Abstract
In this work we report the characterization of plasmid pCTX-M360, isolated from a Klebsiella pneumoniae strain from China and encoding the CTX-M-3 extended-spectrum β-lactamase. Sequence analysis of pCTX-M360 revealed extensive similarity with pEL60 and pCTX-M3, two other enterobacterial plasmids of the IncL/M incompatibility group. Compared to pEL60, pCTX-M360 contains several insertions but lacks most of a 27-kb insert found in pCTX-M3, suggesting that it could be an evolutionary intermediate between pEL60 and pCTX-M3.
Extended-spectrum β-lactamases (ESBLs) are bacterial enzymes that hydrolyze expanded-spectrum cephalosporins and monobactams (13). Infections by ESBL-producing bacteria remain a major threat for patients (13). One ESBL family, CTX-M β-lactamases, is reported worldwide, and they have become the most prevalent ESBLs since the report of the first case in 1986 (1, 2, 3, 10). More than 80 variants of CTX-M-type β-lactamases have been identified in clinical isolates (http://www.lahey.org/Studies/other.asp). These enzymes are classified into five groups according to their amino acid compositions: CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 (15). The corresponding genes coding for the CTX-M-type β-lactamase reside mostly on transferable plasmids, and the chromosomal β-lactamase genes of Kluyvera spp. are believed to be the origin of these genes (4, 9, 16). Recently, plasmids carrying various blaCTX-M genes have been increasingly studied, allowing a better understanding of the mechanisms of dissemination of these genes (5, 7, 12, 17). Despite this progress, more information is still needed to elucidate the complete pedigree of these plasmids (3).
The opportunistic pathogen Klebsiella pneumoniae is one of the major ESBL-producing organisms. We have isolated a multidrug-resistant K. pneumoniae strain from a patient in the intensive care unit in the First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China, in 2003 (L. Luo and Y. J. Lu, unpublished data). A plasmid, designated pCTX-M360, was isolated from this strain. To determine the sequence of pCTX-M360, 5 μg of purified pCTX-M360 was sheared using a Hydroshear apparatus (GeneMachines, San Carlos, CA). To construct a library for sequencing analysis, sheared DNA was fractionated and fragments of 1.5 kb to 3.0 kb were treated with T4 DNA polymerase (Takara, Dalian, China) and ligated to pUC118 digested with HincII (New England Biolabs, Ipswich, MA). The ligated DNA was transformed into Escherichia coli strain DH5α. DNA inserts from a total of 1,004 clones were sequenced by MegaBACE1000 (Applied Biosystems, Foster City, CA) from both ends to achieve approximately 7.5-fold coverage of the plasmid. All sequences were assembled into seven contigs and autofinished using SeqMAN (DNAStar, Madison, WI). Finally, the few remaining gaps on the plasmid map were closed by sequencing the DNA generated by PCR. ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to locate the potential open reading frames (ORFs) on the plasmid. Vector NTI Advance 9.0 (Invitrogen, Carlsbad, CA) was applied to calculate the GC content of the DNA sequence. Potential overlapping or closely clustered ORFs were inspected manually. BLASTx (version 2.0) was used to compare predicted ORFs with the NCBI database to identify potential homologs. Mobile elements and repetitive sequences were first predicted by DNAMAN (Lynnon Biosoft, Vaudreuil-Dorion, Canada) and then identified by searching the NCBI database.
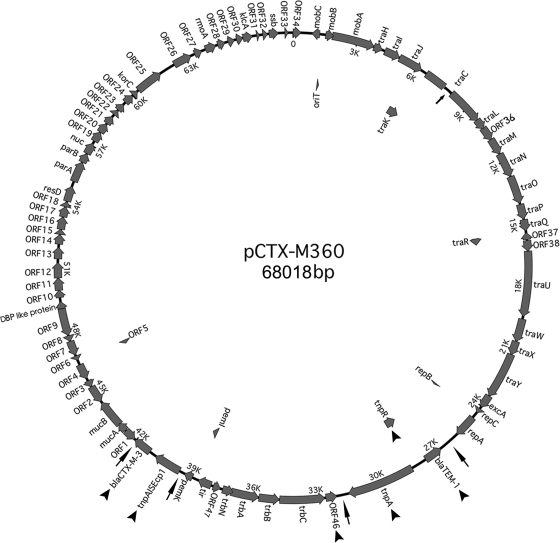
Our analysis revealed that pCTX-M360 is a circular molecule composed of 68,018 bp with a mean GC content of 50.4% (Fig. 1). Eighty-four predicted ORFs were identified, of which 37 encode proteins sharing high similarity to proteins of unknown functions, 46 encode proteins highly similar to proteins with known functions, and one shares low similarity (<30%) to a gene of known function (see Table S1 in the supplemental material).
FIG. 1.
Physical map of pCTX-M360. The hypothetical proteins, predicted ORFs, and their directions inferred from DNA and protein sequence comparisons are indicated by thick arrows. The genes which are absent from pEL60 are marked by arrowheads. The position of the deletion within the traC gene is shown by an arrow inside the circle. Arrows outside the circle indicate the boundaries of Tn2 and of the ISEcp1-blaCTX-M-3 transposition unit.
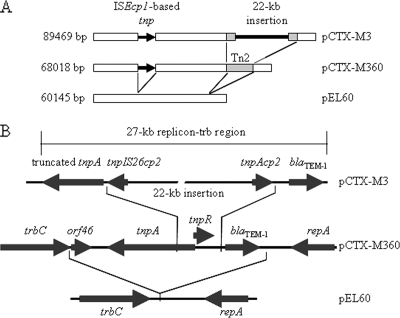
Sequence alignment showed that pCTX-M360 shares extensive similarity with pCTX-M3 (accession no. AF550415) and pEL60 (accession no. AY422214), at both the sequence and the gene organization levels (Fig. 2A). The overall sequence identities between common parts of pCTX-M360 and any of these two plasmids are higher than 98%. The blaCTX-M-3-bearing plasmid pCTX-M3 was isolated originally from Citrobacter freundii in 1996 in Poland and subsequently from other enterobacteria in the same country (1, 2, 8). This plasmid is believed to have been responsible for the quick and extensive spread of the CTX-M-3 ESBL in the family Enterobacteriaceae (8). On the other hand, pEL60 is a highly conjugative plasmid isolated from a plant pathogen, Erwinia amylovora, which causes fire blight in apples and pears (6). Previous investigations revealed that pCTX-M3 and pEL60 are of the IncL/M incompatibility group and suggested that pCTX-M3 might be derived from pEL60 in a multistep process (8, 11).
FIG. 2.
Comparisons of pEL60, pCTX-M360, and pCTX-M3. (A) Overview of the three plasmids; schematic showing the conserved regions and insertions. The regions conserved among the three plasmids are in white, the ISEcp1-based transposon regions are shown by an arrow, the Tn2-like transposon regions are in gray, and the 22-kb insertion is shown by a thick black line. (B) The organization of the regions between trbC and repA among pEL60, pCTX-M360, and pCTX-M3. Arrows indicate the ORFs and their directions of expression. orf46 is a putative transcriptional regulator gene which is also found in pCTX-M3 but absent in pEL60. Note that the lengths of the plasmids, the ORFs, the 22-kb insertion, and the 27-kb replicon-trb region are not to scale.
pCTX-M360 contains two complete transposable elements that are absent on pEL60. One is the ISEcp1-based transposon harboring a blaCTX-M-3 gene located 128 bp from ISEcp1 and terminating at the ISEcp1 alternative right inverted repeat within the Kluyvera ascorbata orf477 sequence (positions 39876 to 42925) (8, 14). Another element is a typical Tn2-type transposon consisting of blaTEM-1, tnpR, and tnpA genes (positions 27071 to 32020) (Fig. 2). Moreover, the intact traC (pri) gene on pEL60 has a 180-bp-long deletion and a short insertion on pCTX-M360. These data strongly suggest that pCTX-M360 was probably derived from pEL60 via several insertion events.
Similarly to pCTX-M360, pCTX-M3 also contains the Tn2-type transposon and the ISEcp1-based transposon (identity rate, >99%), which are inserted at the same places as in pCTX-M360, suggesting a close phylogenetic relationship between these two plasmids. The most significant difference between pCTX-M360 and pCTX-M3 is that pCTX-M3 contains a large DNA segment inserted into the Tn2-like element which, together with this transposon, was described as a “27-kb replicon-trb region” compared to pEL60 (Fig. 2B). The segment located between the blaTEM-1 and tnpA genes of the Tn2-like transposon is 22.4 kb (positions 57991 to 80414 of pCTX-M3) and harbors the majority of the pCTX-M3 mobile elements and resistance genes (Fig. 2B). It appears to have been generated via one or more insertion events that caused also a deletion of tnpR and the 5′ end of tnpA of the Tn2-like transposon, both of which are intact on pCTX-M360 (Fig. 2B). These observations suggest that pCTX-M3 is probably a derivative of pCTX-M360.
In conclusion, our studies suggest that pCTX-M360 was probably derived from pEL60 and may have been a progenitor of pCTX-M3, being possibly an intermediate between those plasmids.
Nucleotide sequence accession number.
The sequence of pCTX-M360 has been deposited in GenBank under accession no. EU938349.
Supplementary Material
Acknowledgments
We thank Zhao-qing Luo at Purdue University for his critical reading of the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (60736028) and the Guangdong Provincial Natural Foundation of China (06201654) to Y.-J.L.
Footnotes
Published ahead of print on 14 September 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baraniak, A., E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2002. Two different extended-spectrum β-lactamases (ESBLs) in one of the first ESBL-producing Salmonella isolates in Poland. J. Clin. Microbiol. 40:1095-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Decousser, J. W., L. Poirel, and P. Nordmann. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Do Carmo Filho, J. R., R. M. Silva, M. Castanheira, M. C. Tognim, A. C. Gales, and H. S. Sader. 2008. Prevalence and genetic characterization of blaCTX-M among Klebsiella pneumoniae isolates collected in an intensive care unit in Brazil. J. Chemother. 20:600-603. [DOI] [PubMed] [Google Scholar]
- 6.Foster, G. C., G. C. McGhee, A. L. Jones, and G. W. Sundin. 2004. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.García-Fernández, A., A. Cloeckaert, A. Bertini, K. Praud, B. Doublet, F.-X. Weill, and A. Carattoli. 2007. Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 51:4177-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gołębiewski, M., I. Kern-Zdanowicz, M. Zienkiewicz, M. Adamczyk, J. Żyliǹska, A. Baraniak, M. Gniadkowski, J. Bardowski, and P. Cegłowski. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 11.Mierzejewska, J., A. Kulińska, and G. Jagura-Burdzy. 2007. Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid 57:95-107. [DOI] [PubMed] [Google Scholar]
- 12.Novais, A., R. Cantón, R. Moreira, L. Peixe, F. Baquero, and T. M. Coque. 2007. Emergence and dissemination of Enterobacteriaceae isolates producing CTX-M-1-like enzymes in Spain are associated with IncFII (CTX-M-15) and broad-host-range (CTX-M-1, -3, and -32) plasmids. Antimicrob. Agents Chemother. 51:796-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez, F., A. Endimiani, K. M. Hujer, and R. A. Bonomo. 2007. The continuing challenge of ESBLs. Curr. Opin. Pharmacol. 7:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez, M. M., P. Power, M. Radice, C. Vay, A. Famiglietti, M. Galleni, J. A. Ayala, and G. Gutkind. 2004. Chromosome-encoded CTX-M-3 from Kluyvera ascorbata: a possible origin of plasmid-borne CTX-M-1-derived cefotaximases. Antimicrob. Agents Chemother. 48:4895-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossolini, G. M., M. M. D'Andrea, and C. Mugnaioli. 2008. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin. Microbiol. Infect. 14(Suppl. 1):33-41. [DOI] [PubMed] [Google Scholar]
- 16.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 17.Zong, Z., S. R. Partridge, L. Thomas, and J. R. Iredell. 2008. Dominance of blaCTX-M within an Australian extended-spectrum β-lactamase gene pool. Antimicrob. Agents Chemother. 52:4198-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.