Abstract
The long-term impact of field-deployed genetically modified trees on soil mutualistic organisms is not well known. This study aimed at evaluating the impact of poplars transformed with a binary vector containing the selectable nptII marker and β-glucuronidase reporter genes on ectomycorrhizal (EM) fungi 8 years after field deployment. We generated 2,229 fungal internal transcribed spacer (ITS) PCR products from 1,150 EM root tips and 1,079 fungal soil clones obtained from the organic and mineral soil horizons within the rhizosphere of three control and three transformed poplars. Fifty EM fungal operational taxonomic units were identified from the 1,706 EM fungal ITS amplicons retrieved. Rarefaction curves from both the root tips and soil clones were close to saturation, indicating that most of the EM species present were recovered. Based on qualitative and/or quantitative α- and β-diversity measurements, statistical analyses did not reveal significant differences between EM fungal communities associated with transformed poplars and the untransformed controls. However, EM communities recovered from the root tips and soil cloning analyses differed significantly from each other. We found no evidence of difference in the EM fungal community structure linked to the long-term presence of the transgenic poplars studied, and we showed that coupling root tip analysis with a soil DNA cloning strategy is a complementary approach to better document EM fungal diversity.
The poplar has become a model tree species in genetic engineering as it can easily be transformed and clonally propagated and has a small genome size (7, 77, 80). Tree growth, agronomic traits, and timber quality can be improved through genetic engineering (61), thereby avoiding the long reproductive cycles of conventional breeding (47, 59, 83). However, concerns have arisen about the potential impact of genetically modified (GM) trees on the environment (10). The potential environmental hazards linked to GM trees differ from those associated with transgenic crop plants at both spatial and temporal scales (84) because trees are long-lived perennials, unlike annual crop plants. They display several biotic interactions with soil microbial communities such as bacteria and fungi. Interactions between GM trees and these communities could result in exposure to the expression of new traits over several decades, a period longer than those for GM crop plants.
Impact studies of GM plants on nontarget organisms usually focus on the potential risk linked to transgene expression (expected effects) that confers a genetic advantage to the transformed plant rather than on unforeseen (pleiotropic) effects from transgene insertion or the expression of other transgene components such as selection markers or reporter genes. The nptII gene, encoding neomycin phosphotransferase II (EC 2.7.1.95), and the GUS gene, encoding β-glucuronidase (GUS; EC 3.2.1.31), are frequently used for genetic selection of transformed cells and for monitoring transgene presence and expression during transgenic plant lifetime (76). The products of the nptII and GUS genes have been subjected to safety assessment studies and were shown to be nondeleterious to human and animal health (21, 23, 27, 51). Nevertheless, pleiotropic effects in crop plants transformed with the nptII and GUS genes have been observed (2, 15, 17, 43). Pleiotropic effects from GM trees coexpressing such selectable markers have also been recorded. For example, Pasonen et al. (56) showed a significant decrease in the number of root tips colonized by Paxillus involutus associated with a line of chitinase-transformed silver birch in vitro. Similar results have been observed in vivo with P. involutus associated with a line of lignin-modified silver birches (72).
Many trees in temperate, boreal, tropical, and subtropical forests establish mutualistic interactions with ectomycorrhizal (EM) fungi (42, 66, 67, 68). EM fungi are a polyphyletic group comprising over 5,000 species (49) that play key roles in biogeochemical soil processes and plant health. They represent one-third of the total microbial biomass in the soil of boreal forests (32). Fine roots colonized by EM fungi, also called EM root tips or ectomycorrhizae, display a fungal mantle from which extends the extraradical mycelium to prospect the soil for nutrient uptake. These two anatomical parts can be sampled for EM fungus molecular identification, but some studies have highlighted dissimilarities between the EM fungal diversity recorded in root tip sampling and that recorded in extraradical mycelium sampling (26, 37, 39).
Given the potential cumulative effects caused by the presence and stable constitutive expression of transgenes over years on GM tree fitness and on the environment, impact studies of GM trees require long-term field trials (5, 72, 84). In this study, we investigated the potential long-term impact on the EM fungal community of hybrid poplars transformed with the binary vector containing the selectable nptII marker and GUS reporter genes, field deployed for 8 years. This plantation was part of the first confined field trial of transgenic trees in Canada. Hybrid poplars constitutively expressed the nptII gene for kanamycin resistance driven by the NOS promoter (30). The activity of the NOS promoter has been shown to increase in the lower part of transgenic tobacco plants (4). Such a vertical gradient has also been observed in transgenic hybrid poplars, where the NOS promoter activity was 2.4-fold higher in roots than in leaves (87).
As no direct negative impact of nptII or GUS gene expression on fungal organisms has been reported in the literature, we first tested the null hypothesis (H0) that the EM fungal community recorded from transgenic poplars was similar to that from untransformed poplars. Second, since the EM fungal diversity picture can be influenced by the sampling method, we contrasted the EM fungal community recovered from root tips with that recorded in soil cloning analyses. Internal transcribed spacer (ITS) sequences from the nuclear rRNA were produced from both EM root tips and extraradical mycelia to compare the EM fungal communities associated with three control and three transgenic poplars. EM fungal communities were characterized by measuring the usual qualitative and quantitative EM species diversity within each community (α-diversity) and then estimating the nucleotide diversity between EM communities in relation to EM phylotype relative abundances (quantitative β-diversity).
MATERIALS AND METHODS
Field site and sampling strategy.
The study site was located at the Valcartier Research Station (Natural Resources Canada, Canadian Forest Service), and the plantation area was 540 m2 (0.05 ha). Geographic, climatic, and pedologic features of the plot are summarized in Table S1 in the supplemental material. The site belongs to the balsam fir-white birch bioclimatic domain. The plot is surrounded mainly by mature white pine (Pinus strobus L.) and red pine (Pinus resinosa Ait.) stands. It was the first trial with outplanted transgenic trees in Canada, and many precautions were taken to avoid transgenic root escape and contact with the neighboring forest. The transformed poplars and controls were deprived of physical contact with their surroundings by a ditch and a geotextile membrane at the bottom of the plot. The underground part of the plot was composed of superposed horizons of gravel, sand (40 cm), and organic soil (20 cm). Poplar clone 5339 (Populus alba L. × Populus grandidentata Michx.) trees were transformed using the binary vector pRT210, containing the wound-inducible promoter from proteinase inhibitor II (pin2) that drives the expression of the GUS reporter gene and the nptII gene driven by the NOS promoter for plant kanamycin selection (30). Transgenic and control poplars were planted in August 1997. Two guard rows of nontransgenic Populus nigra L. (clone 3051) trees surrounded the plantation. The experimental plantation system consisted of 10 blocks; two repetitions of each cloning line were randomly distributed within each block, with a 1-m by 1-m tree spacing. In early October 2005, a total of 24 soil cores (5 cm in diameter, 30 cm in length) and 24 root samples were collected from the three control and three transgenic poplars belonging to the same transformed line from three separate blocks. To limit biases in recovering EM fungal diversity due to a patchy EM distribution, the four cardinal points around each tree were sampled at 30 cm from the trunk. Each soil core presented a 20-cm-thick organic horizon (OH), very dark and compact, and a 10-cm-thick mineral horizon (MH), sandy and gravelly, at the bottom. From each core, around 20 cm3 of organic and mineral soil was subsampled and sieved. Root samples were collected at the same cardinal points from the OH and tracked to ensure that they were linked to the targeted tree. Soil cores and root samples were kept at −80°C until processing.
Soil analyses.
Moisture, organic matter, total C, primary nutrients (N, P, and K), secondary nutrients (Mg and Ca), micronutrients (Zn), H2O pH, cation exchange capacity, and texture were measured from the OH of each of the 24 cores collected in the plantation on a CNS-2000 analyzer (Leco Corp., St. Joseph, MI). Table S1 in the supplemental material presents the average values of these parameters computed from the 24 soil subsamples.
DNA extraction and amplification.
Soil core subsamples were wet sieved (mesh size: 1 mm) to remove rocks and root fragments. Two hundred fifty milligrams (wet weight) was used for total genomic DNA extraction using the UltraClean soil DNA extraction kit (Mo Bio Laboratories, Solana Beach, CA) according to the manufacturer's instructions, except that the final elution volume was 50 μl rather than 100 μl. From each root sample, a minimum of three thin roots, 5 to 10 cm in length, were analyzed for root tip isolation. All the root tips observed (one per aggregate) were sampled along the thin root to obtain a collection of 96 root tips per cardinal point. Root tips were individually frozen in liquid nitrogen and ground in a 1.5-ml Eppendorf tube with a polypropylene micropestle. Ground tissues were resuspended in 30 μl of ultrapure DNase/RNase-free distilled water (Gibco, New York, NY), then added to 150 μl of 15% Chelex 100 (Bio-Rad) suspension with proteinase K (1.6 μg μl−1) (Invitrogen, Carlsbad, CA). Samples were incubated for 2 h at 65°C, then for 20 min at 95°C to inactivate proteinase K, and centrifuged at 1,000 rpm for 5 min. One microliter of the supernatant was used as the genomic DNA template for PCR. ITS regions were amplified using the ITS1-F/ITS4 primer set (88) to sequence the root tip data set and to build soil clone libraries from the OH and MH. The PCR mixture was made up of 1× PCR buffer, 1.6 mM MgCl2, 1.25 mM of each deoxynucleotide triphosphate, 25 μg of bovine serum albumin (Sigma, St. Louis, MO), 0.5 μM of each primer, and 1 unit of Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany), in a total volume of 25 μl. Thermal cycling conditions to amplify root tips and white colonies from each soil clone library were as follows: initial denaturation at 95°C for 2 min; 37 cycles at 94°C for 45 s, 58°C for 1 min, and 72°C for 1 min 30 s; and a final elongation at 72°C for 10 min. The annealing temperature and the number of cycles were decreased to 50°C and 30, respectively, for the precloning PCR step to reach low-stringency conditions. PCRs were done on a PTC-200 (MJ Research Inc., Waltham, MA).
Library construction and sequencing.
PCR products were purified with the QIAquick PCR purification kit and cloned with the PCR Cloning Plus kit (Qiagen, Rockville, MD) according to the manufacturer's instructions. Fivefold molar excesses of PCR products were incubated for 2 h at 14°C with the pdrive cloning vector. After an overnight incubation at 37°C, 24 white bacterial colonies from each core subsample were spiked and transferred into the 25-μl PCR mixture for amplification as described above. Preliminary data indicated a low EM species richness in the soil; thus, we screened soil clone amplicons by restriction fragment length polymorphism (RFLP) analysis prior to sequencing. Enzymatic digestion was performed with HhaI and AluI at 37°C for 2 h. The RFLP mixture was made up of 1× PCR buffer, 2.5 units of each endonuclease, 1 μg of bovine serum albumin, and 1 μl of PCR product, in a total volume of 10 μl. Enzymes were inactivated at 80°C for 20 min. We systematically sequenced 10% of amplicons belonging to the same restriction profile observed on each agarose gel. Sequencing was performed on a 96-capillary 3730xl DNA analyzer at the Genomic Sequencing and Genotyping Platform of the Centre hospitalier de l'Université Laval Research Centre (Québec, Canada).
Bioinformatic analyses and clustering.
ITS sequences were edited and assembled with Sequencher, version 4.6 (GeneCodes, Ann Arbor, MI). The similarity threshold for sequences belonging to the same operational taxonomic unit (OTU) was set to 98%, which corresponds to values used by O'Brien et al. (54) (97%) and Arnold et al. (6) (95 and 99%) to serve as a proxy for “species.” Each OTU consensus sequence was identified with the closest sequences found in the NCBI GenBank database using BLAST (3). PCR-generated chimeric sequences were determined from BLAST hits displaying conspicuous incongruence between the ITS1 and ITS2 regions and were excluded from the data sets. Sequences were aligned with MUSCLE software, version 3.5 (20), with two iterations. Sequences were clustered with those retrieved from the GenBank database by running a neighbor-joining analysis in PAUP version 4.0b10 (78). Significant support in clusters displayed by the neighbor-joining trees was assessed with 1,000 bootstrap resamplings. Ascomycetes and basidiomycetes were analyzed separately. The number of OTUs, rarefaction curves, richness estimators, and Shannon and Simpson species diversity indices (73, 74) were computed with DOTUR (70). Similarities between the EM fungal communities recorded from control and transgenic poplars and the different sampling methods were compared with the nonparametric maximum likelihood estimator θ (NPMLE), based on proportions of species shared and not shared between samples (89) computed with the SONS program (71).
Statistical analyses.
Comparisons between control and transformed poplars were based on a matrix made up of the EM fungal OTU relative abundance recorded from the root tip, the OH, and the MH data sets and observed at each of the 24 points sampled. Normal distribution and homogeneity of variance of the data used for comparisons were assessed with a Shapiro-Wilk normality test and an F test. Synthetic descriptors computed for control and transformed poplars, such as the observed EM fungal species richness, the Chao richness estimator index (11), and the Shannon species index, were compared with a two-sample permutation test (10,000 permutations).
A canonical redundancy analysis (RDA) (62) was performed to investigate the variation in EM fungal composition recorded from the two data sets among the 24 points sampled as a function of the transformed-poplar treatment and soil chemical variables. The raw relative abundance matrix of EM fungal OTUs was transformed with the Hellinger asymmetric association coefficient (63). The explanatory variables selected for the RDA were tested to be uncorrelated using the Pearson coefficient. The explanatory variables considered were the transformation event (defined as a binary variable (0 or 1 for objects associated with control and transformed poplars, respectively) and the soil chemical variables Ca, K, N2, Zn, and H2O pH. The remaining soil chemical variables were not considered, as they were correlated. Finally, we tested the significance of the explanatory variables by running an analysis of variance-like permutation test (1,000 permutations; α = 0.05, β = 0.01).
EM fungal communities from (i) control and transformed poplars and (ii) root tip sampling and soil cloning were also investigated with a double principal coordinate analysis (DPCoA) (58). DPCoA displays the first two orthogonal principal axes, based on the relation between an OTU dissimilarity matrix (one consensus sequence per OTU was used as input) and the corresponding abundance matrix. The Rao diversity index is computed from the dissimilarity matrix and is decomposed into between-community and within-community diversities (58). It takes into consideration the pairwise genetic distance between OTUs; the diversity decreases when OTUs composing a sample are similar and increases when they are more distant.
To assess the significance of the differences between EM fungal communities associated with control and transformed poplars or recovered according to the different sampling methods, point coordinates given by the DPCoA in the S-dimensional space (44) were analyzed with an RDA and the corresponding analysis of variance test statistic F (1,000 permutations). Two explanatory variables were considered: the sampling method and the transformation event, all defined as binary variables. All the statistical analyses were performed using the R statistical language (65). Clustering analysis, RDA, and DPCoA were computed with the vegan 1.8-6 (J. Oksanen, R. Kindt, P. Legendre, B. O'Hara, G. L. Simpson, and M. H. H. Stevens, vegan: community ecology package, R package version 1.11-4 [http://vegan.r-forge.r-project.org], 2008) and ade4 (12) packages.
Nucleotide sequence accession numbers.
Nucleotide sequences were deposited in the NCBI GenBank database and are registered under accession numbers EU554677 to EU555003 and FJ626911 to FJ626949.
RESULTS
Fungal baseline description.
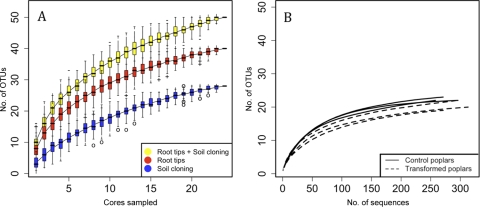
Fungal ITS data obtained with the ITS1-F/ITS4 primer set were produced from 1,150 EM root tips identified by sequencing (≈48 root tips sequenced per cardinal point) and 1,079 extraradical soil clones (≈45 clones sampled per cardinal point) from the OH and MH, identified either by sequencing (53.1% of the soil clone amplicons) or by PCR-RFLP. Eighty-four OTUs were recovered from these two ITS data sets. Clustering the 84 fungal OTUs with the closest sequences available in the NCBI GenBank database showed that 50 OTUs were related to EM fungal groups (Table 1; see Fig S1A and B in the supplemental material). The rarefaction curves for each sampling method (Fig. 1A) and each sampled tree (Fig. 1B) appeared to level off. The Chao and bootstrap estimators confirmed that we achieved EM OTU richness saturation for each data set, with values close to the observed EM species richness (Table 2).
TABLE 1.
Closest sequences recorded from the NCBI GenBank database matching the 84 fungal OTUs (2,229 ITS sequences identified) recorded from the root tip and soil cloning analyses
| BLAST match | Contig/singleton | Accession no. | Coverage (%) | Similarity (%) | EM OTUb | No. of ITS sequences from: |
||
|---|---|---|---|---|---|---|---|---|
| RTe | OH | MH | ||||||
| Uncultured EM Cortinarius | Contig 01 | AY748857 | 100 | 99 | Y | 478 | 395 | 33 |
| Acremonium (strictum)a | Contig 02 | AY138846 | 100 | 100 | N | —c | 25 | 440 |
| Cortinarius atrocoeruleus | Contig 03 | AY083178 | 100 | 99 | Y | 102 | 1 | — |
| Russula emetica | Contig 04 | AY061673 | 100 | 99 | Y | 91 | — | — |
| Hebeloma mesophaeum | Contig 05 | AB211272 | 100 | 99 | Y | 65 | 2 | 1 |
| Laccaria sp. | Contig 06 | AJ534899 | 100 | 100 | Y | 34 | 12 | 4 |
| Inocybe lacera phylotype 1 | Contig 07 | AB211269 | 99 | 96 | Y | 23 | 8 | 3 |
| Inocybe lacera phylotype 1 | Contig 08 | AY750157 | 99 | 99 | Y | 7 | 1 | 21 |
| Hymenogaster arenarius | Contig 09 | DQ328124 | 99 | 91 | Y | 26 | 3 | 10 |
| Uncultured Sebacina isolate | Contig 10 | EU668270 | 100 | 88 | Y | 29 | 3 | 9 |
| Inocybe radiata | Contig 11 | EU819490 | 100 | 83 | Y | 42 | 2 | — |
| Cortinarius favrei | Contig 12 | AF325575 | 100 | 99 | Y | 30 | — | — |
| Uncultured EM Thelephoraceae isolate | Contig 13 | AJ549971 | 100 | 97 | Y | 28 | — | — |
| Inocybe lacera phylotype 2 | Contig 14 | AB211269 | 99 | 94 | Y | 14 | 3 | 1 |
| Cortinarius paleaceus | Contig 15 | FJ039709 | 100 | 96 | Y | — | 11 | 1 |
| Uncultured EM Thelephoraceae isolate | Contig 16 | AY748885 | 100 | 99 | Y | 23 | — | — |
| Cortinarius parvannulatus | Contig 17 | AY669664 | 100 | 99 | Y | 18 | — | 1 |
| Phialophora finlandia | Contig 18 | AF486119 | 100 | 98 | Y | 15 | — | 4 |
| Saccharomyces cerevisiae | Contig 19 | AB212260 | 100 | 99 | N | — | — | 11 |
| Uncultured EM Inocybe isolate | Contig 20 | AY310824 | 99 | 96 | Y | 14 | 5 | — |
| Uncultured EM Pezizales isolate | Contig 21 | AY634125 | 100 | 100 | Y | 14 | — | — |
| Leccinum duriusculum | Contig 22 | AF454577 | 100 | 93 | Y | 10 | — | 1 |
| Uncultured EM Inocybe isolate | Contig 23 | AY310829 | 100 | 85 | Y | 11 | — | — |
| Uncultured EM Tomentella isolate | Contig 24 | EF619832 | 100 | 98 | Y | 8 | — | — |
| Uncultured EM Tuber isolate | Contig 25 | AY748861 | 100 | 99 | Y | 8 | — | — |
| Peziza depressa | Contig 26 | DQ200837 | 100 | 96 | Y | 7 | — | — |
| Cryptococcus (fuscescens)a | Contig 27 | AF145319 | 100 | 95 | N | — | — | 5 |
| Uncultured EM Tomentella isolate | Contig 28 | EF218833 | 100 | 99 | Y | 6 | — | — |
| Inocybe lacera phylotype 3 | Contig 29 | AB211269 | 99 | 94 | Y | — | 1 | 1 |
| Uncultured mycorrhizal fungus | Contig 30 | AY394904 | 100 | 90 | N | 2 | 3 | — |
| Peziza ostracoderma | Contig 31 | EU819461 | 100 | 97 | Y | 4 | 1 | — |
| Uncultured EM fungus | Contig 32 | AY587740 | 91 | 99 | Y | 5 | — | — |
| Uncultured Pezizales isolate | Contig 33 | DQ469743 | 100 | 98 | Y | — | — | 5 |
| Uncultured Thelephoraceae isolate | Contig 34 | AM181385 | 93 | 99 | Y | 5 | — | — |
| Cortinarius hemitrichus | Contig 35 | AY669680 | 100 | 99 | Y | — | 2 | — |
| Cortinarius cf. saniosusd | Contig 36 | DQ102683 | 100 | 94 | Y | 4 | — | — |
| Mortierella sp. | Contig 37 | EU877758 | 100 | 100 | N | — | 4 | — |
| Uncultured fungus (Pezizales) | Contig 38 | DQ414728 | 100 | 95 | Y | — | — | 4 |
| Scleroderma bovista | Contig 39 | AB211267 | 100 | 90 | Y | — | — | 3 |
| Uncultured fungus | Contig 40 | EF434064 | 100 | 94 | N | — | 4 | — |
| Uncultured EM Tuber isolate | Contig 41 | AY634113 | 100 | 99 | Y | 4 | — | — |
| Hymenogaster glacialis | Contig 42 | AF325634 | 100 | 99 | Y | 3 | — | — |
| Inocybe calospora | Contig 43 | AF325665 | 100 | 94 | Y | 3 | — | — |
| Tomentella sp. | Contig 44 | AB211278 | 100 | 93 | Y | — | 1 | 1 |
| Uncultured EM Tomentella isolate | Contig 45 | EF411108 | 96 | 93 | Y | 3 | — | — |
| Uncultured EM Tuber isolate | Contig 46 | AY634174 | 100 | 99 | Y | 3 | — | — |
| Hebeloma albocolossum | Contig 47 | AY308583 | 100 | 98 | Y | 2 | — | — |
| Inocybe lacera phylotype 2 | Contig 48 | AY750157 | 99 | 99 | Y | — | — | 2 |
| Paecilomyces sp. | Contig 49 | DQ191963 | 100 | 93 | N | — | 2 | — |
| Uncultured mycorrhizal fungus | Contig 50 | AY656939 | 100 | 86 | Y | — | — | 2 |
| Serendipita vermifera | Contig 51 | DQ520096 | 98 | 93 | Y | 2 | — | — |
| Uncultured Sebacina isolate | Contig 52 | EU668270 | 100 | 90 | Y | 2 | — | — |
| Mortierellaceae sp. | OH 01F02 | FJ025208 | 87 | 82 | N | — | 1 | — |
| Gigaspora margarita | OH 09F18 | U15692 | 100 | 87 | N | — | 1 | — |
| Mortierella macrocystis | OH 41F30 | AJ878782 | 35 | 91 | N | — | 1 | — |
| Uncultured fungus | OH 41F32 | EF619892 | 100 | 87 | N | — | 1 | — |
| Peziza depressa | OH 53F09 | DQ200837 | 100 | 94 | Y | — | 1 | — |
| Venturia hystrioides | OH 53F15 | EU035459 | 100 | 98 | N | — | 1 | — |
| Hypocrea crassa | OH 53F23 | EU280067 | 100 | 100 | N | — | 1 | — |
| Ganoderma applanatum | OH 57F33 | AY884179 | 100 | 100 | N | — | 1 | — |
| Uncultured fungus | OH 61F03 | AF504878 | 53 | 91 | N | — | 1 | — |
| Lambertella tubulosa | OH 61F10 | EF029195 | 100 | 98 | N | — | 1 | — |
| Tetracladium marchalianum | OH 61F11 | FJ000360 | 100 | 94 | N | — | 1 | — |
| Uncultured EM fungus | OH 61F16 | EF484935 | 100 | 99 | Y | — | 1 | — |
| Fusarium oxysporum | OH 65F14 | EU364863 | 100 | 100 | N | — | 1 | — |
| Glomerella lagenaria | OH 69F17 | AJ301970 | 100 | 95 | N | — | 1 | — |
| Leaf litter ascomycete | OH 77F05 | AF502859 | 93 | 98 | N | — | 1 | — |
| Entophlyctis helioformis | OH 77F21 | AY997048 | 41 | 82 | N | — | 1 | — |
| Trichoderma hamatum | OH 81F19 | EU595036 | 100 | 100 | N | — | 1 | — |
| Nectria sp. | OH 81F29 | DQ317342 | 100 | 97 | N | — | 1 | — |
| Uncultured fungus | MH 34a10 | AY969872 | 98 | 99 | N | — | — | 1 |
| Uncultured fungus | MH 34a12 | DQ388863 | 98 | 92 | N | — | — | 1 |
| Uncultured fungus | MH 78a25 | DQ388863 | 85 | 90 | N | — | — | 1 |
| Geomyces vinaceus | MH 34a19 | AJ608972 | 100 | 100 | N | — | — | 1 |
| Uncultured fungus | MH 34a26 | AY969871 | 98 | 98 | N | — | — | 1 |
| Wilcoxina mikolae | MH 34a22 | DQ069000 | 100 | 99 | N | — | — | 1 |
| Helotiales sp. | MH 38a13 | DQ914730 | 100 | 93 | N | — | — | 1 |
| Uncultured ascomycete | MH 78a02 | AY969669 | 90 | 93 | N | — | — | 1 |
| Rhodotorula mucilaginosa | MH 86a27 | AF444614 | 100 | 100 | N | — | — | 1 |
| Peziza badia | RT A2N49 | DQ384574 | 100 | 99 | Y | 1 | — | — |
| Leptosphaeria sp. | RT A3N22 | DQ093683 | 99 | 99 | N | 1 | — | — |
| Uncultured EM Pezizales isolate | RT A3O60 | AJ893241 | 98 | 99 | Y | 1 | — | — |
| Hebeloma psammophilum | RT A3O95 | AY312980 | 100 | 96 | Y | 1 | — | — |
| Leptodontidium orchidicola | RT T5E32 | AF486133 | 100 | 100 | N | 1 | — | — |
Parentheses indicate that different species belonging to the same genus presented exactly the same matches with our environmental sequences.
Y, yes; N, no. For EM identification based on neighbor-joining trees, see the supplemental material.
—, no sequence recorded.
A Cortinarius species that looks like Cortinarius saniosus.
RT, root tips.
FIG. 1.
EM OTU accumulation curves according to (A) the sampling method (root tip analysis or extraradical mycelium cloning) and (B) poplar type (control poplars, solid lines; transgenic poplars, dashed lines) by pooling data from the root tip data set and soil clone libraries.
TABLE 2.
Impact of the sampling strategy on the fungal and EM diversities recorded at the Valcartier plantation
| Data set | No. of ITS amplicons |
No. of OTUs |
EM frequency (%) | Chao index |
Bootstrap estimate |
Hc |
1/Dd |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fgb | EM | Fg | EM | Fg | EM | Fg | EM | Fg | EM | Fg | EM | ||
| Root tips | 1,150 | 1,146 | 42 | 39 | 99.6 | 42.2 | 39.2 | 43.3 | 40.5 | 2.45 | 2.44 | 2.45 | 5.08 |
| Soil cloninga | 1,079 | 560 | 58 | 26 | 51.9 | 127 | 30.1 | 71.9 | 31.2 | 1.63 | 1.26 | 2.94 | 1.75 |
| OH | 507 | 453 | 39 | 18 | 89.3 | 92.2 | 25.2 | 49.6 | 23.2 | 1.20 | 0.77 | 1.64 | 1.35 |
| MH | 572 | 107 | 31 | 19 | 61.3 | 71.0 | 26.0 | 37.3 | 22.0 | 1.16 | 2.28 | 1.66 | 6.66 |
OH and MH pooled data sets.
Fg, fungi.
Shannon diversity index.
Simpson reciprocal index. The value starts at 1 for the lowest species diversity.
Comparisons of EM communities between transgenic poplars and controls.
Rarefaction curves plotted for control and transformed poplars (Fig. 1B) displayed similar levels of EM fungal species richness. By a permutation test (10,000 permutations), no statistical difference between control and transformed poplars was found for the species richness observed (P = 0.4), i.e., the species richness estimated with either the Chao index (P = 0.3) or the Shannon species index (P = 1).
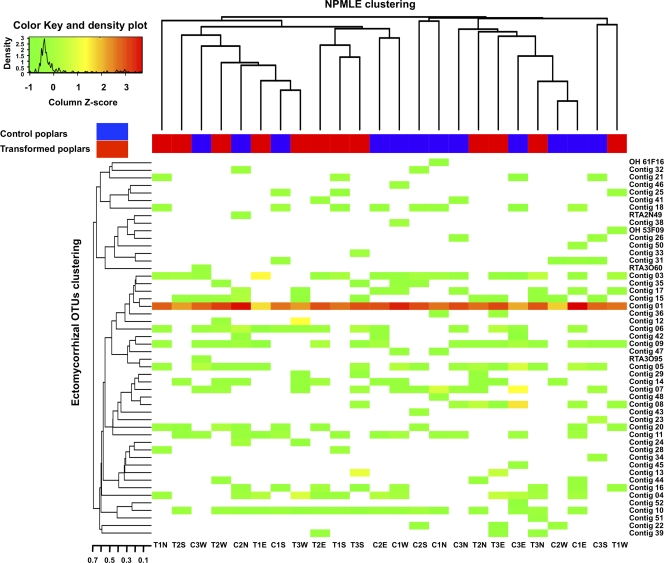
The heat map in Fig. 2 shows the distribution among the 24 cardinal points sampled around the three control and three transgenic poplars of the 50 EM fungal OTUs recovered by pooling the two ITS data sets. A single OTU, identified as an uncultured EM Cortinarius sp., was recovered at each sampled point with a mean frequency of 59% (minimum, 17.6; maximum, 86). Other OTUs had a patchy distribution, with a mean frequency of 0.80% (minimum, 0.04; maximum, 4.77). The NPMLE clustering (Fig. 2, top), based on the Euclidean distance of the NPMLE computed at each cardinal point, showed an absence of partitioning in relation to control and transgenic poplars.
FIG. 2.
Heat map distribution based on the relative abundance of the 50 EM OTUs recorded from the root tip data set and soil clone libraries. Shown is hierarchical clustering of the OTUs and of pairwise NPMLE between control and transgenic poplars. Sampled points are identified at the bottom by a “C” for control poplars and a “T” for transgenic poplars, followed by the number of the tree sampled and the first letter of the corresponding cardinal position.
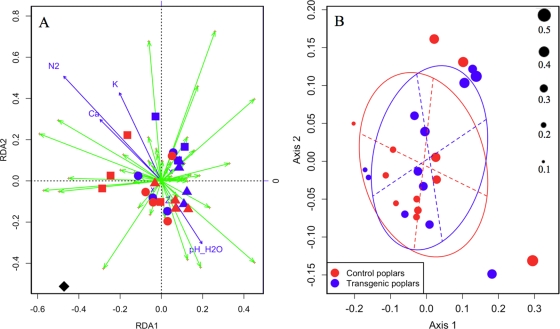
The RDA (Fig. 3A) showed no apparent clustering of the 24 points sampled according to the treatment or the soil chemical variables considered (Ca, K, N2, Zn, and pH). The combination of these six explanatory variables explained 8.2% of the variance, and their effect was not significant (pseudo-F statistic under H0 = 0.11). Analysis using the treatment as the unique explanatory variable showed that the treatment explained 1.6% of the total variance (pseudo-F statistic under H0 = 0.2). Divergence between EM fungal communities from transformed poplars and controls based on phylotype dissimilarities in relation to their relative abundances was also investigated using a DPCoA (Fig. 3B). The DPCoA showed no more difference, in regard to the relation between the dissimilarity of the 50 EM fungal phylotypes and their relative abundances in the 24 soil samples analyzed, between and within control and transformed poplars. Decomposition of the Rao diversity showed that the differences between EM communities recorded from the 24 points sampled represented 7.8% of the total diversity. An RDA performed on the DPCoA point coordinates showed that the difference between the sampled points from control and transgenic poplars was not significant (pseudo-F statistic under H0 = 0.89), indicating that EM fungal species occurring in soil samples associated with transformed poplars were as genetically diversified as those recorded in soil samples from control poplars.
FIG. 3.
(A) Redundancy analysis (scaling, 1) of the 24 points sampled in the poplar plantation, based on 1,706 EM fungal ITS amplicons identified and six explanatory variables. Points represented by the same plot symbol belong to one of the four cardinal points from the same tree. Blue and red points are associated with control and transformed poplar, respectively. Green and blue arrows represent the 50 EM OTUs identified and the five quantitative explanatory variables. The binary explanatory variable is identified with a black diamond. The first two axes explain 14.6% of the total variation. (B) Bubble map from the DPCoA. Point size is proportional to the Rao diversity index computed for each point sampled. The diversity scale is shown on the right side of the figure. Ellipses indicate the distribution of soil samples per treatment.
Root tip sampling versus soil cloning.
The sequencing of 1,150 root tips yielded 42 OTUs, of which 39 were identified to be EM fungi. Shannon and Simpson species diversity indices were the highest for the root tip data set. The identification by sequencing or PCR-RFLP of 1,079 soil clones produced 58 OTUs, of which 26 were determined to be EM fungi (Tables 1 and 2). Root tip and soil cloning data sets shared only 15 EM fungal OTUs (Table 1).
Members of the Cortinariaceae family were most abundant in the EM community. They were sampled from 632 root tips out of 1,146 (55.3%), representing five OTUs of this family. Among them, a single OTU, matching an “uncultured Cortinarius” sequence in GenBank, represented 478 out of the 1,146 (41.7%) root tips sampled. Most OTUs from the Cortinariaceae, Hymenogastraceae, Hydnangiaceae, and Inocybaceae families were recovered from both root tips and soil clone libraries (see Fig. S1B in the supplemental material). This contrasts with the Thelephorales and Pezizales, clade I, for which six out of seven and five out of six OTUs, respectively, were found only on root tips. In all, 24 EM fungal OTUs were exclusive to the root tip data set and had a relative abundance of <1%, except Cortinarius atrocoeruleus (8.9%, the second most abundant OTU within the root tip data set), Russula emetica (7.9%, the third most abundant OTU), Cortinarius favrei (2.6%), and an uncultured EM Thelephoraceae species (2.4%).
The soil clone data set was made by pooling libraries from the OH and MH. The MH clone library had a much lower number of EM ITS amplicons than the OH library (Table 2). A large majority of the ITS amplicons from the MH clone library (77.8%) were identified as Acremonium spp., a genus of mitosporic Hypocreales (contig 02) (Table 1).
Decomposition of the Rao diversity from the double principal coordinate analysis showed that there was a 20.8% difference between EM communities recorded from the 24 root tip data sets and the 24 soil clone libraries. There was a significant relationship between phylotype distribution and the sampling method used (pseudo-F statistic = 0.001), while the difference between points from control and transformed poplars was still not significant (pseudo-F statistic under H0 = 0.96). This showed that the sampling method had an influence on the EM diversity recovered while the treatment did not.
DISCUSSION
Transgenic poplar impact on an EM fungal community.
This study of a long-established transgenic poplar plantation is the most exhaustive assessment of transgenic tree impact on the diversity of the EM fungal community to date. After 8 years of mycorrhiza-GM host interaction, qualitative and/or quantitative α- and β-diversity measurements show no difference in EM fungal community structure between poplars transformed with the binary vector containing the selectable nptII marker and GUS reporter genes and the controls.
Studies monitoring the impact of field-deployed transgenic trees on mycorrhizal fungi are scarce and not extensively documented. Kaldorf et al. (36) found no difference in mycorrhizal colonization and diversity (15 EM fungi identified) between the changed phytohormone balance in rolC-transformed aspens and controls after the trees were field deployed for 3 years, but one of the four most common EM species recorded was significantly less abundant when associated with one transformed line. Vauramo et al. (85) showed that fungal biomass in the leaf litter associated with chitinase-transformed silver birches was not different from that for controls in a field trial that lasted 11 months. Newhouse et al. (53) field deployed American elms transformed to express the synthetic antimicrobial peptide ESF39A in an attempt to improve their resistance to Dutch elm disease caused by the fungus Ophiostoma novo-ulmi. Whereas staining from O. novo-ulmi was significantly reduced on transgenic sapwood, they noted that transgenic and wild-type elms had similar mycorrhizal colonization rates after 3 months of field deployment. Similar results have also been observed in vitro. Hampp et al. (29) did not detect any difference in the formation and morphology of the ectomycorrhiza Amanita muscaria associated with transgenic aspens expressing a hygromycin marker gene and indoleacetic acid-biosynthetic genes. Pasonen et al. (56) investigated the ability of silver birches constitutively expressing the nptII marker gene and the sugar beet chitinase IV gene to form normal ectomycorrhizae with Paxillus involutus. Although one transformed line showed a significant decrease in the number of root tips due to a reduced root system, they concluded that the morphology of mycorrhizae and mycorrhization efficiency were not altered. Similarly, Seppänen et al. (72) found that silver birches with a GM lignin biosynthesis pathway containing the nptII gene for kanamycin selection exhibited a significant pleiotropic effect, decreasing the root biomass; however, the association between the EM fungus Paxillus involutus and transgenic silver birches was not affected.
Selection of transformed tissues based on a selectable marker expressing antibiotic resistance in genetically engineered plants is widely used. The nptII gene is the most frequently used antibiotic-resistant marker in GM plants (24, 48), and among the above-mentioned studies, all the transgenic trees developed were kanamycin or hygromycin resistant. The hazard of using antibiotic-resistant markers for the environment lies particularly in potential horizontal gene transfer from GM plants to soil bacteria. The likelihood of shifts in natural soil microorganism communities due to the emergence of new resistant bacterial strains from horizontal gene transfer between GM plants and soil bacteria is considered almost null (16) or offset by antibiotic prescription in clinical practice (24). Although mycorrhiza helper bacteria modulate mycorrhizal symbiosis (see reference 22 for a review), the impact of antibiotic-resistant markers on the mycorrhizal community as a consequence is hardly to be expected, and the absence of effect on an EM fungal community exposed for over 8 years to the expression of the nptII gene supports the absence of impact from the nptII gene when used as a selectable marker system in plants.
EM fungal species richness.
After extensive root tip and soil analyses, we recovered 84 fungal OTUs (“species”), of which 50 were EM fungi. Based on the rarefaction curves depicted in Fig. 1A, this is an accurate estimate of fungal diversity at the study site. The original plantation site was devoid of established roots in the soil, which play a major role in seedling mycorrhizal colonization (18). Therefore, the transgenic and control trees could be colonized only by wind- or animal-carried spores or vegetative tissues from the neighboring forest or from the soil used at the time of planting. In this highly artificial community, the combination of two sampling approaches with a high sequencing effort led to the recovery of a higher level of EM fungal diversity than that in most of the previous studies focusing on EM fungal communities associated with forest trees in plantations or natural stands. This agrees with the point raised by Horton and Bruns (33), who suggested the use of different methods and increased direct sequencing to better represent EM fungal diversity. They reviewed 14 studies investigating belowground EM fungal diversity that did not manage to saturate EM fungal species accumulation curves. The average number of soil samples collected was 35, and species richness averaged 33, except for the study with the highest sampling effort (198 soil samples), which recorded 200 morphologically distinct EM fungi. On the other hand, EM fungal species richness averaged 44 (minimum, 21; maximum, 79) in other studies using molecular tools mainly based on the identification of root tips with ITS-RFLP and denaturating gradient gel electrophoresis or soil cloning (9, 14, 18, 19, 25, 28, 34, 37, 38, 41, 50, 52, 60, 79, 82, 86). Some studies investigating EM fungal species richness observed values at least twofold greater than those observed in this study. However, those studies were conducted on sites with higher ecological complexity and/or for a longer time period with more-intense sampling (35, 45, 46, 54, 55, 69, 75, 81).
Methodological considerations.
We showed that the observed differences among EM fungal communities were more influenced by the sampling method (root tips versus sieved soil) than by the presence of transformed poplars over an 8-year period. It is faster and easier to increase the sampling effort by soil fungal clone sequencing than by root tip sequencing, and although the species recovered were not demonstrated to function as EM fungi, this method could recover a level of species richness not previously reported by sporocarp or root tip samplings. In our site, root tips provided 24 EM fungal OTUs not observed in the soil clone data set. Despite our attempts to minimize PCR artifacts by performing amplification in low-stringency conditions, a PCR bias toward the dominant Cortinarius OTU could be expected, as it would outcompete rare EM fungal species during primer annealing. The consequence would be a statistical saturation of EM fungal diversity but not a biological saturation, as shown by the EM fungal species richness recovered from the root tip data set. Nevertheless, the soil clone data set brought 11 new OTUs compared with root tip sampling, which represents an EM fungal species richness increase of 22%, while root tip rarefaction curves tend to be saturated.
Our results also revealed that investigating EM extraradical mycelia failed to detect the four common species recorded on root tips. This suggests that these four species—two Cortinarius, one Russulales, and one Thelephoraceae species—may produce limited extraradical mycelium in the soil, thus affecting their detection by soil cloning. It contrasts with results from Agerer (1), who classified species from the Cortinarius genus in the medium-distance exploration type, featuring extended contact with the soil. Parrent and Vilgalys (55) also noticed disparities between EM fungal communities recovered from root tips and from extraradical mycelium and observed that some species occurring at a relatively high frequency as extraradical mycelium or fruiting bodies were rarely or not detectable as root tips. This highlights the different foraging types and ecological roles of EM fungi (1) and explains why the detection of some EM fungal species would depend on the sampling method, an issue already raised by Koide et al. (37).
In conclusion, changes in the EM fungal community associated for 8 years with transformed and untransformed poplars were not detectable, while the analysis of root tips and extraradical mycelium cloning provided contrasting views of the EM fungal community colonizing the experimental poplar stand. The present study is in line with the trend according to which no major effect from transgenic trees on mycorrhizal, soil fungal, or soil bacterial communities from the GM trees studied so far has been observed. Up until now, most of the studies investigating the effect of transgenic trees or transgenic crops on soil microbial communities have observed changes attributable to new traits from GM organisms that are smaller than changes attributable to other factors such as soil type, plant genotype, and stand sites (8, 16, 40, 64). However, transformations leading to deleterious effects on trees (13, 57, 72) or nontarget organisms (31, 36) have been reported, but they involved pleiotropic effects that depend on each unique insertion event. Therefore, future impact studies of GM trees will have to test over a long-term period every transgenic line extensively deployed before large-scale propagation to better evaluate the likelihood of consequences due to transgenic trees for mycorrhizal symbioses and what the consequences are.
Supplementary Material
Acknowledgments
We thank Denis Lachance for the experimental design of the control and transformed poplars and field deployment. We are grateful to Marie-Josée Bergeron and Philippe Tanguay for insightful comments in the course of this study and on an earlier draft of the manuscript, and we thank the Centre de bio-informatique et de biologie computationnelle (Université Laval, QC, Canada) for bioinformatic assistance.
This work was supported by grants from the Canadian Biotechnology Strategic Fund.
Footnotes
Published ahead of print on 2 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Agerer, R. 2001. Exploration types of ectomycorrhizae—a proposal to classify ectomycorrhizal mycelial systems according to their patterns of differentiation and putative ecological importance. Mycorrhiza 11:107-114. [Google Scholar]
- 2.Alla, S., A. Cherqui, L. Kaiser, H. Azzouz, B. S. Sangwann-Norreel, and P. Giordanengo. 2003. Effects of potato plants expressing the nptII-gus fusion marker genes on reproduction, longevity, and host-finding of the peach-potato aphid, Myzus persicae. Entomol. Exp. Appl. 106:95-102. [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.An, G., M. A Costa, A. Mitra, S.-B. Ha, and L. Márton. 1988. Organ-specific and developmental regulation of the nopaline synthase promoter in transgenic tobacco plants. Plant Physiol. 88:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andow, D. A., and C. Zwahlen. 2006. Assessing environmental risks of transgenic plants. Ecol. Lett. 9:196-214. [DOI] [PubMed] [Google Scholar]
- 6.Arnold, A. E., D. A. Henk, R. L. Eells, F. Lutzoni, and R. Vilgalys. 2007. Diversity and phylogenetic affinities of foliar fungal endophytes in loblolly pine inferred by culturing and environmental PCR. Mycologia 99:185-206. [DOI] [PubMed] [Google Scholar]
- 7.Boerjan, W. 2005. Biotechnology and the domestication of forest trees. Curr. Opin. Biotechnol. 16:159-166. [DOI] [PubMed] [Google Scholar]
- 8.Bradley, K. L., J. E. Hancock, C. P. Giardina, and K. S. Pregitzer. 2007. Soil microbial community responses to altered lignin biosynthesis in Populus tremuloides vary among three distinct soils. Plant Soil 294:185-201. [Google Scholar]
- 9.Burke, D. J., K. J. Martin, P. T. Rygiewiczc, and M. A. Topa. 2005. Ectomycorrhizal fungi identification in single and pooled root samples: terminal restriction fragment length polymorphism (TRFLP) and morphotyping compared. Soil Biol. Biochem. 37:1683-1694. [Google Scholar]
- 10.Campbell, F. T., and R. Asante-Owusu. 2001. GE trees: proceed only with caution, p. 158-167. In S. H. Strauss and H. D. Bradshaw (ed.), Tree biology in the new millenium. Proceedings of the First International Symposium on Ecological and Societal Aspects of Transgenic Plantations. Oregon State University, Corvallis.
- 11.Chao, A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 12.Chessel, D., A. B. Dufour, and J. Thioulouse. 2004. The ade4 package-I: one-table methods. R News 4:5-10. [Google Scholar]
- 13.Coleman, H. D., T. Canam, K. Kyu-Young, D. D. Ellis, and S. D. Mansfield. 2007. Over-expression of UDP-gluclose pyrophosphorylase in hybrid poplar affects carbon allocation. J. Exp. Bot. 58:4257-4268. [DOI] [PubMed] [Google Scholar]
- 14.Cullings, K. W., D. R. Vogler, V. T. Parker, and S. K. Finley. 2000. Ectomycorrhizal specificity patterns in a mixed Pinus contorta and Picea engelmannii forest in Yellowstone National Park. Appl. Environ. Microbiol. 66:4988-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dale, P. J., and H. C. McPartlan. 1992. Field performance of transgenic potato plants compared with controls regenerated from tuber discs and shoot cuttings. Theor. Appl. Genet. 84:585-591. [DOI] [PubMed] [Google Scholar]
- 16.Demanèche, S., H. Sanguin, J. Poté, E. Navarro, D. Bernillon, P. Mavingui, W. Wildi, T. M. Vogel, and P. Simonet. 2008. Antibiotic-resistant soil bacteria in transgenic plant fields. Proc. Natl. Acad. Sci. USA 105:3957-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Turck, S., P. Giordanengo, A. Cherqui, C. Ducrocq-Assaf, and B. S. Sangwan-Norreel. 2002. Transgenic potato plants expressing the nptII-gus marker genes affect survival and development of the Colorado potato beetle. Plant Sci. 162:373-380. [Google Scholar]
- 18.Dickie, I. A., B. Xu, and R. B. Koide. 2002. Vertical niche differentiation of ectomycorrhizal hyphae in soil as shown by T-RFLP analysis. New Phytol. 156:527-535. [DOI] [PubMed] [Google Scholar]
- 19.Dickie, I. A., and P. B. Reich. 2005. Ectomycorrhizal fungal communities at forest edges. J. Ecol. 93:244-255. [Google Scholar]
- 20.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flavell, R. B., E. Dart, R. L. Fuchs, and R. T. Fraley. 1992. Selectable marker genes: safe for plants? Bio/Technology 10:141-144. [DOI] [PubMed] [Google Scholar]
- 22.Frey-Klett, P., J. Garbaye, and M. Tarkka. 2007. The mycorrhiza helper bacteria revisited. New Phytol. 176:22-36. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs, R. L., J. E. Ream, B. G. Hammond, M. W. Naylor, R. M. Leimgruber, and S. A. Berberich. 1993. Safety assessment of the neomycin phosphotransferase II (NPTII) protein. Bio/Technology 11:1543-1547. [DOI] [PubMed] [Google Scholar]
- 24.Gay, P. B., and S. H. Gillespie. 2005. Antibiotic resistance markers in genetically modified plants: a risk to human health? Lancet Infect. Dis. 5:637-646. [DOI] [PubMed] [Google Scholar]
- 25.Gehring, C. A., T. C. Theimer, T. G. Whitham, and P. Keim. 1998. Ectomycorrhizal fungal community structure of pinyon pines growing in two environmental extremes. Ecology 79:1562-1572. [Google Scholar]
- 26.Genney, D. R., I. C. Anderson, and I. J. Alexander. 2006. Fine-scale distribution of pine ectomycorrhizas and their extramatrical mycelium. New Phytol. 170:381-390. [DOI] [PubMed] [Google Scholar]
- 27.Gilissen, L. J. W., P. L. J. Metz, W. J. Stiekema, and J.-P. Nap. 1998. Biosafety of E. coli β-glucuronidase (GUS) in plants. Transgenic Res. 7:157-163. [DOI] [PubMed] [Google Scholar]
- 28.Grogan, P., J. Baar, and T. D. Bruns. 2000. Below-ground ectomycorrhizal community structure in a recently burned bishop pine forest. J. Ecol. 88:1051-1062. [Google Scholar]
- 29.Hampp, R., M. Ecke, C. Schaeffer, T. Wallenda, A. Wingler, I. Kottke, and B. Sundberg. 1996. Axenic mycorrhization of wild type and transgenic hybrid aspen expressing T-DNA indoleacetic acid-biosynthetic genes. Trees 11:59-64. [Google Scholar]
- 30.Hay, I., M.-J. Morency, and A. Séguin. 2002. Assessing the persistence of DNA in decomposing leaves of genetically modified poplar trees. Can. J. For. Res. 32:977-982. [Google Scholar]
- 31.Hjälten, J., A. Lindau, A. Wennström, P. Blomberg, J. Witzell, V. Hurry, and L. Ericson. 2007. Unintentional changes of defence traits in GM trees can influence plant-herbivore interactions. Basic Appl. Ecol. 8:434-443. [Google Scholar]
- 32.Högberg, M. N., and P. Högberg. 2002. Extramatrical ectomycorrhizal mycelium contributes one-third of microbial biomass and produces, together with associated roots, half the dissolved organic carbon in a forest soil. New Phytol. 154:791-795. [DOI] [PubMed] [Google Scholar]
- 33.Horton, T. R., and T. D. Bruns. 2001. The molecular revolution in ectomycorrhizal ecology: peeking into the black-box. Mol. Ecol. 10:1855-1871. [DOI] [PubMed] [Google Scholar]
- 34.Horton, T. R., R. Molina, and K. Hood. 2005. Douglas-fir ectomycorrhizae in 40- and 400-year-old stands: mycobiont availability to late successional western hemlock. Mycorrhiza 15:393-403. [DOI] [PubMed] [Google Scholar]
- 35.Ishida, T. A., K. Nara, and T. Hogetsu. 2007. Host effects on ectomycorrhizal fungal communities: insight from eight host species in mixed conifer-broadleaf forests. New Phytol. 174:430-440. [DOI] [PubMed] [Google Scholar]
- 36.Kaldorf, M., M. Fladung, H.-J. Muhs, and F. Buscot. 2002. Mycorrhizal colonization of transgenic aspen in a field trial. Planta 214:653-660. [DOI] [PubMed] [Google Scholar]
- 37.Koide, R. T., B. Xu, and J. Sharda. 2005. Contrasting below-ground views of an ectomycorrhizal fungal community. New Phytol. 166:251-262. [DOI] [PubMed] [Google Scholar]
- 38.Korkama, T., A. Pakkanen, and T. Pennanen. 2006. Ectomycorrhizal community structure varies among Norway spruce (Picea abies) clones. New Phytol. 171:815-824. [DOI] [PubMed] [Google Scholar]
- 39.Korkama, T., H. Fritze, A. Pakkanen, and T. Pennanen. 2007. Interactions between extraradical ectomycorrhizal mycelia, microbes associated with the mycelia and growth rate of Norway spruce (Picea abies) clones. New Phytol. 173:798-807. [DOI] [PubMed] [Google Scholar]
- 40.Lamarche, J., and R. C. Hamelin. 2007. No evidence of an impact on the rhizosphere diazotroph community by the expression of Bacillus thuringiensis Cry1Ab toxin by Bt white spruce. Appl. Environ. Microbiol. 73:6577-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landeweert, R., P. Leeflang, T. W. Kuyper, E. Hoffland, A. Rosling, K. Wernars, and E. Smit. 2003. Molecular identification of ectomycorrhizal mycelium in soil horizons. Appl. Environ. Microbiol. 69:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leake, J., D. Johnson, D. Donnelly, G. Muckle, L. Boddy, and D. Read. 2004. Networks of power and influence: the role of mycorrhizal mycelium in controlling plant communities and agroecosystem functioning. Can. J. Bot. 82:1016-1045. [Google Scholar]
- 43.Lecardonnel, A., G. Prévost, A. Beaujean, R. S. Sangwan, and B. S. Sangwan-Norreel. 1999. Genetic transformation of potato with nptII-gus marker genes enhances foliage consumption by Colorado potato beetle larvae. Mol. Breed. 5:441-451. [Google Scholar]
- 44.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd English edition. Elsevier Science B.V., Amsterdam, The Netherlands.
- 45.Luoma, D. L., C. A. Stockdale, R. Molina, and J. L. Eberhart. 2006. The spatial influence of Pseudotsuga menziesii retention trees on ectomycorrhiza diversity. Can. J. For. Res. 36:2561-2573. [Google Scholar]
- 46.Lynch, M. D. J., and R. G. Thorn. 2006. Diversity of basidiomycetes in Michigan agricultural soils. Appl. Environ. Microbiol. 72:7050-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathews, J. H., and M. M. Campbell. 2000. The advantages and disadvantages of the application of genetic engineering to forest trees: a discussion. Forestry 73:371-380. [Google Scholar]
- 48.Miki, B., and S. McHugh. 2004. Selectable marker genes in transgenic plants: applications, alternatives and biosafety. J. Biotechnol. 107:193-232. [DOI] [PubMed] [Google Scholar]
- 49.Molina, R., H. Massicotte, and J. M. Trappe. 1992. Specificity phenomena in mycorrhizal symbioses: community-ecological consequences and practical implications, p. 357-423. In M. F. Allen (ed.), Mycorrhizal functioning: an integrative plant-fungal process. Chapman and Hall, New York, NY.
- 50.Moser, A. M., C. A. Petersen, J. A. D'Allura, and D. Southworth. 2005. Comparison of ectomycorrhizas of Quercus garryana (Fagaceae) on serpentine and non-serpentine soils in southwestern Oregon. Am. J. Bot. 92:224-230. [DOI] [PubMed] [Google Scholar]
- 51.Nap, J. P., J. Bijvoet, and W. J. Stiekema. 1992. Biosafety of kanamycin-resistant transgenic plants. Transgenic Res. 1:239-249. [DOI] [PubMed] [Google Scholar]
- 52.Nara, K., H. Nakaya, B. Y. Wu, Z. H. Zhou, and T. Hogetsu. 2003. Underground primary succession of ectomycorrhizal fungi in a volcanic desert on Mount Fuji. New Phytol. 159:743-756. [DOI] [PubMed] [Google Scholar]
- 53.Newhouse, A. E., F. Schrodt, H. Liang, C. A. Maynard, and W. A. Powell. 2007. Transgenic American elm shows reduced Dutch elm disease symptoms and normal mycorrhizal colonization. Plant Cell. Rep. 26:977-987. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien, H. E., J. L. Parrent, J. A. Jackson, J.-M. Moncalvo, and R. Vilgalys. 2005. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microbiol. 71:5544-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parrent, J. L., and R. Vilgalys. 2007. Biomass and compositional responses of ectomycorrhizal fungal hyphae to elevated CO2 and nitrogen fertilization. New Phytol. 176:164-174. [DOI] [PubMed] [Google Scholar]
- 56.Pasonen, H.-L., Y. Degefu, J. Brumós, K. Lohtander, A. Pappinen, S. Timonen, and S. K. Seppänen. 2005. Transgenic Betula pendula expressing sugar beet chitinase IV forms normal ectomycorrhizae with Paxillus involutus in vitro. Scand. J. For. Res. 20:385-392. [Google Scholar]
- 57.Pasonen, H.-L., L. Vihervuori, S.-K. Seppänen, P. Lyytikäinen-Saarenmaa, T. Ylioja, K. von Weissenberg, and A. Pappinen. 2008. Field performance of chitinase transgenic silver birch (Betula pendula Roth): growth and adaptive traits. Trees 22:413-421. [Google Scholar]
- 58.Pavoine, S., A.-B. Dufour, and D. Chessel. 2004. From dissimilarities among species to dissimilarities among communities: a double principal coordinate analysis. J. Theor. Biol. 228:523-537. [DOI] [PubMed] [Google Scholar]
- 59.Peña, L., and A. Séguin. 2001. Recent advances in the genetic transformation of trees. Trends. Biotechnol. 19:500-506. [DOI] [PubMed] [Google Scholar]
- 60.Peter, M., F. Ayer, S. Egli, and R. Honegger. 2001. Above- and below-ground community structure of ectomycorrhizal fungi in three Norway spruce (Picea abies) stands in Switzerland. Can. J. Bot. 79:1134-1151. [Google Scholar]
- 61.Pullman, G. S., J. Cairney, and G. Peter. 1998. Clonal forestry and genetic engineering: forest biotechnology—where we stand and future prospects and impacts. Tappi J. 81:57-64. [Google Scholar]
- 62.Rao, C. R. 1964. The use and interpretation of principal component analysis in applied research. Sankhyaá A. 26:329-358. [Google Scholar]
- 63.Rao, C. R. 1995. A review of canonical coordinates and an alternative to correspondence analysis using Hellinger distance. Questió 19:23-63. [Google Scholar]
- 64.Rasche, F., H. Velvis, C. Zachow, G. Berg, J. D. van Elsas, and A. Sessitsch. 2006. Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J. Appl. Ecol. 43:555-566. [Google Scholar]
- 65.R Development Core Team. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. www.R-project.org.
- 66.Read, D. J. 1991. Mycorrhizas in ecosystems. Experientia 47:376-391. [Google Scholar]
- 67.Read, D. J., and J. Perez-Moreno. 2003. Mycorrhizas and nutrient cycling in ecosystems—a journey towards relevance? New Phytol. 157:475-492. [DOI] [PubMed] [Google Scholar]
- 68.Read, D. J., J. R. Leake, and J. Perez-Moreno. 2004. Mycorrhizal fungi as drivers of ecosystem processes in heathland and boreal forest biomes. Can. J. Bot. 82:1243-1263. [Google Scholar]
- 69.Richard, F., S. Millot, M. Gardes, and M.-A. Selosse. 2005. Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth Mediterranean forest dominated by Quercus ilex. New Phytol. 166:1011-1023. [DOI] [PubMed] [Google Scholar]
- 70.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seppänen, S.-K., H.-L. Pasonen, S. Vauramo, J. Vahala, M. Toikka, I. Kilpeläinen, H. Setälä, T. H. Teeri, S. Timonen, and A. Pappinen. 2007. Decomposition of the leaf litter and mycorrhiza forming ability of silver birch with a genetically modified lignin biosynthesis pathway. Appl. Soil. Ecol. 36:100-106. [Google Scholar]
- 73.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. University of Illinois Press, Urbana.
- 74.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 75.Smith, J. E., D. McKay, C. G. Niwa, W. G. Thies, G. Brenner, and J. W. Spatafora. 2004. Short-term effects of seasonal prescribed burning on the ectomycorrhizal fungal community and fine root biomass in ponderosa pine stands in the blue mountains of Oregon. Can. J. For. Res. 34:2477-2491. [Google Scholar]
- 76.Stewart, C. N., Jr. 2005. Monitoring the presence and expression of transgenes in living plants. Trends Plant Sci. 10:390-396. [DOI] [PubMed] [Google Scholar]
- 77.Strauss, S. H., A. M. Brunner, V. B. Busov, C. Ma, and R. Meilan. 2004. Ten lessons from 15 years of transgenic Populus research. Forestry 77:455-465. [Google Scholar]
- 78.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (* and other methods), release 4.B10. Sinauer Associates, Sunderland, MA.
- 79.Taylor, D. L., and T. D. Bruns. 1999. Community structure of ectomycorrhizal fungi in a Pinus muricata forest: minimal overlap between the mature forest and resistant propagule communities. Mol. Ecol. 8:1837-1850. [DOI] [PubMed] [Google Scholar]
- 80.Taylor, G. 2002. Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 90:681-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tedersoo, L., T. Suvi, E. Larsson, and U. Kõljalg. 2006. Diversity and community structure of ectomycorrhizal fungi in a wooded meadow. Mycol. Res. 110:734-748. [DOI] [PubMed] [Google Scholar]
- 82.Toljander, J. F., U. Eberhardt, Y. K. Toljander, L. R. Paul, and A. F. S. Taylor. 2006. Species composition of an ectomycorrhizal fungal community along a local nutrient gradient in a boreal forest. New Phytol. 170:873-883. [DOI] [PubMed] [Google Scholar]
- 83.Tzfira, T., A. Zuker, and A. Altman. 1998. Forest-tree biotechnology: genetic transformation and its application to future forests. Trends Biotechnol. 16:439-446. [Google Scholar]
- 84.van Frankenhuyzen, K., and T. Beardmore. 2004. Current status and environmental impact of transgenic forest trees. Can. J. For. Res. 34:1163-1180. [Google Scholar]
- 85.Vauramo, S., H. L. Pasonen, A. Pappinen, and H. Setala. 2006. Decomposition of leaf litter from chitinase transgenic silver birch (Betula pendula) and effects on decomposer populations in a field trial. Appl. Soil. Ecol. 32:338-349. [Google Scholar]
- 86.Walker, J. F., O. K. Miller, Jr., and J. L. Horton. 2005. Hyperdiversity of ectomycorrhizal fungus assemblages on oak seedlings in mixed forests in the southern Appalachian Mountains. Mol. Ecol. 14:829-838. [DOI] [PubMed] [Google Scholar]
- 87.Wei, H., R. Meilan, A. M. Brunner, J. S. Skinner, C. Ma, and S. H. Strauss. 2006. Transgenic sterility in Populus: expression properties of the poplar PTLF, Agrobacterium NOS and two minimal 35S promoters in vegetative tissues. Tree Physiol. 26:401-410. [DOI] [PubMed] [Google Scholar]
- 88.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press Inc., New York, NY.
- 89.Yue, J. C., and M. K. Clayton. 2005. A similarity measure based on species proportions. Commun. Stat.-Theory Methods 34:2123-2131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.