Abstract
Aflatrem is a potent tremorgenic toxin produced by the soil fungus Aspergillus flavus, and a member of a structurally diverse group of fungal secondary metabolites known as indole-diterpenes. Gene clusters for indole-diterpene biosynthesis have recently been described in several species of filamentous fungi. A search of Aspergillus complete genome sequence data identified putative aflatrem gene clusters in the genomes of A. flavus and Aspergillus oryzae. In both species the genes for aflatrem biosynthesis cluster at two discrete loci; the first, ATM1, is telomere proximal on chromosome 5 and contains a cluster of three genes, atmG, atmC, and atmM, and the second, ATM2, is telomere distal on chromosome 7 and contains five genes, atmD, atmQ, atmB, atmA, and atmP. Reverse transcriptase PCR in A. flavus demonstrated that aflatrem biosynthesis transcript levels increased with the onset of aflatrem production. Transfer of atmP and atmQ into Penicillium paxilli paxP and paxQ deletion mutants, known to accumulate paxilline intermediates paspaline and 13-desoxypaxilline, respectively, showed that AtmP is a functional homolog of PaxP and that AtmQ utilizes 13-desoxypaxilline as a substrate to synthesize aflatrem pathway-specific intermediates, paspalicine and paspalinine. We propose a scheme for aflatrem biosynthesis in A. flavus based on these reconstitution experiments in P. paxilli and identification of putative intermediates in wild-type cultures of A. flavus.
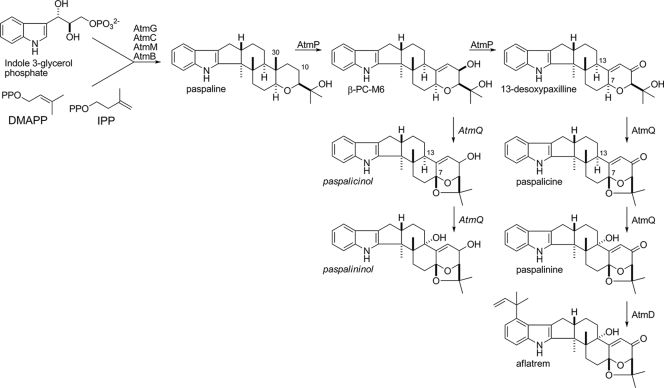
The soil fungus Aspergillus flavus is an opportunistic pathogen that can infect a variety of plant and animal hosts including humans (31). Despite identification of a large number of secondary metabolite genes and gene clusters in the genome of A. flavus (25) and the closely related Aspergillus oryzae genome (21), few of these pathways have been characterized. Aflatrem and its isomer β-aflatrem are indole-diterpenes produced by A. flavus (9, 10, 32). Indole-diterpenes are a structurally diverse group of secondary metabolites produced by disparate members of the Eurotiomycete and Sordariomycete classes of filamentous fungi including Aspergillus and Penicillium spp. of the former and Epichloë and Neotyphodium spp. of the latter (28). The majority of metabolites in this group share a common structural core consisting of a tetracyclic diterpene derived from geranylgeranyldiphosphate (GGPP) (7), fused to an indole moiety, most likely derived from indole 3-glycerol phosphate (5). These precursors combine to form paspaline, the first stable indole-diterpene required for paxilline biosynthesis (22, 28, 29). More complex indole-diterpenes generally consist of a paxilline-like core modified with a variety of decorations including different patterns of prenylation, hydroxylation, epoxidation, acetylation, and ring rearrangements (24). Aflatrem and its isomer β-aflatrem both consist of a paxilline-like core with an additional prenyl group on the indole moiety and an acetal group on the diterpene skeleton as shown in Fig. 1.
FIG. 1.
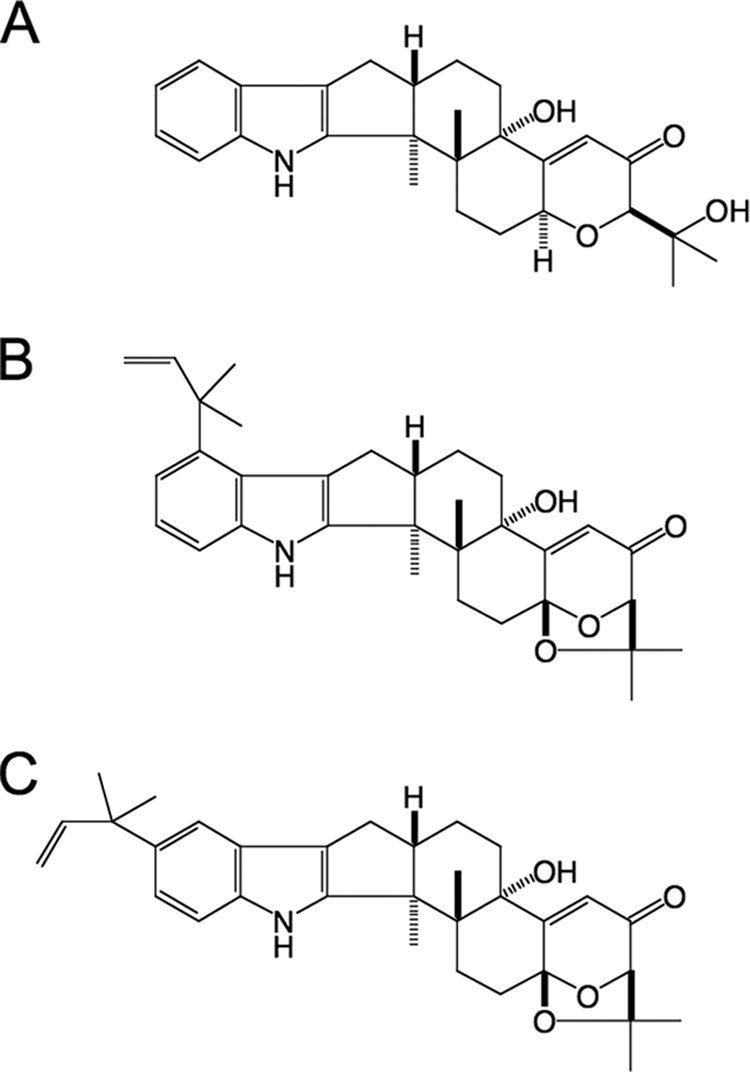
Chemical structures of paxilline (A), aflatrem (B), and β-aflatrem (C).
Like many other indole-diterpenes, aflatrem is a potent mammalian tremorgen (9, 10). The mechanisms by which tremorgenicity is effected are not yet fully understood but may be related to known neurological effects of aflatrem which include modulation of neurotransmitter release in the central nervous system (36, 38) and inhibition of BK channels in the peripheral nervous system (17).
The adoption of Penicillium paxilli as a model experimental system for investigating indole-diterpene biosynthesis has enabled characterization of the genes required for paxilline biosynthesis, paving the way for the identification and characterization of indole-diterpene biosynthesis gene clusters in less tractable fungi such as Neotyphodium lolii and Epichloë festucae (17, 42, 43). Paxilline biosynthesis in P. paxilli requires the products of seven clustered genes (paxG, paxA, paxM, paxB, paxC, paxP, and paxQ) (28, 29). The products of four of these genes, a GGPP synthase (PaxG), a prenyltransferase (PaxC), a flavin adenine dinucleotide (FAD)-dependent monooxygenase (PaxM), and a protein of unknown function (PaxB), are all required for the biosynthesis of paspaline (29). This product is subsequently converted to paxilline by the sequential action of two cytochrome P450 monooxygenases, PaxP and PaxQ (22, 28). The structural similarity of paxilline and aflatrem (Fig. 1) suggests that biosynthesis of these compounds proceeds via similar pathways involving the products of orthologous genes. By comparison with the pax gene cluster in P. paxilli, a complete aflatrem biosynthesis cluster would therefore be expected to contain homologs of all seven paxilline biosynthesis genes, plus a small number of aflatrem-specific genes necessary for additional modifications. Cloning of a secondary metabolite GGPP synthase-encoding gene and chromosome walking identified an atm locus (ATM1) containing a cluster of three putative genes for aflatrem production in A. flavus NRRL6541 (44). This locus contains homologs of paxG, paxM, and paxC, designated atmG, atmM, and atmC, respectively. Furthermore, it was demonstrated that atmM is a functional ortholog of paxM and that the induction of atm gene expression corresponded to the onset of aflatrem biosynthesis. Although ATM1 included candidate genes for some of the early steps required for aflatrem biosynthesis, homologs of four genes known to be necessary for later steps in paxilline synthesis in P. paxilli (paxP, paxQ, paxA, and paxB) were not identified.
Complete genome sequences are now available for several different aspergilli including A. flavus and its close relative A. oryzae (21, 25), enabling rapid identification of genes and gene clusters and predictions of associated biosynthesis potential. The aims of this study were to (i) search the Aspergillus genome databases for putative genes required for indole-diterpene biosynthesis, (ii) characterize the aflatrem biosynthesis pathway in A. flavus by the identification of biosynthesis intermediates, and (iii) propose a scheme for aflatrem biosynthesis based on the chemical and genetic data. This study identified two ATM loci in A. flavus on separate chromosomes and demonstrated through metabolic engineering of P. paxilli that the proposed aflatrem biosynthesis steps from paspaline through paspalinine can be reconstituted in this fungus.
MATERIALS AND METHODS
Fungal strains and growth conditions.
Cultures of A. flavus strains NRRL6541 and NRRL3357 were maintained on 2.4% Difco potato dextrose agar (Becton Dickinson, Maryland) plates or as spore suspensions in 10% (vol/vol) glycerol at −80°C. For aflatrem production, 100-ml Erlenmeyer flasks containing 25 ml of YEPGA medium (22) were inoculated with 5 × 106 spores and grown with shaking (200 rpm) for 48 h at 30°C. Aliquots of 2 ml were used to inoculate 250-ml Erlenmeyer flasks containing 50 ml of aflatrem production medium (44), which were grown without agitation at 29°C in the dark until harvesting. The mycelial mat that formed on the surface of the liquid was retrieved using a spatula and washed in Milli-Q water (Millipore, Massachusetts). Approximately 250 mg of mycelium was collected for RNA preparation, and the remainder was freeze-dried for indole-diterpene analysis. Fungal samples were stored at −80°C prior to drying or analysis. For isolation of genomic DNA, 25 ml of CDYE medium (41) was inoculated with 5 × 106 spores and grown with shaking (200 rpm) for 48 h at 30°C. Mycelium was harvested, washed in Milli-Q water (Millipore), and freeze-dried. Strains of P. paxilli were routinely grown in aspergillus complete medium at 22°C for 4 to 6 days as previously described (29). Penicillium cultures were grown in submerged liquid culture for isolation of genomic DNA, preparation of protoplasts, and indole-diterpene analysis as previously described (29).
Isolation, PCR amplification, and sequencing of genomic DNA.
Genomic DNA was isolated from freeze-dried mycelia by using the method of Yoder (40). Genomic DNA was amplified using the TripleMaster PCR system (Eppendorf, Hamburg, Germany). Reactions were each performed in a 50-μl volume that contained 1× High Fidelity buffer with a final concentration of 4 mM magnesium acetate, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 2 U TripleMaster polymerase mix, a 400 nM concentration of each primer, and 50 ng of genomic DNA. Thermal cycling was performed in a Mastercycler gradient thermocycler (Eppendorf) under the following conditions: 2 min at 94°C followed by 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 3 min, with a final elongation for 10 min at 72°C. Primers used for amplification of atm cluster sequences are shown in Table 1. PCR products were purified using the Wizard SV gel and PCR cleanup system (Promega, Wisconsin) prior to sequencing. At least two independent PCR mixtures were combined and sequenced directly on both strands using the dideoxynucleotide chain termination method with the Big-Dye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems, California) and separated using an ABI3730 genetic analyzer (Applied Biosystems).
TABLE 1.
Oligonucleotide primers used for amplification of gDNA and cDNA
| Target | Forward primer |
Reverse primer |
Product size (bp) |
|||
|---|---|---|---|---|---|---|
| Name | Sequence | Name | Sequence | Genomic | cDNA | |
| Sequence gap | 3357SG1f | GCAAGCACGAGTCGACAAGGTGTTGGA | 3357SG2r | GTTTCGGTCGCCACATAATATCCGACAA | 2,963 | |
| ATM1 | Atm1F1 | CCAGGCACATCTTGTCCGACTCTGGAA | Atm1R1 | CAAATCCAGCACACCGCAGCTTGGCTA | 4,589 | |
| Atm2F1 | CATGTCTTGCTCGGGATAGTGCATCA | Atm2R1 | ACACGACTGCAGTCACTTGTGATGCCA | 5,362 | ||
| Atm2F2 | GAAGGAGGTGATGGTAGATGAATGACA | Atm2R2 | TACGGAGTCCATGTTGTAGAAAGGCTT | 5,073 | ||
| ATM2 | Atm2F3 | GAGCGATCCGTAGACCATGCCGACATA | Atm2R3 | TTCTTCGGGAGACTGGGTCGGCTTTCA | 4,957 | |
| Atm2F4 | AACATCTCGAACGGGATGGACAGCGTT | Atm2R4 | TGCTGTACTGCTATAATGAGAGTGTCA | 5,140 | ||
| KTR2 | CAACAAAATAGCATGATCCAACGCATG | Atm2R5 | CAATCAGGTCGTGGACACAGACATCGT | 6,096 | ||
| atmP | NcoI-atmPF1 | ATAGCCATGGACAAATTGACCGCCA | EcoRI-atmPR1 | TCACGAATTCACCGTAATCGAGGCAGA | 1,976 | |
| atmQ | NcoI-atmQF1 | TCGTCCATGGATCGACTATTGGAGAGAA | EcoRI-atmQR1 | TCAAGAATTCTCCGTGTCTTGGTTATTGA | 2,380 | |
| AF108 | AF108F1 | TGGCCAGCTGACTATTGAAGA | AF108R2 | GGCAGTATTATCCCAGTCGTT | 810 | 591 |
| AF109 | AF109F1 | GGAACGCTTCCACGAACAACA | AF109R1 | GGATCCCCTTGGAAGTTACTA | 832 | 780 |
| AF110 | AF110F1 | ACTCTCGCTCTTGCTCTCCAT | AF110R1 | CGCATTAAGCTCCAACACCTT | 1,031 | 1,031 |
| atmG | AF25 | TGCATCCTCGCTCTCTTCAA | 3357SG3f | CTTGGGGCGATTGAGCTCT | 534 | 380 |
| atmC | atmCF | TCGGATATTATGTGGCGACC | atmCR | GTTGCCGCCTCTGTTGCCTT | 888 | 776 |
| atmM | AF79 | GTTATCCGCTCTGAGATGTG | AF80 | CTGTCATAGACGTAACCTCC | 443 | 375 |
| atmD | atmDR1 | GGTAATCCCGTACATCCATAG | Atm2-1R7 | GACGGAACACCCGCCGAAG | 883 | 883 |
| atmQ | Atm2-2F3 | GAAGACGAACGTTCCTTCG | Atm2-2R4 | GCTGCTAAGTTTTGCAGCG | 640 | 479 |
| atmB | atmBF1 | TGGACGGATTTGGCTCATCAC | Atm2-2F7 | GCTGTGTATATGACGCCCA | 280 | 280 |
| atmA | Atm2-3F2 | GCCACCATGTCGGCTTATC | Atm2-3R3 | GAGAGACTGGATCTTAGAC | 617 | 551 |
| atmP | Atm2-3F4 | TCCGTCCGAGGTAGAGAAT | atmPR1 | CTAGGCGGAGGAAAACCTCAT | 751 | 530 |
| β-Tubulin | AFtub1 | CTTCTTCATGGTTGGCTTCG | AFtub3 | GGTGGAGGACATCTTGAGAC | 369 | 307 |
Preparation of complementation constructs and transformation of P. paxilli.
Genomic DNA containing atmP was amplified from A. flavus strain NRRL6541 by using Platinum pfx50 polymerase (Invitrogen) with primers NcoI-atmPF1 and EcoRI-atmPR1 for atmP and NcoI-atmQF1 and EcoRI-atmQR1 for atmQ (Table 1). Reactions were performed in 50 μl containing 1× pfx50 buffer, 200 μM of each dNTP, 2 U polymerase, 400 nM of each primer, and 50 ng of genomic DNA under the following conditions: 94°C for 2 min followed by 30 cycles of 94°C for 15 s and 68°C for 75 s, with a final elongation for 10 min at 68°C. The base vector pPN1851, which contains 850 bp of the promoter region for P. paxilli paxM (43), and the atmP and atmQ amplification products were each digested with restriction enzymes EcoRI and NcoI and purified as described for PCR products (above). The purified digestion products were ligated together so that atmP and atmQ were cloned separately into pPN1851 to produce pCP5 (atmP) and pCQ16 (atmQ) under the control of the paxM promoter. Protoplasts of P. paxilli paxP and paxQ deletion mutant strains LMP1 and LMQ226 were prepared and cotransformed with 5 μg of either pCP5 (LMP1) or pCQ16 (LMQ226) and 5 μg of pII99 as described previously (29). Transformants were selected on regeneration medium supplemented with 150 μg/ml Geneticin (Roche Applied Science).
Bioinformatics and genomic sequences.
Sequence reads were analyzed using the PHRED algorithm and assembled into contigs using the PHRAP algorithm in MacVector Assembler 1.1 (MacVector Inc., 2007) to a minimum confidence score of PHRED 40 for every nucleotide position. Database searches were performed at the Broad Aspergillus Comparative Database website (http://www.broad.mit.edu/annotation/genome/aspergillus_group/MultiHome.html) using tBLASTn against Aspergillus comparative genomic sequence and at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/) using BLASTp against the nonredundant protein and Swiss-Prot databases. Identity and similarity scores were calculated after ClustalW alignment of sequences using MacVector version 9.5 (MacVector Inc.). Genome annotations and numbering are given in accordance with the Broad Aspergillus Comparative Database. Genome sequence for A. flavus NRRL3357 is available at www.aspergillusflavus.org. Genomic regions for ATM1 and syntenic regions from other genomes include A. flavus NRRL3357 contig 9 positions 1914683 to 1949498; A. oryzae contig 17 positions 1730789 to 1767501; Aspergillus niger contig 5 positions 2137819 to 2151260, contig 11 positions 1578481 to 1568987, contig 19 positions 432796 to 435876, and contig 4 positions 703107 to 698299; Aspergillus clavatus contig 78 positions 890720 to 895084; Neosartorya fischeri contig 576 positions 394080 to 408036 and contig 578 positions 655741 to 650224; Aspergillus fumigatus chromosome 7 positions 164760 to 178480; Aspergillus terreus supercontig 9 positions 218784 to 231776; and Aspergillus nidulans contig 112 positions 177193 to 190274. For ATM2, genomic regions include A. flavus NRRL3357 contig 5 positions 1938653 to 1963908, A. oryzae contig 22 positions 1879902 to 1905024, A. clavatus contig 87 positions 633315 to 623528, A. fumigatus chromosome 2 positions 2418299 to 2411172, N. fischeri contig 580 positions 3775271 to 3766934, A. terreus supercontig 15 positions 968338 to 975158 and 996821 to 998736, A. nidulans contig 104 positions 182943 to 177146, and A. niger contig 18 positions 575746 to 580865.
Indole-diterpene analysis.
Freeze-dried fungal biomasses collected from stationary aflatrem production medium cultures (A. flavus) or submerged liquid cultures (P. paxilli) were each homogenized in 30 ml of a 2:1 mixture of chloroform-methanol to extract the indole-diterpenes. For semiquantitative analysis of aflatrem, equal weights (14.6 g ± 1 g) of A. flavus biomass were used. After being mixed for 1 h, samples were centrifuged at 18,000 × g in a bench top centrifuge for 10 min to pellet the insoluble material. A 2-ml aliquot of each extract was evaporated under a stream of nitrogen gas and then redissolved in 1.0 ml of methanol. Samples of 10 μl were analyzed by reverse-phase high-performance liquid chromatography (HPLC) using a Dionex Summit (Dionex Corporation, California) with a Luna (Phenomenex, California) C18 column (4.6 × 250 mm, 5 μm). Methanol-water (85:15) was routinely used as the mobile phase with a flow rate of 1.5 ml/min. For P. paxilli extracts from the atmQ complementation experiments, the mobile phase was methanol-water (75:25) with a flow rate of 1.2 ml/min. All samples and solvents were filtered through 0.45-μm nylon filters (Millipore) before analysis. Eluted products were analyzed by UV spectrophotometry at either 230 or 280 nm. Three major A. flavus indole-diterpene peaks were collected and analyzed by liquid chromatography-mass spectrometry (LC-MS) along with whole-cell extracts from A. flavus wild-type and atmP- and atmQ-containing derivatives of P. paxilli paxP and paxQ deletion mutants. LC-tandem MS (LC-MS/MS) analysis was performed on a Thermo Finnigan Surveyor (Thermo Finnigan, California) HPLC system equipped with a Luna (Phenomenex) C18 column (150 × 2 mm, 5 μm) at a flow rate of 200 μl/min with a solvent gradient starting with acetonitrile-water (60:40) in 0.1% formic acid and increasing to 95% acetonitrile over 30 min followed by a column wash with 99% acetonitrile. Mass spectra were determined with a linear ion trap mass spectrometer (Thermo LTQ; Thermo Finnigan, California) using electrospray ionization in positive mode. The spray voltage was 5.0 kV, and the capillary temperature was 275°C; sheath gas, auxiliary gas, and sweep gas were set to 20, 5, and 10 (arbitrary units), respectively, and other parameters were optimized automatically while infusing paxilline at 10 μl/min. The mass spectrometer either was set up in data-dependent mode, collecting fragmentation data on the predominant ions in the chromatogram, or was set up in single-reaction-monitoring mode, collecting and fragmenting selected ions to determine their presence or absence.
RNA isolation and cDNA synthesis and analysis.
Mycelial samples were stored at −80°C prior to processing. Total RNA was isolated from mycelia using the FastRNA Pro Green kit (QBioGene, California) and FastPrep machine (QBioGene) per the manufacturer's instructions and quantified using a nanophotometer (Implen, Munich, Germany). Samples of RNA were each treated with DNase I for 30 min at 37°C in a 50-μl reaction volume that contained 30 U of DNase I (Roche, Auckland, New Zealand), 1× DNase I buffer (Roche), 2 mM of dithiothreitol, 20 units of RNase inhibitor (Invitrogen, Auckland, New Zealand), and ≤30 μg of RNA. Reactions were stopped by incubating the reaction mixtures at 75°C for 10 min. First-strand cDNA synthesis was performed as follows: a 10.8-μl volume containing 1 μg of DNase-treated RNA and 25 ng of oligo(dT) was incubated at 65°C for 10 min. A reaction premix of 8.2 μl containing 2× reverse transcription buffer (Roche), 10 mM of dithiothreitol, 1 mM of each dNTP, and 8 U of RNase inhibitor (Invitrogen) was then added to the RNA-oligo(dT) premix, followed by 50 U of Expand reverse transcriptase (Roche). Reaction mixtures were incubated at 43°C for 60 min. Resulting cDNA was diluted 1/10 and used for PCR. Gene-specific amplifications of the cDNA were carried out in 25-μl reaction mixtures that contained 1× PCR buffer (Invitrogen) with a final concentration of 4 mM MgCl2, 200 μM of each dNTP, 0.5 U Platinum Taq polymerase (Invitrogen), 200 nM of each primer, and 1 μl of template cDNA. Thermal cycling was performed in a Mastercycler gradient thermocycler (Eppendorf) with the following conditions: 2 min at 94°C followed by 35 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 1 min, with a final elongation for 10 min at 72°C. Primers that were used for amplification of cDNA are shown in Table 1.
Nucleotide sequence accession numbers.
The sequence with nucleotide accession number AY559849 containing A. flavus NRRL6541 ATM1 has been updated to include new sequence flanking the cluster, and sequence for ATM2 has been deposited with accession number AM921700. A 2,772-bp sequence covering the genomic sequence gap in A. flavus NRRL3357 has been deposited under accession number AM911677.
RESULTS
Identification of aflatrem gene clusters in A. flavus and A. oryzae.
A search of Aspergillus complete genome sequence databases using tBLASTn with the amino acid sequences encoded by three linked genes, atmG, atmC, and atmM, proposed to be required for aflatrem production in A. flavus NRRL6541 (44), identified the presence of highly similar sequences in the genomes of A. flavus NRRL3357 and A. oryzae RIB40. A sequence gap of 1,959 bp on contig 9 of the genome sequence of strain NRRL3357 was closed by directly sequencing a PCR product that covered this region. A PCR product was also sequenced to extend the existing ∼39-kb genomic sequence of the ATM1 locus in strain NRRL6541 by a further 4 kb. A comparison of this ∼43-kb sequence in NRRL3357, NRRL6541, and RIB40 showed that the sequences were >97% identical and contained intact copies of atmG, atmC, and atmM.
The ATM locus described above contained putative orthologs of only three of seven genes that are necessary for paxilline biosynthesis in P. paxilli. Searching the A. flavus and A. oryzae complete genome sequences by using the amino acid sequences encoded by the four remaining paxilline biosynthesis genes (paxA, paxB, paxP, and paxQ) identified a second locus, ATM2, that contained homologs of these four genes and another gene from the pax gene cluster (paxD), which is required for a postpaxilline modification (B. Scott et al., unpublished data). Six overlapping PCR products amplified from genomic DNA of strain NRRL6541 were sequenced to provide a contiguous sequence of ∼25 kb covering this region. Comparison of the sequences corresponding between all three genomes revealed >97% identity, with the exception of the intergenic region immediately 3′ of atmD. This ∼4-kb region was >96% identical for NRRL3357 and RIB40 but differed for NRRL6541, particularly over a region of approximately 1.5 kb where NRRL6541 sequence had no detectable similarity with the other two genomes.
Genomic comparisons of predicted genes in and around both clusters across all three strains enabled refinement of gene predictions resulting in the A. flavus/A. oryzae consensus sequences represented in Fig. 2. Known or predicted functions for gene products based on similarity with characterized proteins are detailed in Table 2. The genes atmG, atmC, and atmM at ATM1 are proposed to encode a GGPP synthase, a prenyltransferase, and an FAD-dependent monooxygenase, respectively (44). Genes at ATM2 are putative orthologs of other pax genes and have been designated atmD, atmQ, atmB, atmA, and atmP. atmA and atmB are putative orthologs of paxA and paxB, respectively, and are predicted to encode polytopic membrane proteins whose precise functions in aflatrem biosynthesis are unknown. However, paxA and paxB are both required for paxilline biosynthesis in P. paxilli (Scott et al., unpublished). atmB contained one intron in the same relative position as the intron in paxB, and the protein products of these genes shared 61% amino acid identity (77% similarity). atmA also contained a single intron in the same relative position as the intron in paxA, and amino acid identity between these two proteins was 29% (49% similarity). atmP and atmQ are both predicted to encode cytochrome P450 monooxygenases and are putative orthologs of paxP and paxQ, respectively. The products of each of these genes contained all of the functional domains expected for cytochrome P450s including heme-binding domains identical to PaxP and PaxQ (HFGLGRYAC for AtmP and QFGDGRHTC for AtmQ) (11, 22). atmP contained five introns identical in the same relative positions as the introns in paxP, and the gene products shared 61% identity (70% similarity). atmQ contained eight introns that were identical in the same relative positions as eight of the nine introns in paxQ. The fourth intron 5′ of paxQ does not have an equivalent in atmQ such that the fourth exon of atmQ was equivalent to the fourth and fifth exons of paxQ. The protein products of paxQ and atmQ share 55% amino acid identity (70% similarity). atmD, which is predicted to encode an aromatic prenyltransferase, is a putative ortholog of paxD. Like paxD, atmD was devoid of introns. The predicted protein products share 29% amino acid identity (46% similarity). AtmD was also similar to other characterized aromatic prenyltransferases including the dimethylallyl tryptophan synthase DmaW from Neotyphodium sp. strain Lp1 (24% amino acid identity; 37% similarity) (37) and FGAPT1, a prenyltransferase from A. fumigatus (22% identity; 35% similarity) (35). However, none of the functional motifs that are characteristic of trans-prenyltransferases (20) could be identified in AtmD.
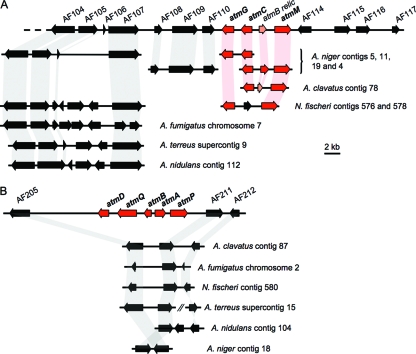
FIG. 2.
Physical maps of ATM1 (A) and ATM2 (B) aflatrem biosynthesis loci in A. flavus and A. oryzae and syntenic regions from other Aspergillus genomes. Arrows represent the positions and transcriptional orientations of genes. The top lines of panels A and B show consensus gene predictions for A. flavus NRRL6541, A. flavus NRRL3357, and A. oryzae RIB40. Genes for other genomes are shown according to annotations as detailed at the Broad Fungal Genome Initiative website with the exception of the atmB-like sequences for A. niger and A. clavatus, for which there were no automated predictions. In some cases, such as for the homolog of gene AF205 in A. fumigatus, the corresponding gene predictions do not match the full-length A. flavus/A. oryzae predictions; however, homology exists over the areas covered by the shaded bands. Proposed aflatrem biosynthesis genes are shown in red. An atmB relic between atmC and atmM at the ATM1 locus matches a similar relic in A. clavatus and an intact atmB-like gene in A. niger.
TABLE 2.
Known and predicted functions of genes at ATM locia
| Gene | Closest characterized match identified by BLASTp |
Predicted function/comment | |||
|---|---|---|---|---|---|
| Species and protein | Accession no. | E value | Function | ||
| AF104 | Pseudomonas putida NahG | AAA25897 | 2e-21 | Salicylate hydroxylase | Aromatic hydroxylase with decarboxylase domain |
| AF105 | Rhodococcus sp. CocE | Q9L9D7 | 2e-17 | Cocaine esterase | Esterase |
| AF106 | Fungal conserved hypothetical | ||||
| AF107 | Xenopus laevis Krox20 | Q08427 | 2e-12 | Transcription factor | Transcription factor |
| AF108 | Homo sapiens BLVRB | NP_990721 | 3e-06 | NADH-flavin reductase | Hydroxylase/dehydrogenase |
| AF109 | Neurospora crassa ACU-15 | PI7000 | 1e-05 | Transcriptional activator—regulates acetate utilization | Fungal conserved hypothetical; transcription factor domains |
| AF110 | Pseudomonas putida NahG | AAA25897 | 1e-15 | Salicylate hydroxylase | Aromatic hydroxylase/monooxygenase |
| atmG | P. paxilliPaxG | AAK11531 | 8e-97 | GGPP synthase | GGPP synthase |
| atmC | P. paxilliPaxC | AAK11529 | 2e-108 | Prenyltransferase | Prenyltransferase |
| atmM | P. paxilliPaxM | AAK11530 | 7e-140 | Monooxygenase | Monooxygenase |
| AF114 | Blumeria graminis | AAL56992 | 5e-29 | Essential for pathogenesis | Pathogenesis/penetration |
| AF115 | Mus musculus Cyp8b1 | NP_034142 | 5e-12 | Cytochrome P450 monooxygenase | Cytochrome P450 monooxygenase |
| AF116 | Fusarium sporotrichioides TRI101 | AAD19745 | 2e-09 | Trichothecene acetyltransferase | Acetyltransferase |
| AF117 | Botrytis elliptica NEP1 | ABB43265 | 4e-84 | Necrosis- and ethylene-inducing protein | Necrosis-inducing protein |
| AF205 | Debaryomyces occidentalis Trk1 | CAB91046 | 1e-112 | Potassium ion transporter | Ion transporter |
| atmD | P. paxilliPaxD | AAK11526 | 9e-56 | Aromatic prenyltransferase | Aromatic prenyltransferase |
| atmQ | P. paxilliPaxQ | AAK11527 | 7e-174 | Cytochrome P450 monooxygenase | Cytochrome P450 monooxygenase |
| atmB | P. paxilliPaxB | 1e-77 | Unknown—required for paxilline biosynthesis | Unknown—required for aflatrem biosynthesis | |
| atmA | P. paxilliPaxA | 9e-30 | Unknown—required for paxilline biosynthesis | Unknown—required for aflatrem biosynthesis | |
| atmP | P. paxilliPaxP | AAK11528 | 0 | Cytochrome P450 monooxygenase | Cytochrome P450 monooxygenase |
| AF211 | S. cerevisiae Jen1p | NP_012705 | 6e-58 | Lactate transporter | Carboxylic acid transport |
| AF212 | A. oryzae AxeA | BAD12626 | 4e-162 | Acetyl xylan esterase | Esterase |
Boldface denotes proposed atm biosynthesis genes.
The amino acid products of all atm genes were found to be at least 95% identical to their orthologs within the three A. flavus/A. oryzae strains investigated. Furthermore, all predicted atm genes in these three genomes appear to have open reading frames capable of encoding functional proteins, with the exception of atmQ in the genome of A. oryzae. The product of this gene contained a single nucleotide insertion in exon 7 that will lead to the synthesis of a nonfunctional protein product.
Seven genes located 3′ of atmG in ATM1 are predicted to encode two aromatic hydroxylases/monooxygenases (AF104 and AF110), an esterase (AF105), a conserved hypothetical protein of unknown function (AF106), two transcription factors (AF107 and AF109), and a dehydrogenase (AF108) (Fig. 2). Four genes 3′ of atmM are predicted to encode an integral membrane protein involved in pathogenesis (AF114), a cytochrome P450 monooxygenase (AF115), an acetyltransferase (AF116), and a necrosis-inducing protein (AF117). Genes that are predicted to encode an ion transporter (AF205), a carboxylic acid transporter (AF211), and an esterase (AF212) flank the ATM2 locus and, based on their predicted function, are unlikely to be required for aflatrem biosynthesis.
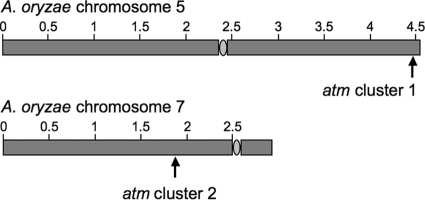
The two ATM loci in A. oryzae are located approximately 85 kb from the end of the short arm of chromosome 5 (ATM1) and close to the middle of chromosome 7 (ATM2) (Fig. 3). The chromosomal location of the two loci in the genome of A. flavus NRRL3357 matches that of A. oryzae RIB40 (25) (G. A. Payne et al., unpublished data). The higher density of secondary metabolite-type genes around ATM1 than around ATM2 is consistent with the observation that genes for secondary metabolism tend to be enriched in telomere-proximal regions of fungal genomes (8, 23).
FIG. 3.
Chromosomal locations of ATM1 (top) and ATM2 (bottom) in A. oryzae. ATM1 is located approximately 85 kb from the end of chromosome 5. Sizes are shown in Mb.
atm cluster remnants in other Aspergillus genomes.
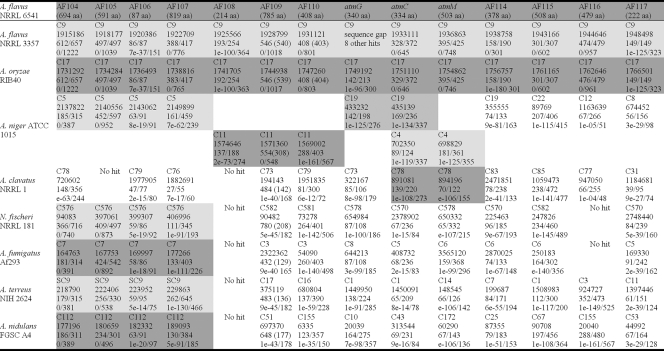
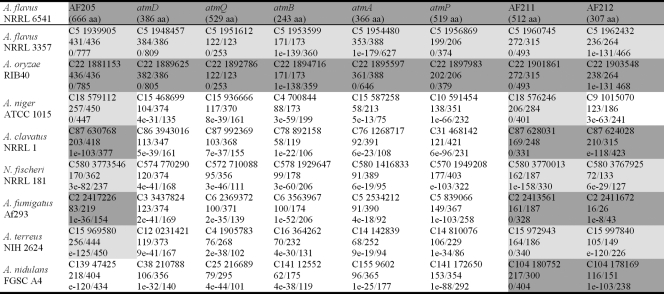
tBLASTn searches identified putative gene homologs in other Aspergillus genomes. Tables 3 and 4 summarize the genomic locations (position on chromosome or contig) and BLAST results for genes in and around ATM1 and ATM2, respectively. Putative homologs that were linked in other Aspergillus genomes are shaded in Tables 3 and 4 and shown in Fig. 2 to illustrate synteny.
TABLE 3.
tBLASTn results for predicted or known translational products of locus ATM1 genes (AF104 to AF117) in NRRL6541 against other Aspergillus genomic sequencesa
Linkage groups within each genome are displayed with similar shading. Each entry contains the following information from top to bottom, respectively: contig or chromosome number, start position on contig, amino acid identity/alignment length using sequence from NRRL6541 as query, and BLAST E value/score.
TABLE 4.
tBLASTn results for predicted translational products of locus ATM2 genes (AF205 to AF212) in NRRL6541 against other Aspergillus genomic sequencesa
Linkage groups within each genome are displayed with similar shading. Each entry contains the following information from top to bottom, respectively: contig or chromosome number and start position on contig, amino acid identity/alignment length using sequence from NRRL6541 as query, and BLAST E value/score.
Genomic regions similar to those containing genes AF104 to AF107 3′ of atmG in A. flavus were identified in all other Aspergillus genomes with the exception of A. clavatus. A lack of conserved synteny with other Aspergillus genomes was evident for genes AF114 to AF117 found 3′ of atmM in A. flavus. Although not linked, homologs of these genes were found in other Aspergillus genomes (Table 3). This observation is consistent with the telomere-proximal location of this locus (Fig. 3) and the increased levels of genomic reorganization associated with these regions (8, 14). Linked copies of AF108, AF109, and AF110 were not identified in any other genome besides A. niger, where a syntenic triad of similar genes was identified. Linked homologs of some atm genes were also identified in the genomes of A. niger, A. clavatus, and N. fischeri (anamorph Aspergillus fischerianus). In A. niger an atmG-atmC pair was detected in the same order and transcriptional orientation as seen in A. flavus/A. oryzae, in addition to a second atmC homolog that was linked to atmB- and atmM-like genes. While the atmC-atmM-like gene pair was syntenic between these genomes, atmB was located in ATM2 in A. flavus and A. oryzae. However, closer examination of the atmC-atmM intergenic region in A. flavus/A. oryzae identified a relic of atmB containing mutations that disrupt the open reading frame. A similar triad was also identified in the genome of A. clavatus, which also contained an atmB-like gene relic. Syntenic homologs of atmG and atmM were identified in the genome of N. fischeri. However, these genes are interrupted by the sequence of an unrelated conserved hypothetical gene.
No synteny was observed in other Aspergillus genomes for genes at ATM2 (Fig. 2B). However, homologs of the genes flanking this locus were identified in all other Aspergillus genomes with similar syntenic organization which continues beyond the 25 kb of sequence represented in Fig. 2B (not shown). In A. flavus and A. oryzae the distance between AF205 and AF211 is approximately 19 kb, compared with approximately 0.5 to 3 kb for other sequenced Aspergillus genomes.
Analysis of atm gene transcript levels.
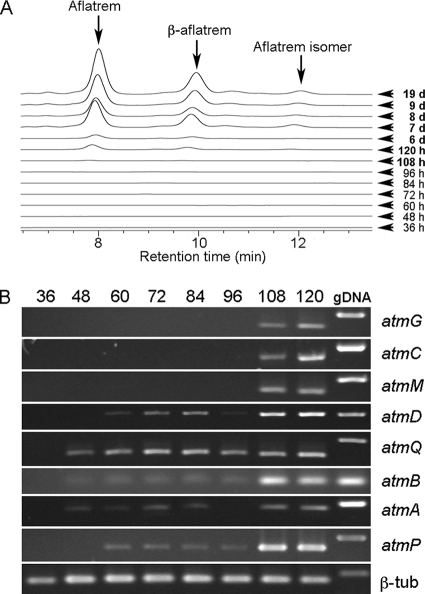
HPLC analysis of extracts of A. flavus NRRL6541 grown in stationary culture for up to 19 days detected increasing aflatrem levels from 108 h postinoculation (Fig. 4A). Peaks representing three different aflatrem isomers were detected (see below for mass spectral evidence), and the two major aflatrem products were present in a 2:1 ratio in all samples. This is consistent with the observation that A. flavus NRRL6541 consistently produces aflatrem and β-aflatrem in a 2:1 ratio (Jan Tkacz, personal communication), indicating that these peaks represented aflatrem and β-aflatrem, respectively. Semiquantitative analysis of steady-state levels of atm transcripts using reverse transcriptase PCR is shown in Fig. 4B. The onset of expression of atmG, atmC, and atmM transcripts at ATM1 correlated with the onset of aflatrem biosynthesis at 108 h postinoculation. While mRNA transcripts at ATM2 were detected much earlier at 48 (atmQ, atmB, and atmA) or 60 (atmD and atmP) h postinoculation, with the exception of atmQ the steady-state levels of the transcripts for the genes at this locus correlated with the onset of aflatrem biosynthesis at 108 h. Low levels of aflatrem were detected by HPLC in 19-day extracts from A. flavus NRRL3357 grown in stationary culture (not shown).
FIG. 4.
Transcript expression analysis of aflatrem biosynthesis genes in A. flavus NRRL6541 (A) Reverse-phase HPLC analysis of indole-diterpenes in culture extracts detected at 280 nm showing the onset of aflatrem production. (B) Reverse transcriptase PCR analysis of steady-state levels of atm transcripts from cultures grown for 36 to 120 h. Total RNA was isolated from fungal mycelia and, following reverse transcription, was amplified by PCR using primers specific for each of the genes shown.
Chemical analysis of indole-diterpene biosynthesis products.
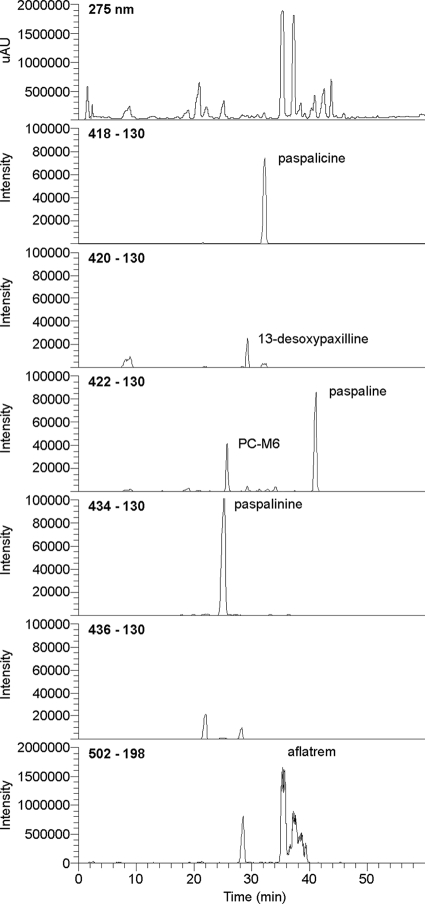
LC-MS analysis identified a range of indole-diterpenes in chemical extracts from 19-day cultures of A. flavus NRRL6541 as shown in Fig. 5. Known indole-diterpenes that were identified using single reaction monitoring included paspalicine (m/z = 418), 13-desoxypaxilline (m/z = 420), PC-M6 (m/z = 422), paspaline (m/z = 422), paspalinine (m/z = 434), and three aflatrem isomers (m/z = 502), two of which were likely to be aflatrem and β-aflatrem. The third aflatrem isomer is likely to be an alternative prenylated derivative of paspalinine. Based on data-dependent fragmentation, we were able to identify novel indole-diterpenes including hydroxyaflatrem (m/z = 518) and a compound that is likely to be the paxitriol version of paspalinine (m/z = 436), which we named paspalininol. Three major indole-diterpene peaks seen in HPLC traces with retention times of 7.9, 9.8, and 11.8 min (Fig. 4A) were purified and analyzed by LC-MS and shown to correspond to aflatrem, β-aflatrem, and a prenylated derivative of paspalinine, respectively. Nineteen-day cultures of A. flavus NRRL3357 contained indole-diterpene profiles similar to that of strain NRRL6541 but at 50- to 100-fold lower levels.
FIG. 5.
Chromatogram showing the elution times for indole-diterpenes detected in A. flavus NRRL6541. The top line shows the UV trace at 275 nm. Subsequent lines show LC-MS/MS traces for 418, 420, 422, 434, 436, and 502 ions containing either 130 or 198 m/z fragments characteristic of indole or prenylated indole moieties, respectively.
Complementation of P. paxilli paxP and paxQ deletion mutants with atmP and atmQ.
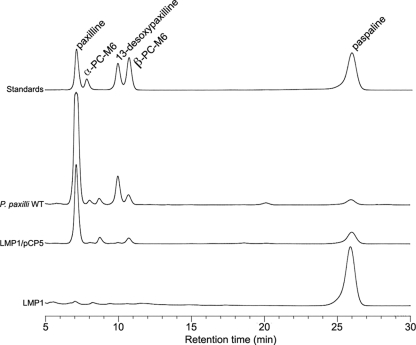
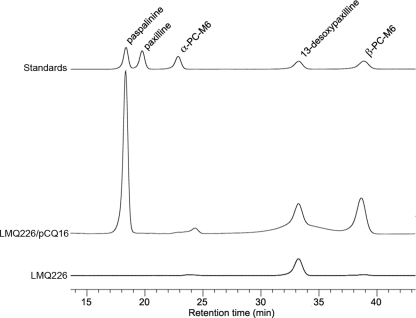
Due to the difficulty of being able to generate a deletion derivative of an atm gene in A. flavus, several steps in the proposed aflatrem biosynthesis pathway were reconstituted in P. paxilli. We previously showed that atmM, from locus ATM1, can complement ΔpaxM to restore paxilline biosynthesis in P. paxilli (44). Using the same approach, we tested whether atmP and atmQ from locus ATM2 could complement ΔpaxP and ΔpaxQ, respectively. In P. paxilli PaxP catalyzes the conversion of paspaline to 13-desoxypaxilline via PC-M6 and PaxQ catalyzes the conversion of 13-desoxypaxilline to paxilline (28). Protoplasts of P. paxilli ΔpaxP (LMP1) and ΔpaxQ (LMQ226) were cotransformed with pII99 (Genr) and pCP5 or pCQ16, plasmids containing the atmP and atmQ genes from A. flavus NRRL6541 under the control of the paxM promoter. Mycelial extracts of Genr transformants found by PCR to contain either atmP (PN2732) or atmQ (PN2733) were then analyzed for the presence of indole-diterpenes by normal-phase thin-layer chromatography (data not shown) and reverse-phase HPLC (Fig. 6 and 7). P. paxilli AtmP-containing transformants were shown to synthesize predominantly paxilline but also contained PC-M6 and 13-desoxypaxilline (Fig. 6). This experiment demonstrates that atmP can complement paxP and that AtmP can convert paspaline to 13-desoxypaxilline. Thin-layer chromatography analysis of extracts of the P. paxilli AtmQ-containing transformants showed that they lacked paxilline but instead had two unidentified indole-diterpenes. HPLC analysis confirmed the presence of indole-diterpenes in these extracts with the predominant compound identified from an authentic standard as paspalinine (18.3 min), but 13-desoxypaxilline (33.3 min) and an unidentified indole-diterpene (38.8 min) were also present (Fig. 7). LC-MS analysis of fractions collected at 18.3 and 38.8 min confirmed that these peaks corresponded to paspalinine (MS1, m/z = 434; MS2, m/z = 419, loss of methyl) and paspalicine (MS1, m/z = 418; MS2, m/z = 403, loss of methyl) (data not shown). This experiment demonstrated that AtmQ is not a functional ortholog of PaxQ. Unlike PaxQ, which catalyzes the conversion of 13-desoxypaxilline to paxilline, AtmQ converts 13-desoxypaxilline to paspalicine and paspalinine, proposed intermediates unique to aflatrem biosynthesis.
FIG. 6.
HPLC analysis of indole-diterpenes from a P. paxilli paxP mutant complemented with atmP. Traces include LMP1 (P. paxilli paxP deletion mutant), LMP1 complemented with pCP5 (atmP complementation construct), wild-type (WT) P. paxilli, and indole-diterpene standards.
FIG. 7.
HPLC analysis of indole-diterpenes from a P. paxilli paxQ mutant containing atmQ. Traces include LMQ226 (P. paxilli paxQ deletion mutant), LMQ226 with pCQ16 (atmQ construct), and indole-diterpene standards.
DISCUSSION
We have identified two loci in A. flavus and A. oryzae that are involved in indole-diterpene biosynthesis. The reconstitution in P. paxilli of biosynthesis steps from paspaline to paspalinine, a proposed intermediate for aflatrem biosynthesis, provides very strong evidence that these two loci encode gene products necessary for the biosynthesis of aflatrem. The genome of A. oryzae is rich in secondary metabolite gene clusters (21), but many of these genes appear not to be expressed under normal growth conditions (1), and we did not detect expression of A. oryzae ATM1 genes (atmG, atmC, and atmM) under growth conditions that induce aflatrem biosynthesis in A. flavus (not shown). Furthermore, the identification in A. oryzae of a frameshift mutation in atmQ suggests that the ability to synthesize aflatrem has been permanently lost by this fungus. Among the apparent ATM locus gene remnants in the genomes of other aspergilli were apparent functional homologs of paxG, paxC, paxB, and paxM in A. niger (Fig. 2). Previously we showed that these four genes are sufficient to mediate paspaline biosynthesis in P. paxilli (29). Whether A. niger synthesizes paspaline or any other indole-diterpenes remains to be demonstrated.
With the advent of whole-genome sequencing, the mechanisms underpinning the evolution and regulation of secondary metabolite gene clusters in filamentous fungi are beginning to be understood. In most cases, the genes for a secondary metabolite pathway are clustered at a single genomic locus (13), and such clusters are enriched in telomere-proximal regions of fungal genomes (8, 13, 14, 23). In both A. flavus and A. oryzae, the atm genes are clustered at two chromosomal loci. The ATM1 locus, containing atmG, atmC, and atmM, is telomere proximal on chromosome 5, while the ATM2 locus, containing atmD, atmQ, atmB, atmA, and atmP, is telomere distal on chromosome 7. Of the two other indole-diterpene gene clusters identified to date, the pax cluster in P. paxilli and the ltm cluster in N. lolii, both are located at a single genomic locus, although the ltm cluster is disrupted by long stretches of AT-rich retrotransposon relic sequences (42, 43). The genes for dothistromin biosynthesis in Dothistroma septosporum are also separated into several miniclusters, although all are located on a 1.3-Mb minichromosome (45). The only other known fungal secondary metabolite biosynthesis pathways for which the genes are located at discrete genomic regions are those for T-toxin biosynthesis in Cochliobolus heterostrophus and for trichothecene biosynthesis in Fusarium spp. In the heterothallic fungus C. heterostrophus, the genes for T-toxin biosynthesis are located on two chromosomes. These genes are inherited together as their respective chromosomes contain reciprocal translocations and form a four-arm linkage group with the T-toxin genes at the translocation breakpoint (18). In Fusarium spp. the genes for trichothecene biosynthesis are split between three coregulated loci including a core cluster of 10 or 11 genes (3), a two-gene minicluster (4), and a third locus with a single trichothecene gene (Tri101) (15). Genes in the outlying miniclusters appear to have been recruited for trichothecene biosynthesis more recently than those in the core cluster, and Tri101 appears to have evolved separately, suggesting that these genes have never been located at a single locus (4, 16). The physical arrangement of atm genes in A. flavus and A. oryzae is likely to have arisen by fragmentation of a single ancestral cluster. In support of this hypothesis, identification of an atmB relic at ATM1 suggests that duplication of atmB preceded or accompanied cluster fragmentation, thus giving rise to a copy of atmB at each ATM locus. Furthermore, the physical position of ATM2 in an otherwise syntenic genomic region (Fig. 2B) suggests a relatively recent insertion event that took place after A. flavus and A. oryzae separated from the other aspergilli.
Enrichment of secondary metabolite gene clusters in telomere-proximal regions of fungal genomes (8, 14, 23) suggests that the rapid structural evolution seen in these regions (8, 14) is a major facilitator of the birth, development, and demise of gene clusters and may thus account for discontinuous phylogenetic distribution of secondary metabolism biosynthesis capability in monophyletic fungal lineages (2). Identification of ATM loci remnants in the genomes of other aspergilli (Fig. 2A) suggests that indole-diterpene biosynthesis ability is an ancestral trait. Subtelomeric genomic reorganization was particularly evident in the sequences surrounding ATM1 where the telomere-distal (atmG) flank displayed a high degree of syntenic conservation with other aspergillus genomes, while there was no detectable synteny for genes on the telomere-proximal (atmM) flank (Fig. 2A). In Saccharomyces cerevisiae genomic comparisons demonstrated that boundaries of subtelomeres (which ranged from ∼7 to ∼52 kb) were defined by differences in number, order, and orientation of genes in telomeric regions (14). The ATM1 locus was ∼86 and ∼102 kb from the chromosome ends and appears to define the subtelomeric boundaries in A. oryzae and A. flavus, respectively.
By analogy with the pathway for paxilline biosynthesis in P. paxilli (22, 28, 29, 41), the identification of biosynthesis intermediates by LC-MS, and genetic complementation experiments involving atmM, atmP, and atmQ, a proposed pathway for aflatrem biosynthesis in A. flavus is presented in Fig. 8. Chemical and genetic evidence suggests that the biosynthesis of paspaline in A. flavus proceeds via the same pathway as in P. paxilli involving AtmG, AtmC, AtmM, and AtmB as orthologs of PaxG, PaxC, PaxM, and PaxB, respectively (29). The precise functions of these genes in paspaline biosynthesis are not yet known. However, it seems likely that synthesis of GGPP by AtmG (a GGPP synthase) precedes condensation of GGPP with indole 3-glycerol phosphate (5), followed by epoxidation and cyclization by AtmM (a FAD-dependent monooxygenase) and AtmC (a prenyltransferase) to produce paspaline. We also propose that AtmB is essential for the biosynthesis of paspaline in A. flavus, although as for the role of PaxB in paspaline biosynthesis by P. paxilli, its biochemical function is unknown (29). We have demonstrated that AtmP, a cytochrome P450 monooxygenase, is a functional ortholog of PaxP in P. paxilli and converts paspaline to 13-desoxypaxilline via PC-M6 by removal of the C-30 methyl group and oxidation at C-10. In contrast, AtmQ is not a functional ortholog of PaxQ. In P. paxilli, PaxQ catalyzes the conversion of 13-desoxypaxilline to paxilline via oxidation at C-13. In contrast, the reconstitution experiments carried out in this study demonstrate that AtmQ is a multifunctional cytochrome P450 monooxygenase that catalyzes the oxidation of 13-desoxypaxilline, first at C-7 to produce paspalicine and then at C-13 to form paspalinine, the latter step being equivalent to the oxidation of 13-desoxypaxilline by PaxQ. PaxQ and AtmQ therefore uniquely define the indole-diterpene biosynthesis capability of P. paxilli and A. flavus, respectively. The C-7 oxidation step may represent either a gain of function in A. flavus (AtmQ) or a loss of function in P. paxilli (PaxQ) compared with its ancestral state. The proposed scheme implicates both AtmP and AtmQ in multiple oxidations at different carbon atoms similar to the multiple steps catalyzed by PaxP in P. paxilli (28). Multifunctional cytochrome P450 enzymes that perform sequential oxidations have also been identified in other fungal secondary metabolism pathways including gibberellin biosynthesis in Fusarium fujikuroi (27, 34) and trichothecene biosynthesis in Fusarium spp. (33). For the latter, an enzyme from Trichothecium roseum is able to perform only three of four oxidations performed by its Fusarium homolog, thus representing a loss or gain of function similar to what we are proposing for PaxQ/AtmQ.
FIG. 8.
Proposed biosynthesis scheme for aflatrem biosynthesis in A. flavus. The scheme shows the proposed steps catalyzed by the atm gene products including the A. flavus-specific major and minor steps catalyzed by AtmQ using 13-desoxypaxilline and β-PC-M6 as the substrate, respectively.
Finally, we propose that AtmD prenylates paspalinine to form aflatrem. AtmD aligned with a newly described group of fungal aromatic prenyltransferases that catalyze prenyl transfer onto indole or an indole moiety (39) and include PaxD, which prenylates the indole group of paxilline in P. paxilli (Scott et al., unpublished), and a dimethylallyl tryptophan synthase from Neotyphodium sp. (37). Unlike trans-prenyltransferases, these proteins typically do not contain a DDXXD substrate-binding motif and their function is not dependent on divalent metal ions. The structure of aflatrem suggests that AtmD functions in a “reverse” manner, i.e., DMAPP is linked via its C-3 as opposed to C-1 or C-2 for “regular” prenyl transfer (35). The identification of three aflatrem isomers which differ in the positions and/or arrangements of the prenyl groups on the indole moiety suggests catalytic promiscuity for AtmD as reported for an aromatic prenyltransferase required for naphterpin biosynthesis in Streptomyces sp., where prenyl transfer occurs preferentially at one of two possible sites on the substrate molecule (19).
Detection of paspalininol (a paxitriol version of paspalinine) suggests a possible side reaction involving the oxidation of PC-M6 at C-13 and C-7 by AtmQ to produce this compound, most likely with paspalicinol as an intermediate, but this paxitriol version of paspalicine was not detected (Fig. 8). C-13 oxidation of PC-M6 has been demonstrated for PaxQ in P. paxilli; however, PaxQ could accept only α- and not β-PC-M6 as a substrate (28). Similar experiments could be carried out by feeding these isomers to the P. paxilli pax gene cluster deletion mutant, containing an ectopically integrated copy of atmP, to determine the substrate stereospecificity of AtmP.
The coordinate increase in the steady-state level of mRNA for seven of the eight atm genes with the onset of aflatrem biosynthesis suggests that the atm genes at both ATM loci are controlled by the same positive regulator, despite their lack of physical proximity. However, the low-level expression of telomere-distal (ATM2) genes at earlier time points when telomere-proximal (ATM1) gene transcripts were not detected suggests that locus-specific regulation may also be important. Transcriptional regulation of secondary metabolite biosynthesis genes in filamentous fungi is mediated by a complex interplay of several regulatory mechanisms. In some cases, such as for aflatoxin and trichothecene biosynthesis, a pathway-specific regulator gene is embedded in the cluster (16, 26). However, no such regulator was identified at either of the ATM gene loci or in the PAX and LTM gene loci (41, 43). AF109, a gene present 3′ of atmG at ATM1, encodes a putative GAL4-type transcription factor, but besides physical proximity to atm genes, there is no evidence to suggest that this gene has any role in aflatrem biosynthesis. Furthermore, this gene forms part of a highly conserved gene triplet also identified in A. niger, suggesting that these three genes are involved in an unrelated process. Differential regulation of the two ATM loci prior to the onset of aflatrem biosynthesis at 108 h may be mediated by a histone deacetylase such as HdaA, which negatively regulates telomere-proximal but not telomere-distal secondary metabolite gene clusters in A. nidulans (30).
The biosynthesis of aflatrem by A. flavus is positively regulated by VeA, which also regulates the production of other secondary metabolites and the formation of sclerotia (6). Deletion of veA results in loss of aflatrem biosynthesis via disruption of transcription of atmG, atmC, and atmM at ATM1 (6). However, the effect of this deletion on expression of genes at ATM2 has not yet been tested. The global regulator LaeA has also been shown to regulate the biosynthesis of aflatrem and several other secondary metabolites in A. flavus, with overexpression of this protein resulting in the production of aflatrem under conditions that do not induce aflatrem biosynthesis in wild-type cultures (12).
Results of this study suggest that all of the structural genes necessary for biosynthesis of aflatrem in A. flavus are clustered at two separate loci. We have demonstrated here roles for both atmP and atmQ in indole-diterpene biosynthesis. Previously we demonstrated that atmM is a functional ortholog of paxM (44). Taken together, these experiments demonstrate that Atm gene products are involved in the synthesis of paspaline and the multistep catalytic steps required for the conversion of paspaline to paspalinine. A comparison of this reconstituted biosynthesis pathway with what is known for indole-diterpene biosynthesis in P. paxilli suggests that structural differences between pax and atm biosynthesis gene products are due to differences in the catalytic abilities of homologous enzymes, and not the action of novel pathway enzymes in one or another species. The organization of atm genes at two discrete loci, one telomere proximal and the other telomere distal, offers insight into the mechanisms underpinning the evolution of secondary metabolite gene clusters and provides a useful experimental model for investigating the positional and general effects of secondary metabolite gene cluster regulation.
Acknowledgments
We thank Sanjay Saikia for technical advice and Chris Miles, Geoff Lane, and Brian Tapper from AgResearch for discussions on the chemistry of indole-diterpene biosynthesis. We also thank Geoff Lane and Karl Fraser for MS analysis of some samples.
This work was funded by the Massey University Research Fund.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Akao, T., M. Sano, O. Yamada, T. Akeno, K. Fujii, K. Goto, S. Ohashi-Kunihiro, K. Takase, M. Yasukawa-Watanabe, K. Yamaguchi, Y. Kurihara, J. Maruyama, P. R. Juvvadi, A. Tanaka, Y. Hata, Y. Koyama, S. Yamaguchi, N. Kitamoto, K. Gomi, K. Abe, M. Takeuchi, T. Kobayashi, H. Horiuchi, K. Kitamoto, Y. Kashiwagi, M. Machida, and O. Akita. 2007. Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res. 14:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berbee, M. L. 2001. The phylogeny of plant and animal pathogens in the Ascomycota. Physiol. Mol. Plant Pathol. 59:165-187. [Google Scholar]
- 3.Brown, D. W., R. B. Dyer, S. F. McCormick, D. F. Kendra, and R. D. Plattner. 2004. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 41:454-462. [DOI] [PubMed] [Google Scholar]
- 4.Brown, D. W., R. H. Proctor, R. B. Dyer, and R. D. Plattner. 2003. Characterization of a Fusarium 2-gene cluster involved in trichothecene C-8 modification. J. Agric. Food Chem. 51:7936-7944. [DOI] [PubMed] [Google Scholar]
- 5.Byrne, K. M., S. K. Smith, and J. G. Ondeyka. 2002. Biosynthesis of nodulisporic acid A: precursor studies. J. Am. Chem. Soc. 124:7055-7060. [DOI] [PubMed] [Google Scholar]
- 6.Duran, R. M., J. W. Cary, and A. M. Calvo. 2007. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73:1158-1168. [DOI] [PubMed] [Google Scholar]
- 7.Fueki, S., T. Tokiwano, H. Toshima, and H. Oikawa. 2004. Biosynthesis of indole diterpenes, emindole, and paxilline: involvement of a common intermediate. Org. Lett. 6:2697-2700. [DOI] [PubMed] [Google Scholar]
- 8.Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman, S. Batzoglou, S. I. Lee, M. Basturkmen, C. C. Spevak, J. Clutterbuck, V. Kapitonov, J. Jurka, C. Scazzocchio, M. Farman, J. Butler, S. Purcell, S. Harris, G. H. Braus, O. Draht, S. Busch, C. D'Enfert, C. Bouchier, G. H. Goldman, D. Bell-Pedersen, S. Griffiths-Jones, J. H. Doonan, J. Yu, K. Vienken, A. Pain, M. Freitag, E. U. Selker, D. B. Archer, M. A. Penalva, B. R. Oakley, M. Momany, T. Tanaka, T. Kumagai, K. Asai, M. Machida, W. C. Nierman, D. W. Denning, M. Caddick, M. Hynes, M. Paoletti, R. Fischer, B. Miller, P. Dyer, M. S. Sachs, S. A. Osmani, and B. W. Birren. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105-1115. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher, R. T., J. Clardy, and B. J. Wilson. 1980. Aflatrem, a tremorgenic toxin from Aspergillus flavus. Tetrahedron Lett. 21:239-242. [Google Scholar]
- 10.Gallagher, R. T., and B. J. Wilson. 1978. Aflatrem, a tremorgenic toxin from Aspergillus flavus. Mycopathology 66:183-185. [DOI] [PubMed] [Google Scholar]
- 11.Graham-Lorence, S. E., and J. A. Peterson. 1996. Structural alignments of P450s and extrapolations to the unknown. Methods Enzymol. 272:315-326. [DOI] [PubMed] [Google Scholar]
- 12.Kale, S. P., L. Milde, M. K. Trapp, J. C. Frisvad, N. P. Keller, and J. W. Bok. 2008. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 45:1422-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 14.Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E. S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241-254. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, M., I. Kaneko, M. Komiyama, A. Takatsuki, H. Koshino, K. Yoneyama, and I. Yamaguchi. 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. J. Biol. Chem. 273:1654-1661. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, M., T. Tokai, N. Takahashi-Ando, S. Ohsato, and M. Fujimura. 2007. Molecular and genetic studies of fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci. Biotechnol. Biochem. 71:2105-2123. [DOI] [PubMed] [Google Scholar]
- 17.Knaus, H.-G., O. B. McManus, S. H. Lee, W. A. Schmalhofer, M. Garcia-Calvo, L. M. H. Helms, M. Sanchez, K. Giangiacomo, J. P. Reuben, A. B. Smith, G. J. Kaczorowski, and M. L. Garcia. 1994. Tremorgenic indole alkaloids potently inhibit smooth muscle high-conductance calcium-activated channels. Biochemistry 33:5819-5828. [DOI] [PubMed] [Google Scholar]
- 18.Kodama, M., M. S. Rose, G. Yang, S. H. Yun, O. C. Yoder, and B. G. Turgeon. 1999. The translocation-associated Tox1 locus of Cochliobolus heterostrophus is two genetic elements on two different chromosomes. Genetics 151:585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuzuyama, T., J. P. Noel, and S. B. Richard. 2005. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435:983-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang, P.-H., T.-P. Ko, and A. H. J. Wang. 2002. Structure, mechanism and function of prenyltransferases. Eur. J. Biochem. 269:3339-3354. [DOI] [PubMed] [Google Scholar]
- 21.Machida, M., K. Asai, M. Sano, T. Tanaka, T. Kumagai, G. Terai, K. Kusumoto, T. Arima, O. Akita, Y. Kashiwagi, K. Abe, K. Gomi, H. Horiuchi, K. Kitamoto, T. Kobayashi, M. Takeuchi, D. W. Denning, J. E. Galagan, W. C. Nierman, J. Yu, D. B. Archer, J. W. Bennett, D. Bhatnagar, T. E. Cleveland, N. D. Fedorova, O. Gotoh, H. Horikawa, A. Hosoyama, M. Ichinomiya, R. Igarashi, K. Iwashita, P. R. Juvvadi, M. Kato, Y. Kato, T. Kin, A. Kokubun, H. Maeda, N. Maeyama, J. Maruyama, H. Nagasaki, T. Nakajima, K. Oda, K. Okada, I. Paulsen, K. Sakamoto, T. Sawano, M. Takahashi, K. Takase, Y. Terabayashi, J. R. Wortman, O. Yamada, Y. Yamagata, H. Anazawa, Y. Hata, Y. Koide, T. Komori, Y. Koyama, T. Minetoki, S. Suharnan, A. Tanaka, K. Isono, S. Kuhara, N. Ogasawara, and H. Kikuchi. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157-1161. [DOI] [PubMed] [Google Scholar]
- 22.McMillan, L. K., R. L. Carr, C. A. Young, J. W. Astin, R. G. T. Lowe, E. J. Parker, G. B. Jameson, S. C. Finch, C. O. Miles, O. B. McManus, W. A. Schmalhofer, M. L. Garcia, G. J. Kaczorowski, M. Goetz, J. S. Tkacz, and B. Scott. 2003. Molecular analysis of two cytochrome P450 monooxygenase genes required for paxilline biosynthesis in Penicillium paxilli, and effects of paxilline intermediates on mammalian maxi-K ion channels. Mol. Gen. Genomics 270:9-23. [DOI] [PubMed] [Google Scholar]
- 23.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. García, M. J. García, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jiménez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafton, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Peñalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodríguez de Córdoba, J. M. Rodríguez-Peña, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 24.Parker, E. J., and D. B. Scott. 2004. Indole-diterpene biosynthesis in ascomycetous fungi, p. 405-426. In Z. An (ed.), Handbook of industrial mycology, vol. 14. Marcel Dekker, New York, NY. [Google Scholar]
- 25.Payne, G. A., W. C. Nierman, J. R. Wortman, B. L. Pritchard, D. Brown, R. A. Dean, D. Bhatnagar, T. E. Cleveland, M. Machida, and J. Yu. 2006. Whole genome comparison of Aspergillus flavus and A. oryzae. Med. Mycol. 44(Suppl. 1):9-11. [DOI] [PubMed] [Google Scholar]
- 26.Price, M. S., J. Yu, W. C. Nierman, H. S. Kim, B. Pritchard, C. A. Jacobus, D. Bhatnagar, T. E. Cleveland, and G. A. Payne. 2006. The aflatoxin pathway regulator AflR induces gene transcription inside and outside of the aflatoxin biosynthetic cluster. FEMS Microbiol. Lett. 255:275-279. [DOI] [PubMed] [Google Scholar]
- 27.Rojas, M. C., P. Hedden, P. Gaskin, and B. Tudzynski. 2001. The P450-1 gene of Gibberella fujikuroi encodes a multifunctional enzyme in gibberellin biosynthesis. Proc. Natl. Acad. Sci. USA 98:5838-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saikia, S., E. J. Parker, A. Koulman, and B. Scott. 2007. Defining paxilline biosynthesis in Penicillium paxilli: functional characterization of two cytochrome P450 monooxygenases. J. Biol. Chem. 282:16829-16837. [DOI] [PubMed] [Google Scholar]
- 29.Saikia, S., E. J. Parker, A. Koulman, and B. Scott. 2006. Four gene products are required for the fungal synthesis of the indole diterpene paspaline. FEBS Lett. 580:1625-1630. [DOI] [PubMed] [Google Scholar]
- 30.Shwab, E. K., J. W. Bok, M. Tribus, J. Galehr, S. Graessle, and N. P. Keller. 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6:1656-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. Leger, R. J., S. E. Screen, and B. Shams-Pirzadeh. 2000. Lack of host specialization in Aspergillus flavus. Appl. Environ. Microbiol. 66:320-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.TePaske, M. R., J. B. Gloer, D. T. Wicklow, and P. F. Dowd. 1992. Aflavarin and β-aflatrem: new anti-insectan metabolites from the sclerotia of Aspergillus flavus. J. Nat. Prod. 55:1080-1086. [Google Scholar]
- 33.Tokai, T., H. Koshino, N. Takahashi-Ando, M. Sato, M. Fujimura, and M. Kimura. 2007. Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem. Biophys. Res. Commun. 353:412-417. [DOI] [PubMed] [Google Scholar]
- 34.Tudzynski, B., P. Hedden, E. Carrera, and P. Gaskin. 2001. The P450-4 gene of Gibberella fujikuroi encodes ent-kaurene oxidase in the gibberellin biosynthesis pathway. Appl. Environ. Microbiol. 67:3514-3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unsöld, I. A., and S. M. Li. 2006. Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus: gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. Chembiochem 7:158-164. [DOI] [PubMed] [Google Scholar]
- 36.Valdes, J. J., J. E. Cameron, and R. J. Cole. 1985. Aflatrem: a tremorgenic mycotoxin with acute neurotoxic effects. Environ. Health Perspect. 62:459-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, J., C. Machado, D. G. Panaccione, H.-F. Tsai, and C. L. Schardl. 2004. The determinant step in ergot alkaloid biosynthesis by an endophyte of perennial ryegrass. Fungal Genet. Biol. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 38.Yao, Y., A. B. Peter, R. Baur, and E. Sigel. 1989. The tremorgen aflatrem is a positive allosteric modulator of the gamma-aminobutyric acid receptor channel in Xenopus oocytes. Mol. Pharmacol. 35:319-323. [PubMed] [Google Scholar]
- 39.Yin, W. B., H. L. Ruan, L. Westrich, A. Grundmann, and S. M. Li. 2007. CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: overproduction, purification and biochemical characterisation. Chembiochem 8:1154-1161. [DOI] [PubMed] [Google Scholar]
- 40.Yoder, O. C. 1988. Cochliobolus heterostrophus, cause of southern corn leaf blight. Adv. Plant Pathol. 6:93-112. [Google Scholar]
- 41.Young, C., L. McMillan, E. Telfer, and B. Scott. 2001. Molecular cloning and genetic analysis of an indole-diterpene gene cluster from Penicillium paxilli. Mol. Microbiol. 39:754-764. [DOI] [PubMed] [Google Scholar]
- 42.Young, C. A., M. K. Bryant, M. J. Christensen, B. A. Tapper, G. T. Bryan, and B. Scott. 2005. Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Gen. Genomics 274:13-29. [DOI] [PubMed] [Google Scholar]
- 43.Young, C. A., S. Felitti, K. Shields, G. Spangenberg, R. D. Johnson, G. T. Bryan, S. Saikia, and B. Scott. 2006. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet. Biol. 43:679-693. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, S., B. J. Monahan, J. S. Tkacz, and B. Scott. 2004. An indole-diterpene gene cluster from Aspergillus flavus. Appl. Environ. Microbiol. 70:6875-6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, S., A. Schwelm, H. Jin, L. J. Collins, and R. E. Bradshaw. 2007. A fragmented aflatoxin-like gene cluster in the forest pathogen Dothistroma septosporum. Fungal Genet. Biol. 44:1342-1354. [DOI] [PubMed] [Google Scholar]