Abstract
Sulfadoxine-pyrimethamine (SP) resistance in Plasmodium falciparum has been widespread across continents, causing the major hurdle of controlling malaria. Resistance is encoded mainly by point mutations in P. falciparum dihydrofolate reductase (pfdhfr) and dihydropteroate synthase (pfdhps) target genes. To study the origin and evolution of pyrimethamine resistance on the Indian subcontinent, microsatellite markers flanking the pfdhfr gene were mapped. Here we describe the characteristics of genetic hitchhiking around the pfdhfr gene among 190 P. falciparum isolates. These isolates were collected from five different geographical regions of India (Uttar Pradesh, Madhya Pradesh, Assam, Orissa, and Andaman and Nicobar Islands) where malarial transmission rates and levels of drug resistance vary across regions. Among the isolates, we observed a significant reduction in genetic variation in the ±20-kb vicinity of the mutant pfdhfr alleles due to hitchhiking. This reduction in genetic diversity was more prominent around quadruple pfdhfr alleles (heterozygosity [He] = 0.23) than around double (He = 0.365) and single (He = 0.465) mutant alleles. Asymmetry in the selective sweep flanking the pfdhfr alleles was observed with regional isolates, emphasizing the drug usage with the parasite population. All the pfdhfr alleles share a single microsatellite haplotype and seem to have originated from a single progenitor similar to that of Southeast Asian (Thailand) pfdhfr mutants. Results of the present study also indicate that the emergence of drug-resistant alleles is a recent phenomenon in India compared to Southeast Asian countries.
Malaria is the major cause of death in tropical and subtropical areas. Among all the four species of Plasmodium, P. falciparum causes the most severe form of malaria, which often leads to death. In the absence of effective vaccines, management of the disease is dependent largely on antimalarial drugs. However, the parasite has developed resistance to most commonly used antimalarial drugs, thus posing a major problem for malaria control programs (14). Emergence of chloroquine resistance in India has required the use of alternative antimalarials, like the sulfadoxine-pyrimethamine (SP) combination, in the recent past. However, the current drug policy in India for treating uncomplicated P. falciparum malaria cases in highly chloroquine-resistant areas prescribes the use of artemisinin combination therapy, consisting of artesunate plus SP (www.nvbdcp.gov.in). Thus, SP drug pressure on the parasite population continues to be high in the country.
SP acts synergistically, targeting the enzymes dihydropteroate synthase and dihydrofolate reductase, which are involved in the folate biosynthesis pathway of the parasite (13). These parasite enzymes and molecular mechanisms underlying antifolate drug resistance have been well characterized (12, 30, 36). Sequencing of the pfdhfr gene from pyrimethamine-resistant and -sensitive parasite isolates identified the important amino acid changes that alter the binding of the drug and hence confer the resistance (12, 30). A Ser-to-Asn change at amino acid position 108 in P. falciparum dihydrofolate reductase is the key event in the development of the resistance and additional mutations at amino acid residues 51, 59, and 164, further increasing the tolerance of the parasite toward the drug (35).
Levels of pyrimethamine resistance differ among the continental population of the parasite due to the presence of various pfdhfr mutant alleles. The triple mutant A16I51R59N108I164 pfdhfr allele is present mostly in Africa and Southeast Asia, while the quadruple mutant A16I51R59N108L164 allele is observed predominantly in Southeast Asia (6, 26). Previously we have reported the A16N51C59N108I164, A16N51R59N108I164, A16I51R59N108I164, A16N51R59N108L164, and A16I51R59N108L164 pfdhfr mutant alleles from India (1-3). Two different pfdhfr triple mutant R50I51C59N108I164 and C50I51C59N108L164 alleles were found in South America (8, 31). The occurrence of Bolivia repeats was found exclusively in South America, which suggested two different evolutionary lineages of dhfr in South America (8, 24).
Microsatellites are simple sequence repeats, abundantly present in the P. falciparum genome, and polymorphism in the number of repeats occurs mainly because of strand slippage during DNA replication (9, 15, 18, 19). These markers, located close to the resistance loci, have been employed to study the emergence and spread of SP resistance alleles in the P. falciparum population (11). There are reports which revealed that the triple mutant pfdhfr allele has three independent origins, one from Southeast Asia (26, 27, 33) and two from South America (8, 23, 24). A majority of the triple mutant alleles found in Africa originated and migrated from Southeast Asia (7, 21, 22, 25, 33). However, minor, independent origins of the triple mutant dhfr allele have been observed (7, 22). Southeast Asian triple and quadruple pfdhfr mutants have a single origin, whereas the African double mutant originated twice (26, 27, 32).
Due to continued drug pressure, the antifolate drug resistance-associated pfdhfr mutations have been reported from India (1-3). However, the spread of SP resistance alleles across the Indian subcontinent has not been investigated. Therefore, in the present study, we determined the microsatellite haplotypes flanking pfdhfr in isolates from five different geographical regions of India (Uttar Pradesh [UP], Madhya Pradesh [MP], Assam, Orissa, and Andaman and Nicobar Islands) having different malaria transmission dynamics and drug resistance levels (Fig. 1). It has been known that the northern state of UP has low levels of malaria transmission and drug resistance, whereas the central state of MP has moderate levels of malaria transmission and drug resistance. On the other hand, the northeastern state of Assam, eastern state of Orissa, and Andaman and Nicobar Islands have high levels of malaria transmission and drug resistance (17). Previously, we reported the presence of different pfdhfr genotypes among isolates from these different regions, which seem to show a relationship to malaria transmission intensities in these areas (2, 3). For example, the quadruple and triple pfdhfr mutations were present only among isolates from the regions where the intensity of malaria transmission is high, i.e., from Andaman and Nicobar Islands, Assam, and Orissa, but not from the low-malaria-transmission area of UP. We report here that the impact levels of selection on hitchhiking differ among the regions of high and low selection at pfdhfr and that there seems to be an independent origin of the pfdhfr mutant alleles in India.
FIG. 1.
Map of India showing sample collection sites.
MATERIALS AND METHODS
PCR amplification and sequencing of microsatellite loci.
Two hundred microliters of heparinized blood samples was collected from the confirmed P. falciparum malaria patients living in different parts of the country, according to institutional ethical guidelines (Fig. 1). Patients were treated according to the described national drug policy of India for the region (www.nvbdcp.gov.in). Clinical isolates were collected from UP (Aligarh and Ghaziabad), Assam (Kamrup), Andaman and Nicobar Islands (Car Nicobar), Orissa (Ganjam and Jagatsingh Pur), and MP (Jabalpur). The majority of the samples were collected during the years 2005 and 2006. Parasite DNA from infected blood was then extracted according to the protocol described previously using an AccuPrep genomic DNA extraction kit according to the manufacturer's instructions (Bioneer Corporation, South Korea) and used for PCR amplification (37). A total of 255 samples were sequenced for the pfmsp1 gene and neutral microsatellite markers (23) to exclude those samples (n = 25) which showed polyclonal infections. The remaining 230 isolates were processed for PCR amplification, and the microsatellites flanking the pfdhfr gene were sequenced. For this, the P. falciparum DNA was subjected to two rounds of PCR to amplify each of the microsatellite loci (primary-nested strategy). The details of the primer sequences are given in Table S1 in the supplemental material. The PCR was performed in a standard reaction mixture as described previously (37). The PCR products were then resolved on a 1.2% agarose gel, and the expected bands were then excised and eluted using an AccuPrep gel purification kit (Bioneer Corporation, South Korea) according to the manufacturer's protocol. The eluted products were then sequenced from both directions using nested primers on an ABI Prism 310 genetic analyzer (PE Applied Biosystems Inc., Foster City, CA) and previously described protocols (4, 37). The nucleotide sequences were then aligned and compared with the sequences of the 3D7 strain of P. falciparum (www.plasmodb.org). The polymorphism in the number of repeats was noted for each microsatellite locus. We also determined the microsatellite haplotypes for Thai strains M18, M25, M39, and M75 with A16N51R59N108I164, A16I51R59N108L164, A16N51R59N108L164, and A16I51R59N108I164 pfdhfr alleles, respectively, of Southeast Asian origin (parasite DNA was kindly provided by T. J. C. Anderson, Southwest Foundation for Biomedical Research, TX). Mutated amino acids are shown in bold.
Microsatellite polymorphism.
A total of 14 microsatellite loci flanking pfdhfr were chosen for the present study. Out of 14 flanking loci, 7 were located upstream (−0.3, −3.9, −5.2, −7.7, −20, −30, and −50) and 7 downstream (+0.3, +1.28, +5.6, +9.8, +23.7, +28.5, and +50) of the region of pfdhfr. Positions are mentioned with respect to the first ATG codon of the pfdhfr gene. Some of the loci (−0.3, −3.9, −7.7, −50, +0.3, +5.6, and +50) were the same as those described previously (26, 29). Out of the 14 loci, 12 of them (−0.3, −3.9, −5.2, −7.7, −20, −30, −50, +0.3, +1.28, +5.6, +28.5, and +50) contained dinucleotide repeats (AT or TA), and two (+9.8 and +23.7) had trinucleotide (ATA) repeats of various lengths. Polymorphism was scored by counting the number of repeats in the sequences of the microsatellite loci. Samples having missing data for more than two loci were not included in the analysis. This scales down the total number of samples analyzed for microsatellites from 230 to 190.
Statistical analysis.
The genetic variation for each microsatellite locus was assessed by calculating expected heterozygosity (He) at each locus using the formula [n/(n − 1)] (1 − Σpi2), where n is the number of samples genotyped for that locus and pi is the frequency of the ith allele. The expected He was then plotted against the distance from pfdhfr to measure the strength of selective sweep among samples collected from different geographical regions by the use of GraphPad Prism4.The Excel microsatellite tool kit was used to compute the He and allele frequencies (28). Various (unique) haplotypes of the microsatellite loci, along with the pfdhfr genotype, were constructed using Arlequin 3.0 (http://cmpg.unibe.ch/software/arlequin3/) (10). Arlequin was also used to calculate genetic differentiation in terms of Fst to analyze the variations in microsatellite loci between the different geographical regions of India and within the pfdhfr alleles. Linkage disequilibrium along microsatellite loci with pfdhfr allele types was measured by calculating the extended haplotype homozygosity (EHH) (34) using the formula EHH = [∑(pi2) − 1/n]/(1 − 1/n), with pi being the relative haplotype frequency and n the sample size. The value of EHH was plotted against the distance from pfdhfr by the use of GraphPad Prism4.
RESULTS
Analysis of pfdhfr alleles.
A total of 190 samples with a single clonal infection from different geographical regions of India, i.e., from the North (UP, n = 34), Northeast (Assam, n = 31), East (Orissa, n = 35), central India (MP, n = 30), and the southern coastal islands of India (Andaman and Nicobar Islands, n = 60), were analyzed. pfdhfr mutations from 94 of these samples had been reported previously (2, 3, 20), while the remaining 96 samples were analyzed in the present study. Briefly, out of 190 samples, the double mutant A16N51R59N108I164 pfdhfr allele was present in all the five regions and was predominant in UP (82%, n = 34), Assam (84%, n = 31), and Orissa (54%, n = 35), whereas the pfdhfr allele with quadruple mutation A16I51R59N108L164 was detected only with isolates from Andaman and Nicobar, where it was highly prevalent (87%, n = 60) (see Table S2 in the supplemental material). The triple mutant A16N51R59N108L164 pfdhfr allele was detected with fewer isolates from Assam (3%, n = 31) and Orissa (6%, n = 35), while the A16I51R59N108I164 mutation was present in those from Assam (13%, n = 31) and MP (33%, n = 30). Isolates with the A16N51C59N108I164 allele were present in isolates from UP (18%, n = 34) and MP (10%, n = 30). The wild-type A16N51C59S108I164 allele was detected only with isolates from Orissa (40%, n = 35) and MP (20%, n = 30). Thus, the Ile-to-Leu mutation at codon 164 was present among isolates collected from highly malarious areas, i.e., Andaman and Nicobar Islands, Assam, and Orissa. These results are similar to those reported previously from our lab by Ahmed and coworkers and seem to show some relationship to SP pressure in these areas (2, 3).
Genetic diversity at the microsatellite loci.
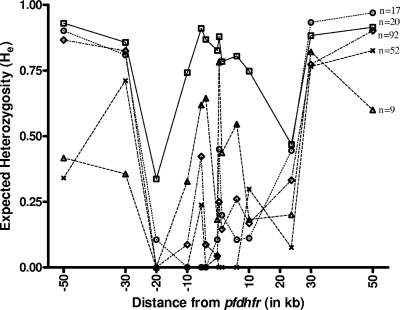
The extent of genetic diversity in the parasite population in terms of expected He for the flanking microsatellites was calculated for all the pfdhfr alleles. The results show that He values for all the pfdhfr alleles were higher for the farthest microsatellite loci than for the nearby loci in the parasite population (Fig. 2). Symmetric reduction in expected He values at the ±20-kb region is due to hitchhiking. Parasite isolates with the wild-type A16N51C59S108I164 allele show high He values at all loci (average He = 0.78). Selective sweep was prominent with the increased level of mutations at pfdhfr (Fig. 2). Parasite isolates with the quadruple mutant A16I51R59N108L164 allele show significant levels of hitchhiking (average He = 0.233) in the flanking microsatellites compared to those with triple (A16I51R59N108I164/A16N51R59N108L164) (average He = 0.363) and double (A16N51R59N108I164) (average He = 0.365) mutant pfdhfr alleles. The parasite population with a single mutant pfdhfr allele (A16N51C59N108I164) shows a high value of genetic diversity in these microsatellites among all pfdhfr mutants (average He = 0.435) (Fig. 2).
FIG. 2.
Genetic variations around the pfdhfr gene among different alleles. Microsatellite markers along the x axis are plotted against the expected He values. He values of Indian P. falciparum isolates having different pfdhfr alleles are shown as the following: □, A16N51C59S108I164; ▵, A16N51C59N108I164; ⋄, A16N51R59N108I164; ○, A16I51R59N108I164/A16N51R59N108L164; ×, A16I51R59N108L164.
Regional variation in hitchhiking around pfdhr alleles.
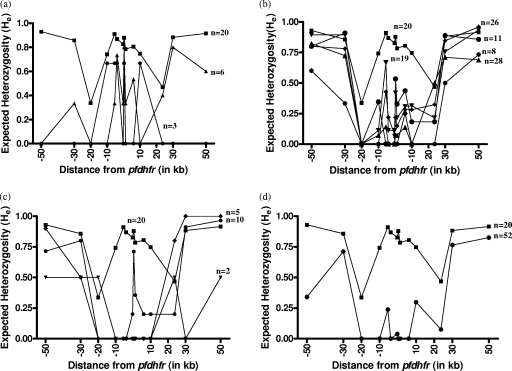
There are regional differences in the selective sweep around pfdhfr alleles among the Indian parasite population. Parasite isolates with double mutant pfdhfr allele A16N51R59N108I164 from Andaman and Nicobar Islands showed more fixation (average He = 0.172) than those with the double mutant pfdhfr allele from UP (average He = 0.281), Assam (average He = 0.382), Orissa (average He = 0.422), and MP (average He = 0.294). Parasite isolates with the quadruple mutant pfdhfr (A16I51R59N108L164), which is the most resistant pyrimethamine allele and found only in Andaman and Nicobar Islands, showed more fixation (up to ±20 kb) (average He = 0.23) (Fig. 3). The single mutant A16N51C59N108I164 pfdhfr allele was found only in isolates from UP and MP and showed more variation in flanking microsatellites than other mutant alleles (UP, average He value of 0.37; MP, He values ranging up to 0.67) (Table 1). An asymmetric selective valley was observed with the parasite population with the mutant pfdhfr allele from Assam, Orissa, and MP (Fig. 3). Microsatellite loci around pfdhfr among Indian isolates were then compared with those of Thai mutants to track the origin of the pyrimethamine resistance in India. Interestingly, the same microsatellite alleles flanking pfdhfr have been observed with parasites of both countries (Table 2).
FIG. 3.
Genetic variations around the pfdhfr gene among different geographical regions. Microsatellite markers along the x axis are plotted against the expected He values. He values of mutant A16N51C59N108I164 (a), A16N51R59N108I164 (b), A16I51R59N108I164/A16N51R59N108L164 (c), and A16I51R59N108L164 (d) pfdhfr alleles from UP (▴), MP (•), Assam (♦), Orissa (▾), and Andaman and Nicobar Islands (*) are shown. He values of isolates having the wild-type A16N51C59S108I16 pfdhfr allele are shown as ▪.
TABLE 1.
Regionwise distribution of expected He at microsatellites around pfdhfr alleles among Indian P. falciparum isolatesa
| Geographical region (no. of isolates)b | pfdhfr mutation (no. of isolates)c |
He at microsatellite locus: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −50 | −30 | −20 | −7.7 | −5.2 | −3.9 | −0.3 | +0.3 | +1.28 | +5.6 | +9.8 | +23.7 | +28.5 | +50 | ||
| UP (34) | A16N51C59N108I164 (6) | 0 | 0.33 | 0 | 0 | 0.33 | 0.73 | 0.33 | 0.8 | 0.33 | 0.53 | 0 | 0.4 | 0.8 | 0.6 |
| A16N51R59N108I164 (28) | 0.823 | 0.724 | 0 | 0.071 | 0.14 | 0 | 0 | 0.071 | 0.071 | 0.138 | 0 | 0.5 | 0.711 | 0.689 | |
| A & N (60) | A16N51R59N108I164 (8) | 0.6 | 0.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.25 | 0 | 0 | 0.5 | 0.733 |
| A16I51R59N108L164 (52) | 0.34 | 0.711 | 0 | 0 | 0.238 | 0 | 0.038 | 0 | 0 | 0 | 0.29 | 0.07 | 0.76 | 0.826 | |
| Assam (31) | A16N51R59N108I164 (26) | 0.8 | 0.78 | 0 | 0 | 0.43 | 0.22 | 0.07 | 0.22 | 0.15 | 0.28 | 0.28 | 0.32 | 0.85 | 0.96 |
| A16I51R59N108I164/A16N51R59N108L164 (5 [4/1]) | 0.9 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.8 | 1 | 1 | |
| Orissa (35) | A16N51C59S108I164 (14) | 0.936 | 0.909 | 0.44 | 0.802 | 0.945 | 0.912 | 0.846 | 0.846 | 0.736 | 0.791 | 0.714 | 0.262 | 0.9 | 0.96 |
| A16N51R59N108I164 (19) | 0.89 | 0.89 | 0 | 0.11 | 0.66 | 0.11 | 0.11 | 0.42 | 0.21 | 0.31 | 0.31 | 0.21 | 0.76 | 0.93 | |
| A16N51R59N108L164 (2) | 0.5 | 0.5 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5 | 0.5 | 0.5 | |
| MP (30) | A16N51C59S108I164 (6) | 0.73 | 0.5 | 0 | 0.53 | 0.6 | 0.8 | 0.6 | 0.8 | 0.33 | 0.6 | 0.8 | 0.73 | 0.67 | 0.7 |
| A16N51C59N108I164 (3) | 0.67 | 0 | 0 | 0.67 | 0.67 | 0.67 | 0 | 0.67 | 0 | 0 | 0 | 0 | 0 | 0.67 | |
| A16N51R59N108I164 (11) | 0.8 | 0.91 | 0 | 0.35 | 0 | 0 | 0 | 0.53 | 0.33 | 0.44 | 0.18 | 0.18 | 0.89 | 0.86 | |
| A16I51R59N108I164 (10) | 0.8 | 0 | 0 | 0 | 0 | 0 | 0.2 | 0.71 | 0.36 | 0.2 | 0.2 | 0.2 | 0.91 | 0.96 | |
Microsatellite loci are the distances from the pfdhfr gene in kb.
n = 190. A & N, Andaman and Nicobar Islands. UP, Uttar Pradesh. MP, Madhya Pradesh.
Mutated amino acids are shown in bold.
TABLE 2.
Microsatellite haplotypes among different pfdhfr genotypes in P. falciparum parasite population
| pfdhfr mutation or group (no. of isolates)a | Haplotype | Microsatellite haplotype at kbb: |
Pic | ||||||
|---|---|---|---|---|---|---|---|---|---|
| −7.7 | −5.2 | −3.9 | −0.3 | 0.3 | +1.28 | +5.6 | |||
| A16N51C59N108I164 (9) | H1 | 10 | 14 | 13 | 17 | 16 | 10 | 11 | 0.11 |
| H2 | 11 | 13 | 13 | 17 | 16 | 10 | 11 | 0.11 | |
| H3 | 10 | 21 | 17 | 17 | 12 | 12 | 11 | 0.11 | |
| H4 | 10 | 21 | 21 | 17 | 14 | 11 | 7 | 0.11 | |
| H5 | 10 | 21 | 17 | 18 | 13 | 11 | 11 | 0.11 | |
| H6 | 10 | 21 | 13 | 17 | 12 | 11 | 11 | 0.11 | |
| H7 | 10 | 14 | 13 | 17 | 16 | 10 | 7 | 0.11 | |
| H8 | 10 | 21 | 17 | 17 | 12 | 11 | 7 | 0.11 | |
| H9 | 10 | 14 | 13 | 17 | 13 | 11 | 7 | 0.11 | |
| A16N51R59N108I164 (92) | H1 | 10 | 14 | 13 | 17 | 16 | 10 | 11 | 0.55 |
| H10 | 10 | 13 | 13 | 17 | 16 | 10 | 11 | 0.11 | |
| H7 | 10 | 14 | 13 | 17 | 16 | 10 | 7 | 0.044 | |
| H11 | 10 | 11 | 13 | 17 | 16 | 10 | 11 | 0.033 | |
| H12 | 10 | 14 | 13 | 17 | 16 | 11 | 11 | 0.033 | |
| H13 | 10 | 14 | 13 | 17 | 17 | 10 | 11 | 0.033 | |
| H14 | 10 | 14 | 13 | 17 | 16 | 10 | 12 | 0.022 | |
| H15 | 10 | 14 | 13 | 17 | 15 | 10 | 11 | 0.022 | |
| H16 | 10 | 11 | 13 | 13 | 12 | 11 | 16 | 0.011 | |
| H17 | 10 | 11 | 18 | 17 | 12 | 11 | 12 | 0.011 | |
| H18 | 10 | 14 | 13 | 17 | 16 | 10 | 16 | 0.011 | |
| H19 | 10 | 11 | 18 | 17 | 16 | 10 | 11 | 0.011 | |
| H20 | 10 | 11 | 23 | 17 | 16 | 10 | 11 | 0.011 | |
| H21 | 10 | 16 | 17 | 17 | 12 | 11 | 7 | 0.011 | |
| H22 | 10 | 13 | 13 | 17 | 15 | 10 | 11 | 0.011 | |
| H23 | 10 | 13 | 13 | 17 | 16 | 10 | 10 | 0.011 | |
| H24 | 10 | 12 | 13 | 17 | 16 | 10 | 11 | 0.011 | |
| H25 | 11 | 14 | 13 | 17 | 16 | 10 | 11 | 0.011 | |
| H26 | 10 | 21 | 13 | 17 | 12 | 10 | 7 | 0.011 | |
| H27 | 19 | 14 | 13 | 17 | 16 | 10 | 11 | 0.011 | |
| H28 | 11 | 14 | 13 | 17 | 17 | 10 | 11 | 0.011 | |
| A16I51R59N108I164 (14) | H1 | 10 | 14 | 13 | 17 | 16 | 10 | 11 | 0.50 |
| H13 | 10 | 14 | 13 | 17 | 17 | 10 | 11 | 0.21 | |
| H29 | 10 | 14 | 13 | 17 | 18 | 10 | 11 | 0.071 | |
| H9 | 10 | 14 | 13 | 17 | 13 | 11 | 7 | 0.071 | |
| H12 | 10 | 14 | 13 | 17 | 16 | 11 | 11 | 0.071 | |
| H30 | 10 | 14 | 13 | 18 | 16 | 10 | 11 | 0.071 | |
| A16N51R59N108L164 (3) | H1 | 10 | 14 | 13 | 17 | 16 | 10 | 11 | 1.00 |
| A16I51R59N108L164 (52) | H1 | 10 | 14 | 13 | 17 | 16 | 10 | 11 | 0.85 |
| H10 | 10 | 13 | 13 | 17 | 16 | 10 | 11 | 0.134 | |
| H31 | 10 | 14 | 13 | 15 | 16 | 10 | 11 | 0.019 | |
| Thai isolates | H1 | 10 | 14 | 13 | 17 | 16 | 10 | 11 | |
Mutated amino acids are shown in bold.
Locations of microsatellites around pfdhfr in kb. Negative positions are upstream of pfdhfr and positive positions are downstream of pfdhfr (20).
Pi is the frequency of the ith haplotype in the population.
Fixation index.
Genetic differentiation in the parasite population across ±50 kb to pfdhfr show moderate Fst values (Fst ≥ 0.26) between the geographical regions (Table 3). The trends were similar when the genetic differentiation was calculated between the dhfr alleles (Fst ≥ 0.344) (Table 4). High Fst values (Fst ≥ 0.5) have been observed when the level of mutation increases from A16N51C59N108I16 to A16I51R59N108L164 in the parasite population (Table 4). This pattern is also depicted among regional distribution of the parasite populations of Andaman and Nicobar Islands, Orissa, and MP (Table 3).
TABLE 3.
Genetic differentiation of dhfr alleles among geographical regionsa
| Geographical region |
Fst for: |
|||
|---|---|---|---|---|
| UP | Assam | Orissa | MP | |
| Assam | 0.0521 | |||
| Orissa | 0.137 | 0.105 | ||
| MP | 0.0758 | 0.117 | 0.0989 | |
| A & N | 0.150 | 0.184 | 0.249 | 0.251 |
A & N, Andaman and Nicobar Islands. UP, Uttar Pradesh. MP, Madhya Pradesh.
TABLE 4.
Genetic differentiation of dhfr alleles among different allelesa
| pfdhfr mutation |
Fst for: |
||
|---|---|---|---|
| A16N51C59N108I164 | A16N51R59 N108I164 | A16I51R59N108I164/A16N51R59N108L164 | |
| A16N51R59N108I164 | 0.3675 | ||
| A16I51R59N108I164/A16N51R59N108L164 | 0.344 | 0.0154 | |
| A16I51R59N108L164 | 0.578 | 0.04 | 0.115 |
Mutated amino acids are shown in bold.
For estimating linkage disequilibrium, EHH was calculated and plotted against the distance from pfdhfr. EHH dropped slowly to zero for all mutants as we moved farther away from the coding region of pfdhfr, whereas sensitive pfdhfr showed consistently low EHH values (Fig. 4).
FIG. 4.
Decline in linkage around the pfdhfr gene (x axis denotes the distance of the microsatellite marker from the pfdhfr gene [in kb], whereas the y axis denotes the value of EHH at that locus). □, A16N51C59S108I164; ▵, A16N51C59N108I164; ⋄, A16N51R59N108I164; ○, A16I51R59N108I164/A16N51R59N108L164; *, A16I51R59N108L164.
DISCUSSION
Introduction of a novel drug into a parasite population causes genomic changes in the parasite DNA to enhance its survival under the drug pressure. These genomic changes are generally the point mutations in the target gene, which further cause the rapid spread of the mutant allele. The spread of the selected allele influences the genetic variation at the flanking loci. This results in reduced variation (“hitchhiking”) and therefore changes in the allele frequency of the microsatellite markers nearest to the locus under selection (5). These markers are used routinely for revealing parasite population structure and transmission dynamics.
Pyrimethamine resistance has been widespread on the Indian subcontinent (1, 2). Genetic variation in terms of expected He has been calculated here for all the loci for the individual pfdhfr alleles among Indian isolates. It was high (He = 0.7 to 0.9) for the wild-type pfdhfr allele, indicating high rates of recombination across the gene (Fig. 2). A single point mutation from Ser to Asn at codon 108 caused moderate pyrimethamine resistance. This event also affects the strength of fixation around the single mutant dhfr allele in Indian isolates. A total of nine microsatellite haplotypes were seen with A16N51C59N108I164 alleles, depicting the multiple origin of the single mutant (Table 2). This is consistent with previous studies reporting that the change from Ser to Asn at codon 108 has evolved multiple times (26, 32). A sequential bottleneck was observed with microsatellite haplotypes when an additional Cys-to-Arg mutation at codon 59, an Asn-to-Ile mutation at 51, and an Ile-to-Leu mutation at codon 164 increased the parasite tolerance to pyrimethamine. Haplotype H1 predominates in the parasite population, whereas other haplotypes are closely related to this haplotype, suggesting that these haplotypes must have originated from the same progenitor H1 through recombination (Table 2). These could not be the novel haplotypes, because the frequency is quite low (7).
With the different geographical regions chosen, we could observe a regional bias among the isolates. The overall expected He was high among the Orissa and MP isolates, as the samples from these regions had high frequencies of the wild-type pfdhfr allele (see Table S2 in the supplemental material). Overall, He values among isolates from UP were moderate, whereas these values were low among Andaman and Nicobar Islands isolates (Table 1). These values are consistent with the fact that Andaman and Nicobar Islands is a high-selection area in terms of SP drug usage compared to UP, leading to the higher-selection events around the pfdhfr allele from Andaman and Nicobar Islands. For example, the double mutant A16N51R59N108I164 pfdhfr allele from Andaman and Nicobar Islands showed more fixation up to ±20 kb (He = 0 to 0.1) than the double mutant allele from any other geographical region. Also, there could be a difference in transit time for the spread of the resistance allele in the population (27). The transit time is fast for the parasite population of Andaman and Nicobar Islands, as it is a strong selection area and recombination events to restore the diversity occur among the resistant pfdhfr alleles. There was no sensitive pfdhfr allele detected among isolates of Andaman and Nicobar Islands, whereas the quadruple mutant A16I51R59N108L164 pfdhfr allele was found only in this region (see Table S2 in the supplemental material).
An asymmetric selective sweep around mutated pfdhfr alleles was observed with isolates from Assam, Orissa, and MP (Fig. 3). Variations were reduced to low levels in upstream loci, whereas downstream loci were significantly polymorphic (Table 1) in these geographic regions. The simulation theory of hitchhiking explains the difference in the levels of genetic variation during strong directional selection, which causes an asymmetric selective sweep around pfdhfr alleles (16). Our results showed consistency with those of previous reports from Southeast Asia (27). Similarly, the triple mutant pfdhfr allele found in central (MP) and eastern (Orissa and Assam) India also showed a greater level of hitchhiking around pfdhfr. This is also consistent with the data obtained from other areas, like Africa and Southeast Asia (7, 26, 32, 33). However, it may be pointed out here that the selective sweep around mutant pfdhfr alleles among Indian isolates was only up to ±20 kb, whereas it went much farther (up to ±100 kb) among isolates from Southeast Asian countries (26), indicating that the emergence of pyrimethamine drug resistance alleles is a recent phenomenon in India. Haplotype H1 of the triple mutant pfdhfr allele shares the lineage with the double mutant A16N51R59N108I164 allele. As the level of mutations increased from double to triple to quadruple in the pfdhfr locus, the change from Ile to Leu at 164 tended to fix up the nearby loci around pfdhfr and show more-extensive linkage disequilibrium around the pfdhfr allele (Fig. 3). This could be explained by differences in fitness levels of the A16I51R59N108L164 allele (27). The A16I51R59N108L164 allele from Andaman and Nicobar Islands was more fit than the A16N51R59N108I164 allele from any other geographical region. The differences in the levels of fitness and selection intensity explain the differences in the genetic diversities among isolates from Andaman and Nicobar Islands and other geographical regions. The haplotype H1 that was shared by a maximum number of isolates showed the single lineage for A16N51R59N108I164 and A16I51R59N108I164/A16N51R59N108L164 mutants (Table 2).
The Fst among pfdhfr alleles suggests that there is more differentiation in pfdhfr alleles from A16N51C59N108I164 to A16I51R59N108L164 (Fst values as high as 0.5), whereas the differentiation for other pfdhfr alleles was less (Fst = 0.01 to 0.1) (Table 4). We also observed a significant level of geographical differentiation among the samples (Table 3). Fst values were high up to 0.25 among MP and Andaman and Nicobar Islands samples because most of the L164 mutations were present in Andaman and Nicobar Islands samples. The lowest Fst values were observed with the alleles of UP, with those from Assam and MP showing similar population structures.
Analyzing the microsatellite haplotypes among geographical regions, we identified regional differences in the Indian P. falciparum population. The difference in the extents of hitchhiking is due to various selection events imposed by SP drug usage. For example, in lower-SP-drug-pressure region of UP, the hitchhiking around pfdhfr mutant alleles in the parasite population reached only up to +10 kb. On the other hand, it reached up to ±20 kb among the isolates from the high-SP-pressure area of Andaman and Nicobar Islands (Fig. 3). It is quite possible that in the future the parasite population with more pfdhfr mutations may move further from high- to low-malaria-transmission regions of the adjacent regions (e.g., from the Northeast to the North). All the mutant pfdhfr alleles in the present study share the predominant haplotype H1, suggesting the possibility that these mutant lineages could have occurred independently in India. However, the most predominant H1 haplotype is also present in the Thai pfdhfr mutants, suggesting that pyrimethamine-resistant alleles of both countries may have a common origin. The alternate possibility of gene flow (particularly of the A16I51R59N108L164 allele) from Thailand to India could also exist, considering the close geographical proximity of the Andaman and Nicobar Islands to Thailand. Nevertheless, analysis of historical samples is required to track the migration of mutant lineages. A strong surveillance system is needed to track the migration of the mutant alleles across the countries through their coasts.
Supplementary Material
Acknowledgments
This work was supported by financial assistance from the Department of Biotechnology (Government of India) and the Indian Council of Medical Research. V.L. received a senior research fellowship from the Council for Scientific and Industrial Research.
We are thankful to T. J. C. Anderson, Southwest Foundation for Biomedical Research, TX, for providing the parasite DNA from Thailand isolates of P. falciparum. The facility of the Bio-Technology Information System (BTIS) of the Biotechnology Department is gratefully acknowledged. We thank Shalini Narang for preparing the manuscript.
Footnotes
Published ahead of print on 28 September 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ahmed, A., D. Bararia, S. Vinayak, M. Yameen, S. Biswas, V. Dev., A. Kumar, M. A. Ansari, and Y. D. Sharma. 2004. Plasmodium falciparum isolates in India exhibit a progressive increase in mutations associated with sulfadoxine-pyrimethamine resistance. Antimicrob. Agents Chemother. 48:879-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, A., M. K. Das, V. Dev, M. A. Saifi, Wajihullah, and Y. D. Sharma. 2006. Quadruple mutations in dihydrofolate reductase of Plasmodium falciparum isolates from Car Nicobar Island, India. Antimicrob. Agents Chemother. 50:1546-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed, A., V. Lumb, M. K. Das, V. Dev, Wajihullah, and Y. D. Sharma. 2006. Prevalence of mutations associated with higher levels of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Car Nicobar Island and Assam, India. Antimicrob. Agents Chemother. 50:3934-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alam, M. T., R. Agarwal, and Y. D. Sharma. 2007. Extensive heterozygosity at four microsatellite loci flanking Plasmodium vivax dihydrofolate reductase gene. Mol. Biochem. Parasitol. 153:178-185. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, T. J. 2004. Mapping drug resistance genes in Plasmodium falciparum by genome-wide association. Curr. Drug Targets Infect. Disord. 4:65-78. [DOI] [PubMed] [Google Scholar]
- 6.Bwijo, B., A. Kaneko, M. Takechi, I. L. Zungu, Y. Moriyama, J. K. Lum, T. Tsukahara, T. Mita, N. Takahashi, Y. Bergqvist, A. Bjorkman, and T. Kobayakawa. 2003. High prevalence of quintuple mutant dhps/dhfr genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 85:363-373. [DOI] [PubMed] [Google Scholar]
- 7.Certain, L. K., M. Briceno, S. M. Kiara, A. M. Nzila, W. M. Watkins, and C. H. Sibley. 2008. Characteristics of Plasmodium falciparum dhfr haplotypes that confer pyrimethamine resistance, Kilifi, Kenya, 1987-2006. J. Infect. Dis. 197:1743-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortese, J. F., A. Caraballo, C. E. Contreras, and C. V. Plowe. 2002. Origin and dissemination of Plasmodium falciparum drug-resistance mutations in South America. J. Infect. Dis. 186:999-1006. [DOI] [PubMed] [Google Scholar]
- 9.Ellegren, H. 2004. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5:435-445. [DOI] [PubMed] [Google Scholar]
- 10.Excoffier, L., G. Laval, and S. Schneider. 2005. posting date. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. 1:47-50. [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdig, M. T., and X. Z. Su. 2000. Microsatellite markers and genetic mapping in Plasmodium falciparum. Parasitol. Today 16:307-312. [DOI] [PubMed] [Google Scholar]
- 12.Foote, S. J., D. Galatis, and A. F. Cowman. 1990. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc. Natl. Acad. Sci. USA 87:3014-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 14.Hyde, J. E. 2005. Drug-resistant malaria. Trends Parasitol. 21:494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashi, Y., and D. G. King. 2006. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 22:253-259. [DOI] [PubMed] [Google Scholar]
- 16.Kim, Y., and W. Stephan. 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160:765-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, A., N. Valecha, T. Jain, and A. P. Dash. 2007. Burden of malaria in India: retrospective and prospective view. Am. J. Trop. Med. Hyg. 77:69-78. [PubMed] [Google Scholar]
- 18.Lai, Y., and F. Sun. 2003. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 20:2123-2131. [DOI] [PubMed] [Google Scholar]
- 19.Li, Y. C., A. B. Korol, T. Fahima, A. Beiles, and E. Nevo. 2002. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol. Ecol. 11:2453-2465. [DOI] [PubMed] [Google Scholar]
- 20.Lumb, V., M. K. Das, P. Mittra, A. Ahmed, M. Kumar, P. Kaur, A. P. Dash, S. S. Singh, and Y. D. Sharma. 2009. Emergence of an unusual sulfadoxine-pyrimethamine resistance pattern and a novel K540N mutation in dihydropteroate synthetase in Plasmodium falciparum isolates obtained from Car Nicobar Island, India, after the 2004 tsunami. J. Infect. Dis. 199:1064-1073. [DOI] [PubMed] [Google Scholar]
- 21.Maiga, O., A. A. Djimde, V. Hubert, E. Renard, A. Aubouy, F. Kironde, B. Nsimba, K. Koram, O. K. Doumbo, J. Le Bras, and J. Clain. 2007. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J. Infect. Dis. 196:165-172. [DOI] [PubMed] [Google Scholar]
- 22.McCollum, A. M., L. K. Basco, R. Tahar, V. Udhayakumar, and A. A. Escalante. 2008. Hitchhiking and selective sweeps of Plasmodium falciparum sulfadoxine and pyrimethamine resistance alleles in a population from central Africa. Antimicrob. Agents Chemother. 52:4089-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCollum, A. M., K. Mueller, L. Villegas, V. Udhayakumar, and A. A. Escalante. 2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51:2085-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCollum, A. M., A. C. Poe, M. Hamel, C. Huber, Z. Zhou, Y. P. Shi, P. Ouma, J. Vulule, P. Bloland, L. Slutsker, J. W. Barnwell, V. Udhayakumar, and A. A. Escalante. 2006. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J. Infect. Dis. 194:189-197. [DOI] [PubMed] [Google Scholar]
- 25.Mita, T., K. Tanabe, N. Takahashi, R. Culleton, M. Ndounga, M. Dzodzomenyo, W. S. Akhwale, A. Kaneko, and T. Kobayakawa. 2009. Indigenous evolution of Plasmodium falciparum pyrimethamine resistance multiple times in Africa. J. Antimicrob. Chemother. 63:252-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair, S., J. T. Williams, A. Brockman, L. Paiphun, M. Mayxay, P. N. Newton, J. P. Guthmann, F. M. Smithuis, T. T. Hien, N. J. White, F. Nosten, and T. J. Anderson. 2003. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol. Biol. Evol. 20:1526-1536. [DOI] [PubMed] [Google Scholar]
- 27.Nash, D., S. Nair, M. Mayxay, P. N. Newton, J. P. Guthmann, F. Nosten, and T. J. Anderson. 2005. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc. Biol. Sci. 272:1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, S. D. E. 2001. Trypanotolerance in West African cattle and the population genetic effects of selection. Ph.D. thesis. University of Dublin, Dublin, Ireland.
- 29.Pearce, R., A. Malisa, S. P. Kachur, K. Barnes, B. Sharp, and C. Roper. 2005. Reduced variation around drug-resistant dhfr alleles in African Plasmodium falciparum. Mol. Biol. Evol. 22:1834-1844. [DOI] [PubMed] [Google Scholar]
- 30.Peterson, D. S., D. Walliker, and T. E. Wellems. 1988. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. USA 85:9114-9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plowe, C. V., J. F. Cortese, A. Djimde, O. C. Nwanyanwu, W. M. Watkins, P. A. Winstanley, J. G. Estrada-Franco, R. E. Mollinedo, J. C. Avila, J. L. Cespedes, D. Carter, and O. K. Doumbo. 1997. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J. Infect. Dis. 176:1590-1596. [DOI] [PubMed] [Google Scholar]
- 32.Roper, C., R. Pearce, B. Bredenkamp, J. Gumede, C. Drakeley, F. Mosha, D. Chandramohan, and B. Sharp. 2003. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet 361:1174-1181. [DOI] [PubMed] [Google Scholar]
- 33.Roper, C., R. Pearce, S. Nair, B. Sharp, F. Nosten, and T. Anderson. 2004. Intercontinental spread of pyrimethamine-resistant malaria. Science 305:1124. [DOI] [PubMed] [Google Scholar]
- 34.Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. Levine, D. J. Richter, S. F. Schaffner, S. B. Gabriel, J. V. Platko, N. J. Patterson, G. J. McDonald, H. C. Ackerman, S. J. Campbell, D. Altshuler, R. Cooper, D. Kwiatkowski, R. Ward, and E. S. Lander. 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832-837. [DOI] [PubMed] [Google Scholar]
- 35.Sirawaraporn, W., R. Sirawaraporn, A. F. Cowman, Y. Yuthavong, and D. V. Santi. 1990. Heterologous expression of active thymidylate synthase-dihydrofolate reductase from Plasmodium falciparum. Biochemistry 29:10779-10785. [DOI] [PubMed] [Google Scholar]
- 36.Triglia, T., J. G. Menting, C. Wilson, and A. F. Cowman. 1997. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc. Natl. Acad. Sci. USA 94:13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vinayak, S., P. Mittra, and Y. D. Sharma. 2006. Wide variation in microsatellite sequences within each Pfcrt mutant haplotype. Mol. Biochem. Parasitol. 147:101-108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.