Abstract
The emergence of multidrug-resistant bacteria highlights the need for new antibacterial agents. Arminin 1a is a novel antimicrobial peptide discovered during investigations of the epithelial defense of the ancient metazoan Hydra. Following proteolytic processing, the 31-amino-acid-long positively charged C-terminal part of arminin 1a exhibits potent and broad-spectrum activity against bacteria, including multiresistant human pathogenic strains, such as methicillin-resistant Staphylococcus aureus (MRSA) strains (minimal bactericidal concentration, 0.4 μM to 0.8 μM). Ultrastructural observations indicate that bacteria are killed by disruption of the bacterial cell wall. Remarkably, the antibacterial activity of arminin 1a is not affected under the physiological salt conditions of human blood. In addition, arminin 1a is a selective antibacterial agent that does not affect human erythrocyte membranes. Arminin 1a shows no sequence homology to any known antimicrobial peptide. Because of its high level of activity against multiresistant bacterial strains pathogenic for humans, the peptide arminin 1a is a promising template for a new class of antibiotics. Our data suggest that ancient metazoan organisms such as Hydra hold promise for the detection of novel antimicrobial molecules and the treatment of infections caused by multiresistant bacteria.
The improper use of antibiotics has led to a widespread increase in the occurrence of drug-resistant and multidrug-resistant bacteria (22, 28). At the same time, the discovery and development of drugs that are effective against these resistant bacteria have slowed (14). In the case of methicillin-resistant Staphylococcus aureus (MRSA), isolates have spread internationally. While they have traditionally been associated with infections in hospitals, new MRSA strains have recently emerged and have rapidly spread in the community. They cause infections that are acquired by persons who have not been recently hospitalized or undergone a medical procedure (19). These so-called community-acquired MRSA (CA-MRSA) strains are responsible for infections with a particular clinical presentation and are associated with skin and soft tissue infections that occur in previously healthy and young persons (19). Another important group of pathogens which are responsible for common nosocomial infections are enterococci. The acquisition of resistance is a major cause of concern, particularly in Enterococcus faecalis and Enterococcus faecium. Most prominently, resistance to penicillin-ampicillin, aminoglycosides (high-level resistance), and glycopeptides is being reported in an increasing number of enterococcal isolates (19), which obviously limits the therapeutic possibilities.
Given the urgent requirement for new drug targets as a basis for antibiotics with novel mechanisms of action and given the fact that phylogenetically old cnidarians are remarkably well equipped to prevent infectious agents from entering the body, we thought that these organisms offer considerable potential as a source of novel antimicrobial substances. This seems to be appreciated all the more in light of the observation that CA-MRSA has already developed resistance to human antimicrobial peptides (AMPs) (20). In this study, it was shown that USA 500 and USA 300, the two major strains of CA-MRSA, had higher levels of resistance to dermicidin and indolicidin, whereas the levels of resistance to other AMPs were unchanged.
In support of this idea, we have previously shown that Toll-like receptor-mediated antibacterial immune responses in the simple metazoan Hydra magnipapillata include short bactericidal peptides with novel structural features and novel modes of action (5, 16). One example is hydramacin 1, a cationic AMP mostly active against gram-negative bacteria, including multiresistant bacteria pathogenic for humans, such as extended-spectrum beta-lactamase (ESBL)-producing Klebsiella oxytoca and Klebsiella pneumoniae strains (5). The initial step of the bactericidal mechanism of hydramacin seems to be aggregation of the bacteria, which has not been described before (16). From the nuclear magnetic resonance-derived solution structure of hydramacin 1, a model which might explain how hydramacin 1 connects the bacterial membranes could be obtained (16). In contrast to the epitheliopeptide hydramacin 1 (5), which is a prototypical AMP, we have recently purified and characterized the anti-Staphylococcus aureus activity of a Hydra protein extract which turned out to be a kazal-type serine protease inhibitor, kazal2 (3). Like all other members of the kazal gene/protein family, it is expressed in secretory gland cells all over the gastric column in Hydra (3).
Here we report on the discovery of a novel family of genes that display characteristics typical of AMPs but that lack cysteines and any sequence homology to known proteins. One member of this family, arminin 1a, exhibits strong activity against MRSA and vancomycin-resistant enterococci (VRE), indicating that it may be a promising new antibacterial drug candidate that could be used to combat the rise of multidrug-resistant pathogens.
MATERIALS AND METHODS
Animals and culture conditions.
Experiments were carried out with Hydra vulgaris strain AEP and Hydra magnipapillata strain 105. Transgenic animals were generated by using H. vulgaris strain AEP. The animals were cultured by standard procedures at 18°C.
Microorganisms.
The following microorganisms were used in this study: Bacillus megaterium ATCC 14581, Staphylococcus aureus ATCC 12600, MRSA ATCC 33393, MRSA ATCC 43300, MRSA 344, MRSA 355, MRSA 358, vancomycin-resistant strain Enterococcus faecalis ATCC 51299, vancomycin-resistant strain Enterococcus faecium 354, vancomycin-resistant strain Enterococcus faecium 356, vancomycin-resistant strain Enterococcus faecium G70, vancomycin-resistant strain Enterococcus faecium G71, ESBL-producing strain Klebsiella pneumoniae ATCC 700603, ESBL-producing strain Klebsiella pneumoniae CF1, ESBL-producing strain Klebsiella pneumoniae CF7, Escherichia coli Rosetta (Novagen), Escherichia coli DH5α, ESBL-producing strain Escherichia coli E4, and ESBL-producing strain Escherichia coli E9.
Bioinformatics.
Nucleotide, protein, and translated BLAST engines at the NCBI server (1), Compagen (12), and http://hydrazome.metazome.net/cgi-bin/gbrowse/hydra/were used for homology searches. The ClustalW program (6) was used for sequence alignments. The SignalP program was used to predict a possible signal peptide (4).
In situ hybridization.
In situ hybridization was performed with H. magnipapillata strain 105, as described previously (2). The digoxigenin-labeled probe used covers the whole open reading frame of the gene for arminin 1a.
Recombinant and synthetically produced arminin 1a.
Recombinant c-arminin 1a was produced as a fusion protein and consisted of the SUMO protein (SMT3; Saccharomyces cerevisiae [21]) and the C-terminal part of arminin 1a starting with amino acid residue K58. The fusion construct was ligated into pPICHOLI (MoBiTec). The resulting overexpression construct contains a His tag at the N terminus. After the transformation of E. coli Rosetta (Novagen), the resulting clones were grown overnight on low-salt Luria-Bertani (LSLB; 2.5 g/liter NaCl) agar plates containing zeocin (25 μg/ml) and chloramphenicol (34 μg/ml). For each overexpression construct, a fresh clone was picked and grown in 5 ml LSLB medium containing zeocin (25 μg/ml) and chloramphenicol (34 μg/ml) overnight. The overnight culture was diluted 1:100 and was incubated at 37°C for 3 h until it reached an optical density at 600 nm (OD600) of 0.6. Protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 2 mM. Three hours after induction, the bacteria were harvested by centrifugation at 8,000 × g for 10 min and were stored at −80°C. The bacterial pellet was resuspended in 50 mM sodium phosphate buffer, pH 8.0-300 mM NaCl. The suspension was sonicated on ice by using a burst regimen consisting of 10 15-s bursts, with a 15-s cooling period being used between each burst. After centrifugation at 15,000 × g for 30 min, the supernatants were applied to Ni-TED columns (Macherey & Nagel). The columns were washed, and the bound proteins were eluted with 250 mM levamisole in 50 mM sodium phosphate, pH 8.0-300 mM NaCl. The protein concentration was measured with a MicroBCA kit (Pierce). Subsequently, the fusion protein (SUMO-c-arminin 1a) was cleaved by using a SUMO protease kit (Invitrogen). One unit of SUMO protease per 20 μg of fusion protein was used in the digestion reaction, which was performed in 50 mM Tris-HCl, pH 8.0-150 mM NaCl-0.2% Igepal (Nonidet P-40)-1 mM dithiothreitol for 2 to 3 h at 30°C. The complete digestion mixture was subjected to reverse-phase high-pressure liquid chromatography (Äkta purifier; GE Healthcare) with a Vydac C4 column (particle size, 5 μm; 250 mm by 3.2 mm; Grace) with gradient elution starting with 0.1% trifluoroacetic acid (TFA)-1.7% (vol/vol) acetonitrile to 0.1% TFA-59% (vol/vol) acetonitrile (slope, 1% [vol/vol] acetonitrile). Cleaved c-arminin 1a was eluted at 35.3% acetonitrile. These fractions were pooled, dried, and redissolved in 0.01% TFA and further analyzed.
A C-terminally amidated variant of c-arminin 1a (amino acid residues 58 to 88) was chemically synthesized as a trifluoroacetate salt at Caslo Laboratory ApS (Technical University of Denmark) and had a purity of 99.8%.
Antimicrobial activity.
The antimicrobial activity of c-arminin 1a was evaluated as described previously (26). Briefly, test isolates were grown for 2 to 3 h in brain heart infusion broth at 36°C, washed three times in 10 mM sodium phosphate buffer (pH 7.4), supplemented with 1% tryptic soy broth, and adjusted to 104 to 105 bacteria/ml. A 100-μl volume of the bacterial suspension was mixed with 10 μl of c-arminin 1a solution (range of final concentrations tested, 0.0125 to 100 μg/ml) and incubated at 36°C. After 2 h, the numbers of CFU were determined. Bacterial suspensions supplemented with 10 μl of phosphate buffer-1% tryptic soy broth instead of c-arminin 1a served as negative controls. The results are given either as the 90% lethal doses or as the minimal bactericidal concentrations (MBCs; >99.9% killing).
Salt dependency of the activity.
The antimicrobial activity of c-arminin 1a against B. megaterium ATCC 14581, E. coli DH5α, and S. aureus ATCC 12600 in the presence of different salt concentrations was examined by the microdilution susceptibility assay described by Fedders and Leippe (8). The experiments were performed in 10 mM sodium phosphate buffer, pH 7.4, containing sodium chloride at a concentration of 0, 75, 150, or 200 mM. The activity in the presence of 0 mM sodium chloride was set equal to 1. The values were expressed as the mean values (including the standard deviations) of two independent experiments, each of which was performed in duplicate.
Assay for hemolytic activity.
The hemolytic activities of c-arminin 1a and the cytotoxic peptide melittin (Sigma) were determined with human red blood cells, as described by Fedders and Leippe (8). In brief, peptides from stock solutions were serially diluted twofold in 50 mM sodium phosphate-150 mM NaCl, pH 7.4, starting from 250 μM for c-arminin 1a and from 20 μM for melittin. As controls, erythrocytes were incubated with distilled water (maximal lysis), and the buffer was supplemented with the corresponding volume of peptide solvent (spontaneous lysis). The absorbance of the diluted supernatants was measured at 405 nm. The values were expressed as the means of two independent experiments, each of which was performed in duplicate.
Electron microscopy (Staphylococcus aureus ATCC 12600).
An overnight culture (12 to 16 h) of S. aureus ATCC 12600 was diluted 1:1,000 and was grown until an OD600 of 0.1 was reached. The bacteria were washed twice with 10 mM sodium phosphate buffer, pH 7.4, supplemented with 1% tryptic soy broth and were concentrated to an OD600 of 3.0 (∼108 bacteria/ml). The bacterial suspension was incubated with c-arminin 1a at a final concentration of 41 or 410 μM peptide (100 and 1,000 times the MBC, respectively) in a final volume of 100 μl. The incubations were performed for 1.5 h at 37°C on a rocking platform. As a negative control, bacteria were incubated with solvent only. As a growth control, after the incubation, 50 μl of the bacterial suspension was plated on Luria-Bertani agar and the numbers of CFU per ml were determined. The other part of the treated bacterial suspension was fixed for 2 h at a ratio of 1:1 with 10% glutaraldehyde (Fluka) in 10 mM sodium phosphate buffer, pH 7.8. Subsequently, samples were treated and embedded as described previously (16).
RESULTS
Identification of novel antimicrobial peptide arminin 1a.
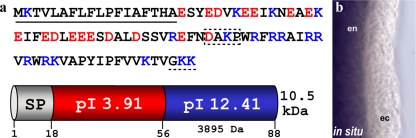
To identify the genes involved in the innate immunity of Hydra magnipapillata, we previously produced cDNA libraries enriched for genes with functions in the immune response by using suppression subtraction hybridization (5). While 65% of the cDNA sequences obtained from this suppression subtraction hybridization showed matches to previously identified proteins by analysis with the BLAST program (5), the remaining 35% of the protein-encoding sequences showed no matches in any database and therefore might represent novel genes. To detect new AMPs within this fraction, we screened for short, secreted, and positively charged peptides. One of the genes which fulfilled these criteria had a strongly positively charged C-terminal region (pI > 12) and a predicted 18-amino-acid residue signal peptide sequence (Fig. 1a). The signal peptide and the cationic C-terminal region are separated by a highly negatively charged N-terminal region (pI < 4). Since detailed searches of other databases for conserved domains or orthologous sequences in genera other than Hydra failed to identify related genes, this sequence encodes a novel gene (Fig. 1a). We have termed this gene and peptide arminin 1a (Fig. 1). Analysis of the Hydra magnipapillata expressed sequence tags as well as the corresponding genome (http://hydrazome.metazome.net/cgi-bin/gbrowse/hydra) indicated that in addition to arminin 1a, there are several more related genes in Hydra magnipapillata (Fig. 2). According to their sequence homology, they can be grouped into three classes (Fig. 2). The two genes most closely related to arminin 1a, termed arminin 1b and arminin 1c, as well as arminin class 2 and class 3 genes, could represent isoforms which originated from recent gene duplication events. Interestingly, sequence alignment of all eight arminin-related peptides revealed that all peptides contain the positively charged C-terminal region. This region is rather nonconserved, whereas the negatively charged N-terminal part shows a high degree of conservation. It is known that a significant part of AMPs are made as precursors. Furthermore, there are examples, e.g., clavanins from Styela clava (29), ranalexin from Rana catesbeiana (7), and human defensin 5 (15), in which the active cationic part is released by cleavage from an anionic region. Unfortunately, there is no algorithm that can be used to predict the cleavages of antimicrobial propeptides. We therefore predicted that the active peptide in the C-terminal region was the longest possible fragment containing no negatively charged residues (c-arminin 1a). Starting with lysine (K58), c-arminin 1a has 31 amino acid residues and a high pI value (Fig. 1), and in addition, it consists of the region with the highest degree of variability among all Hydra arminins (Fig. 2, gray box). In contrast to the majority of AMPs isolated so far (18), c-arminin 1a and arminin 1a do not contain any cysteine residues.
FIG. 1.
(a) Schematic representation and size of the domains characterized by the separation of negatively and positively charged amino acid residues of arminin 1a. Underlined sequence, amino acid signal sequence; box with dashed border, putative cleavage site; dashed underline, putative amidation signal; red amino acids, negatively charged residues; blue amino acids, positively charged residues. The whole peptide has a predicted molecular mass of 10.5 kDa, and the functional cationic C-terminal part (c-arminin 1a) has a predicted molecular mass of 3.9 kDa. (b) In situ hybridization with the gene for arminin 1a showed strong blue staining in the entodermal (en) epithelium along the body column in Hydra. ec, ectodermal.
FIG. 2.
Arminin 1a is a member of a gene family containing eight members. Amino acid sequence alignment with the ClustalW algorithm shows a conserved N-terminal part and a variable C-terminal part (gray box). Nearly all positively charged amino acid residues are within the variable part (compare with Fig. 1a). The homology tree illustrates how arminin family members group together according to their sequence identity (the numbers given at the nodes of the tree are the percentage of identical amino acid residues).
Interestingly, in situ hybridization showed that arminin 1a is expressed in the entodermal epithelium along the body column (Fig. 1b). Since it was shown that the entoderm is the most vulnerable tissue in Hydra (5), this would be the most preferable site of expression of an AMP.
The C-terminal part of arminin 1a has a high degree of activity and broad-spectrum efficacy against multiresistant human pathogens.
To test whether the predicted peptide from the C-terminal part of arminin 1a has antimicrobial activity, we produced recombinant c-arminin 1a in Escherichia coli. When c-arminin 1a was used in liquid growth inhibition assays, we observed a very strong bactericidal activity against Bacillus megaterium ATCC 14581, Escherichia coli DH5α, and Staphylococcus aureus ATCC 12600, indicating that this peptide is a potent component of the Hydra host defense. Uncovering a high degree of bactericidal activity against S. aureus led us to characterize the MBC against S. aureus in more detail. Additionally, it was interesting to see whether the already implied broad-spectrum efficacy against gram-negative and gram-positive bacteria holds true. For this larger screening, synthetically produced c-arminin 1a, which was amidated at the C terminus, was used. We confirmed (Table 1) that c-arminin 1a is highly active against B. megaterium ATCC 14581, E. coli DH5α, and S. aureus ATCC 12600. In addition, c-arminin 1a was found to be capable of killing a number of MRSA strains at very low concentrations of 0.4 to 0.8 μM (Table 1). Moreover, c-arminin 1a was also active against gram-positive vancomycin-resistant Enterococcus faecalis and Enterococcus faecium strains. Interestingly, c-arminin 1a was able to kill gram-negative ESBL-producing members of the Enterobacteriaceae family, such as K. pneumoniae strains and E. coli strains E4 and E9 (Table 1). These observations make c-arminin 1a a potent and broad-spectrum AMP and therefore an interesting lead structure for the design of a novel group of antibiotics.
TABLE 1.
Antimicrobial activity of c-arminin 1a
| Strain group and strain | MBC (μM) | LD90a (μM) |
|---|---|---|
| S. aureus ATCC 12600 | 0.4 | 0.05 |
| E. coli DH5α | 0.2 | 0.1 |
| B. megaterium ATCC 14581 | 0.1 | 0.01 |
| MRSA strains | ||
| ATCC 33393 | 0.40 | 0.20 |
| ATCC 43300 | 0.40 | 0.20 |
| Wild strain 344 | 0.80 | 0.20 |
| Wild strain 355 | 0.80 | 0.20 |
| Wild strain 358 | 0.40 | 0.20 |
| VRE strains | ||
| E. faecalis ATCC 51299 | 1.60 | 0.80 |
| E. faecium wild strain 354 | 0.40 | 0.20 |
| E. faecium wild strain 356 | 0.80 | 0.20 |
| E. faecium wild strain G70 | 0.40 | 0.20 |
| E. faecium wild strain G71 | 0.40 | 0.20 |
| Gram-negative ESBL-producing Enterobacteriaceae strains | ||
| K. pneumoniae ATCC 700603 | 0.80 | 0.20 |
| K. pneumoniae wild strain CF1 | 0.40 | 0.20 |
| K. pneumoniae wild strain CF7 | 0.40 | 0.20 |
| E. coli wild strain E4 | 0.40 | 0.10 |
| E. coli wild strain E9 | 0.20 | 0.05 |
LD90, 90% lethal dose.
c-Arminin 1a is active in the presence of a wide range of salt concentrations.
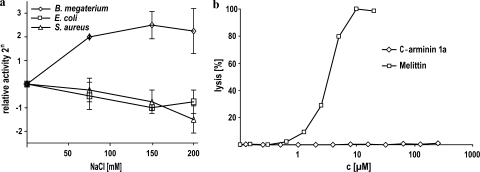
To assess the activity of c-arminin 1a in the presence of different salt concentrations, we used a microdilution susceptibility assay and NaCl concentrations ranging from 0 to 200 mM. As shown in Fig. 3a, the activity of c-arminin 1a against E. coli DH5α and S. aureus ATCC 12600 in the presence of a salt concentration that mimics the physiology of human blood (150 mM) decreased by only a factor of 2, while the activity against B. megaterium ATCC 14581 was more than four times higher than that under low-salt conditions. A further increase in the salt concentration did not significantly affect the activity of c-arminin 1a against E. coli and B. megaterium, while the activity against S. aureus continued to decrease slightly. These data show that c-arminin 1a is still active even under the physiological conditions of human blood. That finding was unexpected, since Hydra is an obligate freshwater animal with nearly no salt tolerance.
FIG. 3.
Activity of c-arminin 1a in the presence of different salt concentrations and on red blood cell membranes. (a) The antimicrobial activity of c-arminin 1a against B. megaterium ATCC 14581 (diamonds), E. coli DH5α (squares), and S. aureus ATCC 12600 (triangles) was tested in the presence of different salt concentrations. Data were obtained from microdilution susceptibility assays performed at pH 7.4 with 0, 75, 150, or 200 mM NaCl. The values are expressed as the mean (including the standard deviation) of the relative activity of two experiments, each of which was performed in duplicate. The activity measured by using buffer without NaCl was set equal to 1. (b) The activity of c-arminin 1a (up to 250 μM) against human red blood cells was tested. The cytotoxic peptide melittin (up to 20 μM) was used as a positive control.
c-Arminin 1a is a selective antibacterial agent that does not affect eukaryotic cells.
Selectivity, or the killing of bacteria at concentrations not harmful to normal eukaryotic cells, is a highly desirable characteristic of an antimicrobial agent. To determine selectivity, c-arminin 1a was evaluated for its toxicity to human red blood cells by using an erythrocyte lysis assay under the physiological conditions of human blood. Red blood cells were incubated with various concentrations of c-arminin 1a in 10 mM phosphate buffer for 1 h. As shown in Fig. 3b, even at concentrations 50 times higher then the MIC, no harmful effects of c-arminin 1a on red blood cells could be detected. As a control, the exposure of red blood cells to the cytotoxic peptide melittin resulted in dose-dependent hemolytic activity.
Morphological analysis of S. aureus exposed to c-arminin 1a.
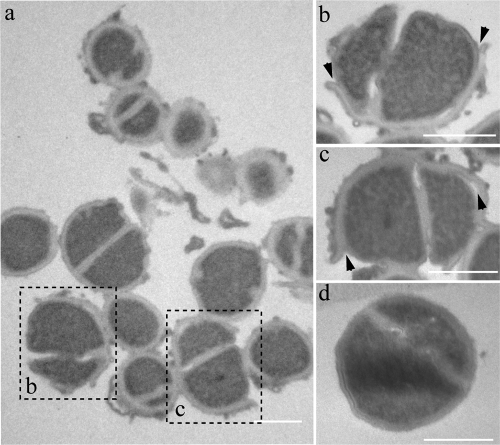
In general, AMPs have different ways of interfering with the mechanisms essential for bacterial survival. The most common and best-studied mode is the integration of AMPs into the bacterial membrane, which leads to membrane destabilization, energy leakage, and finally, the death of the bacterial cell. To understand how exposure to c-arminin 1a leads to the death of the bacterial cell, we examined the morphological changes to S. aureus ATCC 12600 (108 CFU/ml) after treatment with 410 μM of c-arminin 1a for 1.5 h by transmission electron microscopy. As shown in Fig. 4, short-term exposure to arminin 1a leads to the destruction and detachment of the peripheral cell wall of S. aureus ATCC 12600. S. aureus cells exposed to the AMP human beta defensin 3 (HBD3) showed similar cell wall defects (11). Interestingly, HBD3 and c-arminin 1a also share other functional characteristics, such as salt insensitivity and broad-spectrum antimicrobial activity, especially against MRSA and VRE strains (11). Since it is known that HBD3 interferes with the membrane-bound enzyme machinery for cell wall biosynthesis (27), it will be interesting to find out whether c-arminin 1a functions in a similar manner.
FIG. 4.
Changes in morphology of c-arminin 1a-treated S. aureus ATCC 12600. (a) Transmission electron micrograph of S. aureus (108 cells/ml) incubated with c-arminin 1a (410 μM, i.e., 1,000 times the MBC) for 1.5 h. (b and c) Magnifications of two bacterial cells shown in panel a; the arrowheads point to the detachment of the peripheral cell wall. (d) Transmission electron micrograph of S. aureus (108 cells/ml) incubated in 10 mM sodium phosphate buffer, pH 7.4, for 1.5 h as a negative control (intact cells). Bars, 1 μm.
DISCUSSION
The problem of bacterial resistance to antimicrobial drugs is nearly as old as the drugs themselves. With the emergence outside hospitals of multidrug-resistant bacteria that especially affect young persons who have not recently been hospitalized, the need for novel antibacterial substances has never been greater (24).
Probably hundreds, if not even thousands, of AMPs are known to exist in organisms from amoebae to humans. However, very few are as active against MRSA as the AMP arminin 1a (C-terminal part) described in this study. The discovery of the novel peptide arminin 1a derived from the basal metazoan Hydra therefore has several implications. First, c-arminin 1a is bactericidal at nanomolar concentrations against many medically important bacteria. Its selectivity for microbial pathogens and a lack of efficacy against eukaryotic cells make it a promising new drug candidate. Its ability to kill bacteria in the presence of various salt concentrations which mimic the salt concentrations in human blood adds to the attractiveness of this candidate molecule. Second, the absence of cysteines in the peptide strongly facilitates the production of c-arminin 1a in large quantities. Third, the activity of c-arminin 1a is not restricted to gram-negative or gram-positive bacteria. Although the precise mode of action remains to be uncovered, its broad spectrum of activity indicates the wide applicability of c-arminin 1a and its derivatives as new effective antibiotics. So far the activity of c-arminin 1a has been examined by a number of in vitro assays. To develop this peptide into a pharmaceutically useful drug, extensive in vivo evaluations will be required.
Phylogenetically old organisms as drug sources.
In the last 2 years, evidence has accumulated that the innate defense systems of ancient organisms employ both evolutionarily conserved signaling pathways and novel taxon-specific host defense-associated molecules (3, 5, 13, 16, 23). Thus, despite the conservation of the pathogen recognition system and the signaling pathways (5, 25), effector molecules have been modified early during animal evolution and branched off into a variety of unique components in present-day organisms. We have proposed elsewhere (17) that such effector molecules without homologues, classified as orphan or taxonomically restricted genes, in combination with the rewiring of the conserved regulatory networks, allow organisms to make fine adaptations to constantly changing ecological conditions. Why do novel AMPs isolated from organisms only very distantly related to humans and living in completely different environments kill bacteria pathogenic for humans with such a high degree of efficacy? Colonizing microbiota are known to adapt quickly to their hosts (9, 10). Therefore, bacteria closely associated with a particular species may be much less sensitive to the antimicrobial molecules produced by their hosts than to the antimicrobial molecules derived from unrelated organisms.
Taken together, ancient animals such as Hydra and their plethora of highly active novel AMPs appear to be sources of antimicrobial drugs that are effective even against multiresistant bacteria.
Acknowledgments
We thank C. Gelhaus (mass spectrometry) and S. Voss (antimicrobial activity screening) for their excellent experimental support and G. Waetzig and S. Lino for critically reading the manuscript.
The work was supported in part by grants from the Deutsche Forschungsgemeinschaft (grant DFG SFB 617-A1) and grants from the DFG Cluster of Excellence programs The Future Ocean and Inflammation at Interfaces (to T.C.G.B.).
Footnotes
Published ahead of print on 21 September 2009.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Augustin, R., A. Franke, K. Khalturin, R. Kiko, S. Siebert, G. Hemmrich, and T. C. Bosch. 2006. Dickkopf related genes are components of the positional value gradient in Hydra. Dev. Biol. 296:62-70. [DOI] [PubMed] [Google Scholar]
- 3.Augustin, R., S. Siebert, and T. C. Bosch. 2009. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra's innate immune system. Dev. Comp. Immunol. 33:830-837. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bosch, T. C., R. Augustin, F. Anton-Erxleben, S. Fraune, G. Hemmrich, H. Zill, P. Rosenstiel, G. Jacobs, S. Schreiber, M. Leippe, M. Stanisak, J. Grotzinger, S. Jung, R. Podschun, J. Bartels, J. Harder, and J. M. Schroder. 2009. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev. Comp. Immunol. 33:559-569. [DOI] [PubMed] [Google Scholar]
- 6.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, D. P., S. Durell, W. L. Maloy, and M. Zasloff. 1994. Ranalexin. A novel antimicrobial peptide from bullfrog (Rana catesbeiana) skin, structurally related to the bacterial antibiotic, polymyxin. J. Biol. Chem. 269:10849-10855. [PubMed] [Google Scholar]
- 8.Fedders, H., and M. Leippe. 2008. A reverse search for antimicrobial peptides in Ciona intestinalis: identification of a gene family expressed in hemocytes and evaluation of activity. Dev. Comp. Immunol. 32:286-298. [DOI] [PubMed] [Google Scholar]
- 9.Fraune, S., Y. Abe, and T. Bosch. 2009. Disturbing epithelial homeostasis in the metazoan Hydra leads to drastic changes in associated microbiota. Environ. Microbiol. 11:2361-2369. [DOI] [PubMed] [Google Scholar]
- 10.Fraune, S., and T. C. Bosch. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. USA 104:13146-13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 12.Hemmrich, G., and T. C. Bosch. 2008. Compagen, a comparative genomics platform for early branching metazoan animals, reveals early origins of genes regulating stem-cell differentiation. Bioessays 30:1010-1018. [DOI] [PubMed] [Google Scholar]
- 13.Hemmrich, G., D. J. Miller, and T. C. Bosch. 2007. The evolution of immunity: a low-life perspective. Trends Immunol. 28:449-454. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, D. 2003. Exploiting genomics, genetics and chemistry to combat antibiotic resistance. Nat. Rev. Genet. 4:432-441. [DOI] [PubMed] [Google Scholar]
- 15.Jones, D. E., and C. L. Bevins. 1992. Paneth cells of the human small intestine express an antimicrobial peptide gene. J. Biol. Chem. 267:23216-23225. [PubMed] [Google Scholar]
- 16.Jung, S., A. J. Dingley, R. Augustin, F. Anton-Erxleben, M. Stanisak, C. Gelhaus, T. Gutsmann, M. U. Hammer, R. Podschun, A. M. Bonvin, M. Leippe, T. C. Bosch, and J. Grotzinger. 2009. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan Hydra. J. Biol. Chem. 284:1896-1905. [DOI] [PubMed] [Google Scholar]
- 17.Khalturin, K., F. Anton-Erxleben, S. Sassmann, J. Wittlieb, G. Hemmrich, and T. C. Bosch. 2008. A novel gene family controls species-specific morphological traits in Hydra. PLoS Biol. 6:e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, Y., and R. L. Gallo. 2009. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 30:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leclercq, R. 2009. Epidemiological and resistance issues in multidrug-resistant staphylococci and enterococci. Clin. Microbiol. Infect. 15:224-231. [DOI] [PubMed] [Google Scholar]
- 20.Li, M., B. A. Diep, A. E. Villaruz, K. R. Braughton, X. Jiang, F. R. DeLeo, H. F. Chambers, Y. Lu, and M. Otto. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 106:5883-5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, S. J., and M. Hochstrasser. 1999. A new protease required for cell-cycle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 22.Livermore, D. 2004. Can better prescribing turn the tide of resistance? Nat. Rev. Microbiol. 2:73-78. [DOI] [PubMed] [Google Scholar]
- 23.Miller, D. J., G. Hemmrich, E. E. Ball, D. C. Hayward, K. Khalturin, N. Funayama, K. Agata, and T. C. Bosch. 2007. The innate immune repertoire in cnidaria—ancestral complexity and stochastic gene loss. Genome Biol. 8:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne, D. J., M. N. Gwynn, D. J. Holmes, and D. L. Pompliano. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 25.Rosenstiel, P., E. E. R. Philipp, S. Schreiber, and T. C. G. Bosch. 2009. Evolution and function of innate immune receptors—insights from marine invertebrates. J. Innate Immun. 1:291-300. [DOI] [PubMed] [Google Scholar]
- 26.Sahly, H., S. Schubert, J. Harder, P. Rautenberg, U. Ullmann, J. Schroder, and R. Podschun. 2003. Burkholderia is highly resistant to human beta-defensin 3. Antimicrob. Agents Chemother. 47:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sass, V., U. Pag, A. Tossi, G. Bierbaum, and H. G. Sahl. 2008. Mode of action of human beta-defensin 3 against Staphylococcus aureus and transcriptional analysis of responses to defensin challenge. Int. J. Med. Microbiol. 298:619-633. [DOI] [PubMed] [Google Scholar]
- 28.Walsh, C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65-70. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, C., L. Liaw, I. H. Lee, and R. I. Lehrer. 1997. cDNA cloning of clavanins: antimicrobial peptides of tunicate hemocytes. FEBS Lett. 410:490-492. [DOI] [PubMed] [Google Scholar]