Abstract
To detect anoxygenic bacteria containing either type 1 or type 2 photosynthetic reaction centers in a single PCR, we designed a degenerate primer set based on the bchY gene. The new primers were validated in silico using the GenBank nucleotide database as well as by PCR on pure strains and environmental DNA.
Anoxygenic photosynthetic bacteria are diverse and important members of microbial communities (11, 13, 17, 20). There are five bacterial phyla containing anoxygenic phototrophs: Proteobacteria (purple bacteria), Chlorobi (green sulfur bacteria), Chloroflexi (green nonsulfur bacteria), Acidobacteria (“Candidatus Chloracidobacterium thermophilum” [7]), and Firmicutes (heliobacteria). While Heliobacterium modesticaldum, Chlorobi, and “Ca. Chloracidobacterium thermophilum” have a type 1 reaction center (RC1) similar to photosystem I in Cyanobacteria and higher plants, Chloroflexi and Proteobacteria possess a type 2 reaction center (RC2) similar to photosystem II of oxygenic phototrophs (7, 16).
Primers based on pufM, the gene encoding the M subunit of RC2, have been widely used to detect phototrophic purple bacteria (1, 4, 12, 19). However, phototrophic bacteria that do not possess RC2 are not retrieved when pufM is used as the target. Achenbach and coworkers (1) developed primers targeting rRNA genes of Chlorobi, Chloroflexi, and heliobacteria, while Alexander and coworkers (2) have developed primers to specifically detect green sulfur bacteria (Chlorobi) by using 16S rRNA and fmoA as gene targets and applied these primers in environmental studies (3). No currently available primer set can simultaneously target phototrophs containing either RC1 or RC2.
Since it is well established that both RC1- and RC2-containing anoxygenic phototrophs synthesize bacteriochlorophylls (BChls), we searched for a universal anoxygenic photosynthesis gene marker among all enzymes involved in BChl biosynthetic pathways. All known pathways for chlorophyll and BChl biosynthesis branch from the heme biosynthesis pathway at protoporphyrin IX and continue to chlorophyllide a (Chlide a) through the same intermediates (9). Chlide a is the branching point that separates chlorophyll and BChl biosynthetic pathways. Moreover, pathways for the synthesis of different BChls are also split at this stage: chlorophyllide oxidoreductase converts Chlide a to 3-vinyl-bacteriophyllide a, which is the precursor for BChls a, b, and g, while a yet unknown enzyme reduces Chlide a to 3-vinyl-bacteriophyllide d, a precursor for antenna BChls c, d, and e in Chlorobium spp. (9). Since 3-vinyl-bacteriophyllide a is the last common intermediate in the synthesis of BChl a and BChl g, and the latter is the only BChl in heliobacteria (14, 15), chlorophyllide oxidoreductase is the only enzyme that is (i) present in anoxygenic phototrophic bacteria and not in oxygenic phototrophs and (ii) common to all known anoxygenic phototrophic bacterial species (with the exception of “Ca. Chloracidobacterium thermophilum,” where the pathway for BChl synthesis is not yet known). Analyzing multiple alignments of the subunits of chlorophyllide oxidoreductase, we found that only the Y subunit (encoded by the BchY gene) had two conserved regions distinguishing this protein from its closest homologs; therefore, the bchY gene was chosen as a universal marker for anoxygenic photosynthesis.
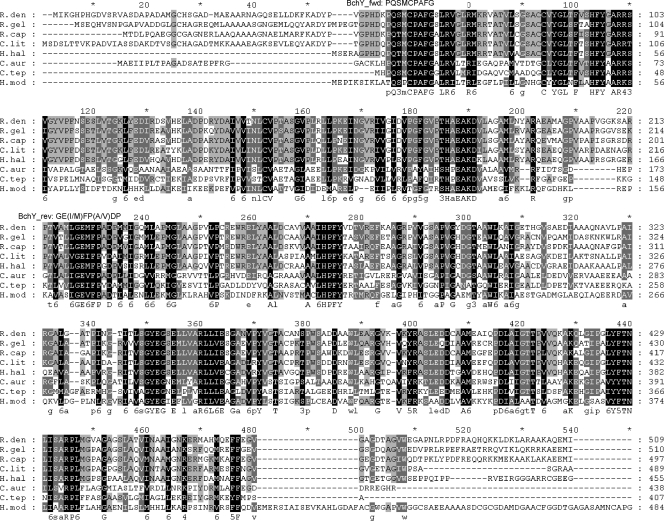
Due to likely codon variations coding identical amino acid sequences in different genomes (19), degenerate BchY primers were designed by reverse translation of two conserved regions of the BchY alignment (Fig. 1): bchY_fwd (5′-CCNCARACNATGTGYCCNGCNTTYGG-3′ [26 bases; 2,048 variants; corresponding amino acid sequence, PQTMCPAFG]) and bchY_rev (5′-GGRTCNRCNGGRAANATYTCNCC-3′ [23 bases; 4,096 variants; corresponding amino acid sequence, GE{I/M}FP{A/ V}DP]). Each primer had no more than two bases deviating from known bchY sequences in the GenBank nr database (except for H. modesticaldum) as well as to environmental BchY variants in the GenBank env_nr database. None of these deviations were located in the 3′ ends of the primers (see Tables S2 and S3 in the supplemental material). These primers, therefore, were predicted to amplify a wide diversity of bchY genes under nonstringent PCR conditions (50 to 52°C annealing temperature). The lengths of the expected PCR products were either 480 bp (for green sulfur, green nonsulfur bacteria, and heliobacteria) or 510 bp (for purple bacteria).
FIG. 1.
Multiple-amino-acid alignment of BchY proteins. Sequence abbreviations: R.den, Roseobacter denitrificans (gi|110677524); R.gel, Rubrivivax gelatinosus (gi|29893484); R.cap, Rhodobacter capsulatus (gi|114868); C.lit, Congregibacter litoralis KT 71 (gi|88706663); H.hal, Halorhodospira halophila (gi|121998388); C.aur, Chloroflexus aurantiacus (gi|163849328); C.tep, Chlorobium tepidum (gi|66576270); and H.mod, Heliobacterium modesticaldum (gi|167629410).
In order to check primer specificity in silico, a screening procedure was developed. Putative primer sites (tags) for both the bchY_fwd and the bchY_rev primers were gathered from the GenBank nucleotide collection (nt) by BLAST with relaxed search conditions; the tags having mismatches at the 3′ end or more than five overall mismatches from their primer were filtered out, and the remaining tags were mapped to their sequences mimicking PCR primer annealing. Fragments ranging from 300 to 700 bp (virtual “PCR products”) were retrieved from GenBank and annotated (see Table S4 in the supplemental material). All bchY genes present in the GenBank nt database were virtually “amplified,” pointing to the robustness of the primers and our in silico PCR analysis. On the other hand, all nonspecific “amplicons” have major deviations from the primer sequences and would likely not be amplified by a real PCR. The same screening procedure was performed against the GenBank environmental nucleotide collection (env_nt) (see Table S5 in the supplemental material), and as in the case with the nt database, only bchY fragments were virtually “amplified.”
The BchY primer set was validated using five key control organisms, including the RC2-containing the purple sulfur bacterium Allochromatium vinosum and the purple nonsulfur bacterium Rhodobacter capsulatus as well as the RC1-containing green sulfur bacterium Chlorobium limicola, green nonsulfur bacterium Chloroflexus aurantiacus, and the heliobacterium H. modesticaldum. Amplifications yielded the predicted products of 510 bp from the purple bacteria and 480 bp from the green sulfur and nonsulfur bacteria and H. modesticaldum. Negative-control Escherichia coli and Synechocystis sp. strain PCC 6803 did not yield amplification products when the bchY primers were used.
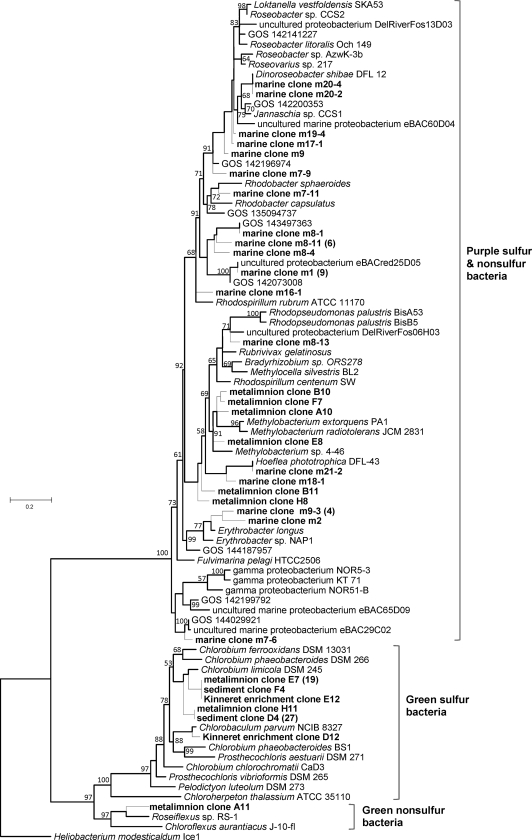
The designed BchY primer set successfully amplified bchY genes from DNA obtained from both marine (East Mediterranean Sea) and freshwater (Lake Kinneret) environments (see Table S6 in the supplemental material for best BLASTX hits for selected sequenced fragments). These habitats were chosen for testing due to the previously reported wide diversity of their anoxygenic phototrophs (8, 10, 18, 19). A phylogenetic tree of bchY gene fragments amplified from both freshwater and marine DNA samples is shown in Fig. 2.
FIG. 2.
BchY phylogenetic tree based on a maximum likelihood tree to which short sequences were added by ARB parsimony. The branches that appeared on the original maximum likelihood tree are shown with thicker lines. Bootstrap values greater than 50% are indicated next to the branches. Sequences obtained in this study are shown in bold. For reasons of clarity, not all BchY sequences retrieved are shown in the tree. For cases in which a BchY fragment was found in more than three clones, the numbers of clones are given in parentheses. Clones m21_2 and m21_3 are identical to the bchY gene of Hoeflea phototrophica strain DFL-43 (6); the m20_2 clone was identical to the bchY gene of Dinoroseobacter shibae (5).
Our study underlines the utility of the bchY gene as a molecular marker for revealing genetic heterogeneity in phototrophic microbial populations. Using both wide-scale bioinformatic analysis and PCR on control strains and naturally occurring microbial community DNA, we have confirmed the specificity and coverage of the proposed degenerate BchY primers.
Nucleotide sequence accession numbers.
The bchY sequences were deposited in GenBank under accession numbers EU854432 (Allochromatium vinosum), EU888421 and EU888422 (Chlorobium enrichments), EU888377 to EU888420, EU888424 to EU888440 (Lake Kinneret), and GQ861394 to GQ861424 (Mediterranean Sea).
Supplementary Material
Acknowledgments
We thank Yosef Yacobi and Werner Eckert, who provided the Lake Kinneret field and cultured samples used in the current study within the framework of their project 932/04, funded by the Israeli Science Foundation. We thank Yuri Wolf (NCBI) for helpful discussion on virtual PCR design.
This work was supported in part by the Department of Health and Human Services intramural program of the National Institutes of Health, National Library of Medicine (N.Y.); grant 434/02 from the Israel Science Foundation (O.B.); a grant from the Israeli Ministry of Science and Technology; an EMBO YIP award (O.B.); and grants NSF-0950550 (M.T.M.) and OCE-0550547 (M.T.S.) from the U.S. National Science Foundation.
Footnotes
Published ahead of print on 2 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Achenbach, L. A., J. Carey, and M. T. Madigan. 2001. Photosynthetic and phylogenetic primers for detection of anoxygenic phototrophs in natural environments. Appl. Environ. Microbiol. 67:2922-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, B., J. H. Andersen, R. P. Cox, and J. F. Imhoff. 2002. Phylogeny of green sulfur bacteria on the basis of gene sequences of 16S rRNA and of the Fenna-Matthews-Olson protein. Arch. Microbiol. 178:131-140. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, B., and J. F. Imhoff. 2006. Communities of green sulfur bacteria in marine and saline habitats analyzed by gene sequences of 16S rRNA and Fenna-Matthews-Olson protein. Int. Microbiol. 9:259-266. [PubMed] [Google Scholar]
- 4.Béjà, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 5.Biebl, H., M. Allgaier, B. J. Tindall, M. Koblizek, H. Lunsdorf, R. Pukall, and I. Wagner-Dobler. 2005. Dinoroseobacter shibae gen. nov., sp. nov., a new aerobic phototrophic bacterium isolated from dinoflagellates. Int. J. Syst. Evol. Microbiol. 55:1089-1096. [DOI] [PubMed] [Google Scholar]
- 6.Biebl, H., B. J. Tindall, R. Pukall, H. Lunsdorf, M. Allgaier, and I. Wagner-Dobler. 2006. Hoeflea phototrophica sp. nov., a novel marine aerobic alphaproteobacterium that forms bacteriochlorophyll a. Int. J. Syst. Evol. Microbiol. 56:821-826. [DOI] [PubMed] [Google Scholar]
- 7.Bryant, D. A., A. M. G. Costas, J. A. Maresca, A. G. M. Chew, C. G. Klatt, M. M. Bateson, L. J. Tallon, J. Hostetler, W. C. Nelson, J. F. Heidelberg, and D. M. Ward. 2007. Candidatus Chloracidobacterium thermophilum: an aerobic phototrophic acidobacterium. Science 317:523-526. [DOI] [PubMed] [Google Scholar]
- 8.Butow, B., and T. Bergstein-Ben Dan. 1992. Occurrence of Rhodopseudomonas palustris and Chlorobium phaeobacteroides blooms in Lake Kinneret. Hydrobiologia 232:193-200. [Google Scholar]
- 9.Chew, A. G. M., and D. A. Bryant. 2007. Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu. Rev. Microbiol. 61:113-129. [DOI] [PubMed] [Google Scholar]
- 10.Eckert, W., T. Frevert, T. Bergstein-Ben Dan, and B. Z. Cavari. 1986. Competitive development of Thiocapsa roseopersicina and Chlorobium phaeobacteroides in Lake Kinneret. Can. J. Microbiol. 32:917-921. [Google Scholar]
- 11.Imhoff, J. F. 2001. True marine and halophilic anoxygenic phototrophic bacteria. Arch. Microbiol. 176:243-254. [DOI] [PubMed] [Google Scholar]
- 12.Karr, E. A., W. M. Sattley, D. O. Jung, M. T. Madigan, and L. A. Achenbach. 2003. Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl. Environ. Microbiol. 69:4910-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morris, R. M., M. S. Rappe, E. Urbach, S. A. Connon, and S. J. Giovannoni. 2004. Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl. Environ. Microbiol. 70:2836-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sattley, W. M., M. T. Madigan, W. D. Swingley, P. C. Cheung, K. M. Clocksin, A. L. Conrad, L. C. Dejesa, B. M. Honchak, D. O. Jung, L. E. Karbach, A. Kurdoglu, S. Lahiri, S. D. Mastrian, L. E. Page, H. L. Taylor, Z. T. Wang, J. Raymond, M. Chen, R. E. Blankenship, and J. W. Touchman. 2008. The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J. Bacteriol. 190:4687-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trost, J. T., and R. E. Blankenship. 1989. Isolation of a photoactive photosynthetic reaction center-core antenna complex from Heliobacillus mobilis. Biochemistry 28:9898-9904. [DOI] [PubMed] [Google Scholar]
- 16.Xiong, J., and C. E. Bauer. 2002. Complex evolution of photosynthesis. Annu. Rev. Plant Biol. 53:503-521. [DOI] [PubMed] [Google Scholar]
- 17.Yan, S., B. M. Fuchs, S. Lenk, J. Harder, J. Wulf, N. Z. Jiao, and R. Amann. 2009. Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria. Syst. Appl. Microbiol. 32:124-139. [DOI] [PubMed] [Google Scholar]
- 18.Yutin, N., O. Béjà, and M. T. Suzuki. 2008. The use of denaturing gradient gel electrophoresis with fully-degenerate pufM primers to monitor aerobic anoxygenic phototrophic assemblages. Limnol. Oceanogr. Methods 6:427-440. [Google Scholar]
- 19.Yutin, N., M. T. Suzuki, and O. Béjà. 2005. Novel primers reveal wider diversity among marine aerobic anoxygenic phototrophs. Appl. Environ. Microbiol. 71:8958-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yutin, N., M. T. Suzuki, H. Teeling, M. Weber, J. C. Venter, D. B. Rusch, and O. Béjà. 2007. Assessing diversity and biogeography of aerobic anoxygenic phototrophic bacteria in surface waters of the Atlantic and Pacific Oceans using the Global Ocean Sampling expedition metagenomes. Environ. Microbiol. 9:1464-1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.