg) is more peripheral to the PTC than the other mutations, making a structural rationale for its effect speculative.
(iii) Ribosomal protein L4 Lys68Gln mutation.
The tip of the extended region of L4 interacts with the 23S rRNA primarily through contacts with bases A2059 to G2061, stabilizing this key region of the PTC through charge neutralization and direct packing interactions in a manner typical for ribosomal protein extensions (33). Specifically, Lys68 interacts with the sugar-phosphate backbone of A2059 and G2061, and together they form a pocket around A2503 (Fig. 4C). The loss of the compensating charge of Lys68 in the Lys68Gln mutant would lead to a deformation of this pocket and a consequent movement of A2503 toward the PTC, impacting the binding of TIA, CHL, LZD, and TR-700, as is reflected by the observed twofold changes in the MIC (Table 3). A2503 is the target of Cfr methylation (32), so the Lys68Gln substitution likely affects an analogous mechanism of crowding the PTC, thereby reducing the affinity of multiple classes of antibiotics.
DISCUSSION
In this study, we show that spontaneous mutations conferring decreased susceptibility to TR-700 occur more than an order of magnitude less frequently than those conferring decreased susceptibility to LZD in strains MSSA 29213 and MRSA 33591, supporting preliminary studies that suggested a very low frequency of resistance to TR-700 (13, 30). A recent study utilizing an inoculum of S. aureus of 2.2 × 109 CFU obtained no growth on plates at 4×, 6×, and 8× the MIC of TR-700 (30). These data are consistent with our study given that the frequencies of spontaneous mutation to decreased susceptibility to TR-700 generated here were in the low 10−10 range and that not all of the TR-700-selected spontaneous mutants demonstrated a greater-than-twofold MIC shift. Similarly, a previous study estimated the spontaneous LZD mutation frequency of S. aureus at <10−9 following no observed growth after plating of 109 CFU on medium containing LZD at 2× the MIC (31). Based on the resistance levels observed in our single-step LZD-selected mutants, 2× the MIC is an appropriate drug concentration; however, given LZD mutation frequencies in the low 10−9 range (in addition to potential strain-to-strain variation and other assay variables), again, the number of CFU plated in that study was insufficient to capture mutation events. The large-format plating assay we employed enabled determination of frequencies of mutation to reduced LZD susceptibility consistent with previous studies where finite frequency values for LZD were generated (35, 62).
Through serial passage experiments, we have shown that S. aureus has a very limited ability to develop decreased susceptibility to TR-700, consistent with a previous TR-700 serial passage study (30). MSSA 29213 had no change in the TR-700 MIC over 30 passages. Interestingly, three different ribosomal mutations were observed in this strain in the spontaneous mutation frequency experiments with TR-700. While replicate serial passage data would be informative, this result could suggest that there are different selective pressures between these two experiments or that TR-700 has additional properties which limit the viability of resistant strains during prolonged drug exposure. Reduced susceptibility to TR-700 was eventually observed in MRSA 33591; however, this required 20 to 30 passages to achieve an MIC of 1 to 2 μg/ml. Depending upon dosing, these antibiotic levels are likely to be clinically achievable.
In the TR-700 serial passage experiment, multiple mutations in 23S rRNA (T2571C/G2576T) were required for the initial stepwise MIC increase for MRSA 33591. This is in sharp contrast to LZD serial passage experiments, where several different single mutations were identified in both strains resulting in two- to fourfold changes in the LZD MIC within five to eight serial passages, including G2447T, G2576T, L3 Gly155Arg, or L4 Lys68Gln mutations. These results are consistent with literature reports showing increased resistance to LZD after five to nine serial passages (31, 49, 52). Through serial passage with LZD, strains with an LZD MIC of ≥8 μg/ml were obtained as early as passage 7 and continued to demonstrate higher levels of resistance over time with additional 23S rRNA gene conversion events and acquisition of L3 mutations, resulting in final MICs at passage 30 of 32 μg/ml (MRSA 33951) and 128 μg/ml (MSSA 29213).
Crystallographic studies of the 50S ribosomal subunit have shown that several ribosomal proteins contain extensions that approach 23S rRNA bases of the PTC. A critical subset of these proteins includes L3, L4, and L22 (3, 24). Although they are not part of the PTC per se, it is likely that mutations of amino acid residues close to the PTC impact its conformation and stability due to modulation of second- and third-shell interactions. Typically, mutations in L4 have been implicated with macrolide resistance (23, 59, 75); however, deletion of L4 residues Lys68 and Gly69 in a clinical isolate of Streptococcus pneumoniae conferred reduced susceptibility to LZD (76). Our isolation of a Lys68Gln mutant is the first report of an L4 mutation in S. aureus associated with resistance through selection with oxazolidinones. Further studies are needed to conclusively link this mutation to the observed increase in LZD resistance; however, a number of other documented L4 mutations involve alterations in amino acids in this region of the protein (18, 23, 56, 76).
Mutations in the rplC gene encoding ribosomal protein L3 were found in a large portion of the mutants selected in our spontaneous mutation frequency experiments. L3 also interacts closely with the PTC in the 50S subunit, and mutations in rplC have been implicated in resistance to the pleuromutilin TIA (6, 7). However, cross-resistance to oxazolidinones was not reported in the previously described Gly155Arg mutant (20, 48), possibly due to strain differences. All of the L3 mutations identified in this study occur within the same region of L3 where established mutations conferring pleuromutilin resistance have been characterized (7, 20, 48, 54). Individually, they lead to two- to fourfold changes in oxazolidinone and TIA MICs and appear to be additive in combination with the 23S rRNA G2447T mutation for resistance to both oxazolidinone and TIA. Given the prevalence of L3 mutations observed here, this class of mutation is certainly worth exploring in any of the numerous uncharacterized laboratory and clinical LZDr isolates previously reported. Directed mutagenesis and heterologous expression studies, however, are needed to conclusively link these mutations to oxazolidinone resistance.
The binding site for the oxazolidinones overlaps the binding sites of several other classes of PTC inhibitors, including the pleuromutilins and phenicols. For example, eperezolid and Rx-01 oxazolidinone compounds have been shown to compete with the binding of CHL (34, 37, 67). Additionally, results of mutational analysis and rRNA footprinting are consistent with the observation of cross-resistance between oxazolidinones and pleuromutilins (36, 41, 63, 77). In this study, cross-resistance to TIA was observed for all of the mutants where 2- to 32-fold changes in the MIC were observed. However, cross-resistance to CHL, as demonstrated by two- to fourfold changes, was only observed for the G2576T and T2571C/G2576T mutants and possibly L4 Lys68Gln and the 23S rRNA two-gene copy T2500A mutant. Similarly, not all L3 mutations confer cross-resistance to pleuromutilins and LZD, including E. coli mutations Arg141Ser, Arg149Ser, and Gly153Arg and S. aureus mutations Lys157Gln and Ser158Leu (48).
Mutations selected on TR-700 with changes in 23S rRNA genes included T2500A and T2571C/G2576T, while LZD-selected mutations in 23S rRNA genes included T2500A, G2447T, and G2576T. However, strains with multiple copies of these mutations demonstrated cross-resistance. MIC analysis of strains with a single 23S rRNA copy of any of these mutations showed that a single G2447T, T2500A, G2576T, or T2571C/G2576T mutation conferred a twofold or smaller increase in the MIC of TR-700 and a two- to fourfold increase in the MIC of LZD. Similarly, two copies of these alleles resulted in two- to fourfold versus four- to eightfold TR-700 and LZD MIC increases, respectively. These data are consistent with previous observations of single-copy and multicopy mutant 23S rRNA alleles in S. aureus (5, 38) and have important implications for the ability to select first-step mutations, as well as higher levels of resistance in the clinic.
While we are the first to report a coupled 23S rRNA mutation involving bases 2571 and 2576, there is a report of mutations in 2571, both independent (G2571C) and coupled (G2571C/A2503G), associated with LZD resistance in S. pneumoniae (12). Others have reported antibiotic resistance mutations in adjacent and pairing bases. A dual 23S rRNA mutation involving C2055A/A2572T was characterized in TIAr Brachyspira (54), and a serial passage-derived E. faecalis LZDr isolate possessed four unique 23S rRNA mutations, including G2576T, C2610G, G2512T, and G2513T (the corresponding base to T2571, which in E. faecalis is a G instead of a T) (57). In the TR-700 serial passage experiment, we were unable to isolate a single-copy mutant possessing either the T2571C or the G2576T mutation alone, so we cannot speculate as to which came first, although it appears that the second mutation was rapidly selected.
Generation of a dual T2571C/G2576T 23S rRNA mutant occurred in TR-700-passaged MRSA 33591, while the same strain passaged in LZD developed the single G2576T mutation. This may be due to differences in the binding of TR-700 and LZD to the PTC region. The tetrazole ring extension on TR-700 is thought to add additional binding interactions with key PTC bases (such as U2585), which may help account for its 4- to 16-fold MIC advantage over LZD (64). The projection of this ring may also necessitate greater perturbation of the PTC via additional and/or unique mutations that would not be required for resistance to LZD. Such increased disruption of the PTC may amplify fitness/resistance tradeoffs documented previously with other 23S rRNA (5, 8, 44) and L4 (76) mutations associated with LZD resistance. Future studies will examine the potential fitness costs associated with the oxazolidinone resistance mechanisms reported here.
Understanding the potential for and basis of antibiotic resistance is a critical component of drug development. Our investigation of such properties under selection with TR-700 in S. aureus failed to generate any mutants with MICs of >2 μg/ml, whereas the same analyses performed with LZD produced a large number of mutants that would be considered clinically resistant (i.e., MICs of >4 μg/ml). TR-700 demonstrated 4- to 16-fold greater potency than LZD against all of the ribosomal mutations documented here, including those specific to selection with TR-700. These data, coupled with the greater potency of TR-700 than LZD against gram-positive pathogens, including Cfr strains, support the continued clinical development of TR-701.
Acknowledgments
We thank Jeffrey Hsiao and Amanda Castellano for their assistance with sequencing data and MIC assays. We also thank Alexander Mankin for useful discussions on the manuscript.
Footnotes
Published ahead of print on 14 September 2009.
REFERENCES
- 1.Ament, P. W., N. Jamshed, and J. P. Horne. 2002. Linezolid: its role in the treatment of Gram-positive, drug-resistant bacterial infections. Am. Fam. Physician 65:663-670. [PubMed] [Google Scholar]
- 2.Baker, C. N., and F. C. Tenover. 1996. Evaluation of Alamar colorimetric broth microdilution susceptibility testing method for staphylococci and enterococci. J. Clin. Microbiol. 34:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 4.Barbachyn, M. R. 2008. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 802.
- 5.Besier, S., A. Ludwig, J. Zander, V. Brade, and T. A. Wichelhaus. 2008. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob. Agents Chemother. 52:1570-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böck, A., F. Turnowsky, and G. Hogenauer. 1982. Tiamulin resistance mutations in Escherichia coli. J. Bacteriol. 151:1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bøsling, J., S. M. Poulsen, B. Vester, and K. S. Long. 2003. Resistance to the peptidyl transferase inhibitor tiamulin caused by mutation of ribosomal protein L3. Antimicrob. Agents Chemother. 47:2892-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourgeois-Nicolaos, N., P. Kharrat, M. J. Butel, and F. Doucet-Populaire. 2008. Abstracts of the 18th European Congress of Clinical Microbiology and Infectious Diseases, Barcelona, Spain, abstr. O345.
- 9.Bryson, V., and W. Szybalski. 1952. Microbial selection. Science 116:45-51. [PubMed] [Google Scholar]
- 10.Burleson, B. S., D. J. Ritchie, S. T. Micek, and W. M. Dunne. 2004. Enterococcus faecalis resistant to linezolid: case series and review of the literature. Pharmacotherapy 24:1225-1231. [DOI] [PubMed] [Google Scholar]
- 11.Campanile, F., D. Bongiorno, S. Borbone, G. Mongelli, M. Baldi, R. Provenzani, and S. Stefani. 2009. Abstracts of the 19th European Congress of Clinical Microbiology and Infectious Diseases, Helsinki, Finland, abstr. P929.
- 12.Carsenti-Dellamonica, H., M. Galimand, F. Vandenbos, C. Pradier, P. M. Roger, B. Dunais, M. Sabah, G. Mancini, and P. Dellamonica. 2005. In vitro selection of mutants of Streptococcus pneumoniae resistant to macrolides and linezolid: relationship with susceptibility to penicillin G or macrolides. J. Antimicrob. Chemother. 56:633-642. [DOI] [PubMed] [Google Scholar]
- 13.Choi, S., T. Son, T. Lee, W. Im, and J. Rhee. 2005. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., abstr. F-1241.
- 14.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, CLSI document M7-A7, seventh ed., vol. 26, no. 2. Clinical and Laboratory Standards Institute, Wayne, PA.
- 15.Davidovich, C., A. Bashan, and A. Yonath. 2008. Structural basis for cross-resistance to ribosomal PTC antibiotics. Proc. Natl. Acad. Sci. USA 105:20665-20670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA.
- 16a.De La Torre, M., M. Sanchez, G. Morales, E. Baos, A. Arribi, N. Garcia, R. Andrade, B. Pelaez, M. P. Pacheco, S. Domingo, J. Conesa, M. Nieto, F. J. Candel, and J. Picazo. 2008. Abstr. 48th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-1835a.
- 17.Engemann, J. J., M. J. Joyce, L. J. Harrell, S. Evans, L. Reller, and D. Sexton. 2003. Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1112.
- 18.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48:3169-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng, J., A. Lupien, H. Gingras, J. Wasserscheid, K. Dewar, D. Legare, and M. Ouellette. 2009. Genome sequencing of linezolid-resistant Streptococcus pneumoniae mutants reveals novel mechanisms of resistance. Genome Res. 19:1214-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentry, D. R., S. F. Rittenhouse, L. McCloskey, and D. J. Holmes. 2007. Stepwise exposure of Staphylococcus aureus to pleuromutilins is associated with stepwise acquisition of mutations in rplC and minimally affects susceptibility to retapamulin. Antimicrob. Agents Chemother. 51:2048-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giessing, A. M., S. S. Jensen, A. Rasmussen, L. H. Hansen, A. Gondela, K. Long, B. Vester, and F. Kirpekar. 2009. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzales, R. D., P. C. Schreckenberger, M. B. Graham, S. Kelkar, K. DenBesten, and J. P. Quinn. 2001. Infections due to vancomycin-resistant Enterococcus faecium resistant to linezolid. Lancet 357:1179. [DOI] [PubMed] [Google Scholar]
- 23.Gregory, S. T., and A. E. Dahlberg. 1999. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23S ribosomal RNA. J. Mol. Biol. 289:827-834. [DOI] [PubMed] [Google Scholar]
- 24.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 25.Hillemann, D., S. Rüsch-Gerdes, and E. Richter. 2008. In vitro-selected linezolid-resistant Mycobacterium tuberculosis mutants. Antimicrob. Agents Chemother. 52:800-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howe, R. A., M. Wootton, A. R. Noel, K. E. Bowker, T. R. Walsh, and A. P. MacGowan. 2003. Activity of AZD2563, a novel oxazolidinone, against Staphylococcus aureus strains with reduced susceptibility to vancomycin or linezolid. Antimicrob. Agents Chemother. 47:3651-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ippolito, J. A., Z. F. Kanyo, D. Wang, F. J. Franceschi, P. B. Moore, T. A. Steitz, and E. M. Duffy. 2008. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 51:3353-3356. [DOI] [PubMed] [Google Scholar]
- 28.Jones, R. N., T. R. Fritsche, H. S. Sader, and J. E. Ross. 2007. LEADER surveillance program results for 2006: an activity and spectrum analysis of linezolid using clinical isolates from the United States (50 medical centers). Diagn. Microbiol. Infect. Dis. 59:309-317. [DOI] [PubMed] [Google Scholar]
- 29.Jones, R. N., T. R. Fritsche, H. S. Sader, and J. E. Ross. 2007. Zyvox annual appraisal of potency and spectrum program results for 2006: an activity and spectrum analysis of linezolid using clinical isolates from 16 countries. Diagn. Microbiol. Infect. Dis. 59:199-209. [DOI] [PubMed] [Google Scholar]
- 30.Jones, R. N., G. J. Moet, H. S. Sader, R. E. Mendes, and M. Castanheira. 2009. TR-700 in vitro activity against and resistance mutation frequencies among Gram-positive pathogens. J. Antimicrob. Chemother. 63:716-720. [DOI] [PubMed] [Google Scholar]
- 31.Kaatz, G. W., and S. M. Seo. 1996. In vitro activities of oxazolidinone compounds U100592 and U100766 against Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 40:799-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehrenberg, C., S. Schwarz, L. Jacobsen, L. H. Hansen, and B. Vester. 2005. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 57:1064-1073. [DOI] [PubMed] [Google Scholar]
- 33.Klein, D. J., P. B. Moore, and T. A. Steitz. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340:141-177. [DOI] [PubMed] [Google Scholar]
- 34.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 35.Kosowska-Shick, K., C. Clark, K. Credito, P. McGhee, B. Dewasse, T. Bogdanovich, and P. C. Appelbaum. 2006. Single- and multistep resistance selection studies on the activity of retapamulin compared to other agents against Staphylococcus aureus and Streptococcus pyogenes. Antimicrob. Agents Chemother. 50:765-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leach, K. L., S. M. Swaney, J. R. Colca, W. G. McDonald, J. R. Blinn, L. M. Thomasco, R. C. Gadwood, D. Shinabarger, L. Xiong, and A. S. Mankin. 2007. The site of action of oxazolidinone antibiotics in living bacteria and in human mitochondria. Mol. Cell 26:393-402. [DOI] [PubMed] [Google Scholar]
- 37.Lin, A. H., R. W. Murray, T. J. Vidmar, and K. R. Marotti. 1997. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob. Agents Chemother. 41:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livermore, D. M., S. Mushtaq, M. Warner, and N. Woodford. 2009. Activity of oxazolidinone TR-700 against linezolid-susceptible and -resistant staphylococci and enterococci. J. Antimicrob. Chemother. 63:713-715. [DOI] [PubMed] [Google Scholar]
- 39.Livermore, D. M., M. Warner, S. Mushtaq, S. North, and N. Woodford. 2007. In vitro activity of the oxazolidinone RWJ-416457 against linezolid-resistant and -susceptible staphylococci and enterococci. Antimicrob. Agents Chemother. 51:1112-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lobritz, M., R. Hutton-Thomas, S. Marshall, and L. B. Rice. 2003. Recombination proficiency influences frequency and locus of mutational resistance to linezolid in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3318-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long, K. S., L. H. Hansen, L. Jakobsen, and B. Vester. 2006. Interaction of pleuromutilin derivatives with the ribosomal peptidyl transferase center. Antimicrob. Agents Chemother. 50:1458-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Long, K. S., J. Poehlsgaard, L. H. Hansen, S. N. Hobbie, E. C. Bottger, and B. Vester. 2009. Single 23S rRNA mutations at the ribosomal peptidyl transferase centre confer resistance to valnemulin and other antibiotics in Mycobacterium smegmatis by perturbation of the drug binding pocket. Mol. Microbiol. 71:1218-1227. [DOI] [PubMed] [Google Scholar]
- 43.Long, K. S., J. Poehlsgaard, C. Kehrenberg, S. Schwarz, and B. Vester. 2006. The cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazur, W., C. Knob, and H. S. Fraimow. 2002. Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1607.
- 45.Meka, V. G., S. K. Pillai, G. Sakoulas, C. Wennersten, L. Venkataraman, P. C. DeGirolami, G. M. Eliopoulos, R. C. Moellering, and H. S. Gold. 2004. Linezolid resistance in sequential Staphylococcus aureus isolates associated with a T2500A mutation in the 23S rRNA gene and loss of a single copy of rRNA. J. Infect. Dis. 190:311-317. [DOI] [PubMed] [Google Scholar]
- 46.Mendes, R. E. 2009. Abstracts of the 19th European Congress of Clinical Microbiology and Infectious Diseases, Helsinki, Finland, abstr. O96. [DOI] [PMC free article] [PubMed]
- 47.Mendes, R. E., M. Sanchez, L. Deshpande, M. De La Torre, and R. N. Jones. 2009. Abstracts of the 19th European Congress of Clinical Microbiology and Infectious Diseases, Helsinki, Finland, abstr. P928.
- 48.Miller, K., C. J. Dunsmore, C. W. Fishwick, and I. Chopra. 2008. Linezolid and tiamulin cross-resistance in Staphylococcus aureus mediated by point mutations in the peptidyl transferase center. Antimicrob. Agents Chemother. 52:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, K., A. J. O'Neill, M. H. Wilcox, E. Ingham, and I. Chopra. 2008. Delayed development of linezolid resistance in Staphylococcus aureus following exposure to low levels of antimicrobial agents. Antimicrob. Agents Chemother. 52:1940-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.North, S. E., M. J. Ellington, A. P. Johnson, D. M. Livermore, and N. Woodford. 2005. Abstr. 45th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1417.
- 51.North, S. E., M. J. Ellington, A. P. Johnson, D. M. Livermore, and N. Woodford. 2005. Abstracts of the 15th European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, Denmark, abstr. P974.
- 52.Overbye, K. M., and D. Mordekhay. 2007. AM-7359—a novel oxazolidinone with low resistance potential and potent activity against drug resistant pathogens. J. Chemother. 19:249-255. [DOI] [PubMed] [Google Scholar]
- 53.Pillai, S. K., G. Sakoulas, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., M. J. Ferraro, and H. S. Gold. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J. Infect. Dis. 186:1603-1607. [DOI] [PubMed] [Google Scholar]
- 54.Pringle, M., J. Poehlsgaard, B. Vester, and K. S. Long. 2004. Mutations in ribosomal protein L3 and 23S ribosomal RNA at the peptidyl transferase centre are associated with reduced susceptibility to tiamulin in Brachyspira spp. isolates. Mol. Microbiol. 54:1295-1306. [DOI] [PubMed] [Google Scholar]
- 55.Prunier, A. L., B. Malbruny, D. Tande, B. Picard, and R. Leclercq. 2002. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob. Agents Chemother. 46:3054-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prunier, A. L., H. N. Trong, D. Tande, C. Segond, and R. Leclercq. 2005. Mutation of L4 ribosomal protein conferring unusual macrolide resistance in two independent clinical isolates of Staphylococcus aureus. Microb. Drug. Resist. 11:18-20. [DOI] [PubMed] [Google Scholar]
- 57.Prystowsky, J., F. Siddiqui, J. Chosay, D. L. Shinabarger, J. Millichap, L. R. Peterson, and G. A. Noskin. 2001. Resistance to linezolid: characterization of mutations in rRNA and comparison of their occurrences in vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 45:2154-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richter, E., S. Rusch-Gerdes, and D. Hillemann. 2007. First linezolid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 51:1534-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts, M. C. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282:147-159. [DOI] [PubMed] [Google Scholar]
- 60.Ruggero, K. A., L. K. Schroeder, P. C. Schreckenberger, A. S. Mankin, and J. P. Quinn. 2003. Nosocomial superinfections due to linezolid-resistant Enterococcus faecalis: evidence for a gene dosage effect on linezolid MICs. Diagn. Microbiol. Infect. Dis. 47:511-513. [DOI] [PubMed] [Google Scholar]
- 61.Reference deleted.
- 62.Sander, P., L. Belova, Y. G. Kidan, P. Pfister, A. S. Mankin, and E. C. Bottger. 2002. Ribosomal and non-ribosomal resistance to oxazolidinones: species-specific idiosyncrasy of ribosomal alterations. Mol. Microbiol. 46:1295-1304. [DOI] [PubMed] [Google Scholar]
- 63.Schlünzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 64.Shaw, K. J., S. Poppe, R. Schaadt, V. Brown-Driver, J. Finn, C. M. Pillar, D. Shinabarger, and G. Zurenko. 2008. In vitro activity of TR-700, the antibacterial moiety of the prodrug TR-701, against linezolid-resistant strains. Antimicrob. Agents Chemother. 52:4442-4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shinabarger, D. L., K. R. Marotti, R. W. Murray, A. H. Lin, E. P. Melchior, S. M. Swaney, D. S. Dunyak, W. F. Demyan, and J. M. Buysse. 1997. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob. Agents Chemother. 41:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinclair, A., C. Arnold, and N. Woodford. 2003. Rapid detection and estimation by pyrosequencing of 23S rRNA genes with a single nucleotide polymorphism conferring linezolid resistance in enterococci. Antimicrob. Agents Chemother. 47:3620-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skripkin, E., T. S. McConnell, J. Devito, L. Lawrence, J. A. Ippolito, E. M. Duffy, J. Sutcliffe, and F. Franceschi. 2008. Rx-01: a new family of oxazolidinones that overcomes ribosome-based linezolid resistance. Antimicrob. Agents Chemother. 52:3550-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swaney, S. M., H. Aoki, M. C. Ganoza, and D. L. Shinabarger. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 42:3251-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swaney, S. M., D. L. Shinabarger, R. D. Schaadt, J. H. Bock, J. L. Slightom, and G. E. Zurenko. 1998. Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-104.
- 70.Toh, S. M., L. Xiong, C. A. Arias, M. V. Villegas, K. Lolans, J. Quinn, and A. S. Mankin. 2007. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 64:1506-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Treviño, M., L. Martinez-Lamas, P. A. Romero-Jung, J. M. Giraldez, J. Alvarez-Escudero, and B. J. Regueiro. 2009. Endemic linezolid-resistant Staphylococcus epidermidis in a critical care unit. Eur. J. Clin. Microbiol. Infect. Dis. 28:527-533. [DOI] [PubMed] [Google Scholar]
- 72.Tsiodras, S., H. S. Gold, G. Sakoulas, G. M. Eliopoulos, C. Wennersten, L. Venkataraman, R. C. Moellering, and M. J. Ferraro. 2001. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 38:207-208. [DOI] [PubMed] [Google Scholar]
- 73.Wilson, D. N., F. Schluenzen, J. M. Harms, A. L. Starosta, S. R. Connell, and P. Fucini. 2008. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. USA 105:13339-13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson, P., J. A. Andrews, R. Charlesworth, R. Walesby, M. Singer, D. J. Farrell, and M. Robbins. 2003. Linezolid resistance in clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 51:186-188. [DOI] [PubMed] [Google Scholar]
- 75.Wittmann, H. G., G. Stoffler, D. Apirion, L. Rosen, K. Tanaka, M. Tamaki, R. Takata, S. Dekio, and E. Otaka. 1973. Biochemical and genetic studies on two different types of erythromycin resistant mutants of Escherichia coli with altered ribosomal proteins. Mol. Gen. Genet. 127:175-189. [DOI] [PubMed] [Google Scholar]
- 76.Wolter, N., A. M. Smith, D. J. Farrell, W. Schaffner, M. Moore, C. G. Whitney, J. H. Jorgensen, and K. P. Klugman. 2005. Novel mechanism of resistance to oxazolidinones, macrolides, and chloramphenicol in ribosomal protein L4 of the pneumococcus. Antimicrob. Agents Chemother. 49:3554-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan, K., L. Madden, A. E. Choudhry, C. S. Voigt, R. A. Copeland, and R. R. Gontarek. 2006. Biochemical characterization of the interactions of the novel pleuromutilin derivative retapamulin with bacterial ribosomes. Antimicrob. Agents Chemother. 50:3875-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zurenko, G. E., W. M. Todd, B. Hafkin, B. Meyers, C. Kauffman, J. Bock, J. Slightom, and D. Shinabarger. 1999. Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 848.
- 79.Zurenko, G. E., B. H. Yagi, R. D. Schaadt, J. W. Allison, J. O. Kilburn, S. E. Glickman, D. K. Hutchinson, M. R. Barbachyn, and S. J. Brickner. 1996. In vitro activities of U-100592 and U-100766, novel oxazolidinone antibacterial agents. Antimicrob. Agents Chemother. 40:839-845. [DOI] [PMC free article] [PubMed] [Google Scholar]