Abstract
Diversity and abundance of ammonia-oxidizing Betaproteobacteria (β-AOB) and archaea (AOA) were investigated in a New England salt marsh at sites dominated by short or tall Spartina alterniflora (SAS and SAT sites, respectively) or Spartina patens (SP site). AOA amoA gene richness was higher than β-AOB amoA richness at SAT and SP, but AOA and β-AOB richness were similar at SAS. β-AOB amoA clone libraries were composed exclusively of Nitrosospira-like amoA genes. AOA amoA genes at SAT and SP were equally distributed between the water column/sediment and soil/sediment clades, while AOA amoA sequences at SAS were primarily affiliated with the water column/sediment clade. At all three site types, AOA were always more abundant than β-AOB based on quantitative PCR of amoA genes. At some sites, we detected 109 AOA amoA gene copies g of sediment−1. Ratios of AOA to β-AOB varied over 2 orders of magnitude among sites and sampling dates. Nevertheless, abundances of AOA and β-AOB amoA genes were highly correlated. Abundance of 16S rRNA genes affiliated with Nitrosopumilus maritimus, Crenarchaeota group I.1b, and pSL12 were positively correlated with AOA amoA abundance, but ratios of amoA to 16S rRNA genes varied among sites. We also observed a significant effect of pH on AOA abundance and a significant salinity effect on both AOA and β-ΑΟΒ abundance. Our results expand the distribution of AOA to salt marshes, and the high numbers of AOA at some sites suggest that salt marsh sediments serve as an important habitat for AOA.
Nitrification, the sequential oxidation of ammonia to nitrite and nitrate, is a critical step in the nitrogen cycle and is mediated by a suite of phylogenetically and physiologically distinct microorganisms. The recent discovery of ammonia oxidation among Archaea (17, 38) has led to a dramatic shift in the current model of nitrification and to new questions of niche differentiation between putative ammonia-oxidizing Archaea (AOA) and the more-well-studied ammonia-oxidizing Betaproteobacteria (β-AOB). Based on surveys of 16S rRNA genes and archaeal amoA genes, it is evident that AOA occupy a wide range of niches (10), suggesting a physiologically diverse group of Archaea. Additionally, in studies where AOA and β-AOB were both targeted, AOA were typically more abundant than their bacterial counterparts (19, 21, 42). However, there are reports of β-AOB outnumbering AOA in estuarine systems (6, 33), suggesting a possible shift in competitive dominance under certain conditions.
Patterns of β-AOB diversity in estuaries have been well characterized and appear to be regulated by similar mechanisms within geographically disparate systems (4, 11, 32). However, AOA distribution and their role in nitrification relative to β-AOB remain to be determined. A few studies have begun to address this question in different estuaries, but no unifying patterns or mechanisms have emerged. Although β-AOB have been well studied along estuarine salinity gradients (1, 3, 4, 7, 11, 13, 22, 33, 39) and recent studies have begun to address AOA in estuaries (1, 6, 22, 32, 33), few have investigated β-AOB in salt marshes (9), and none has included AOA.
In this study, we investigated the distribution and abundance of AOA and β-AOB based on the distribution and abundance of amoA genes in salt marsh sediments dominated by different types of vegetation. Although we equate the presence of archaeal amoA genes with the genetic potential to oxidize ammonia, we acknowledge the possibility that all Archaea that have amoA genes may not all represent functional ammonia oxidizers. Vegetation patterns of New England salt marshes are strongly correlated with marsh elevation and are controlled by a combination of interspecific competition and tolerance to physico-chemical stress (28). The dominant grasses of New England salt marshes are Spartina alterniflora and Spartina patens, which typically grow as pure stands. S. alterniflora is found in two phenotypically distinct but genetically identical forms, a tall and a short growth form (34). The tall S. alterniflora grows to heights of 1 to 2 m and is typically found at the edges of the marsh and along creek banks (SAT sites), while the short-form S. alterniflora may reach heights of only 30 cm and is found in sites (SAS sites) slightly higher on the marsh where soil drainage is limited and conditions are more reduced compared to SAT sites (14). Conversely, S. patens, due to its lower tolerance of salt and more reduced conditions, is found in sites (SP sites) highest on the marsh, in areas that receive less flooding (5). Because the marsh is subjected to daily tidal fluctuations, most sites experience periods of anoxia, the degree of which depends on the marsh elevation. We hypothesized that ammonia-oxidizing communities in areas dominated by different marsh grasses would reflect the different edaphic conditions associated with each type of grass, due to differences in vertical zonation in the marsh.
MATERIALS AND METHODS
Study site and sample collection.
The research was carried out in the Wequetequock-Pawcatuck tidal marsh (locally referred to as Barn Island) of southeastern Connecticut from March to October 2006 (see references 40 and 41 for more complete site descriptions). DNA was extracted from 0 to 2 cm from replicate cores as previously described (23). Pore water salinity, pH, and ammonium levels have been reported elsewhere (23). Pore water nitrate (plus nitrite) was measured by enzymatic reduction of nitrate to nitrite (8), followed by colorimetric determination of nitrite for seawater (35).
Clone library construction.
Clone libraries were constructed from samples collected in March 2006. One clone library was constructed from each site for each gene. β-AOB amoA genes were amplified as described by Bernhard et al. (3). Archaeal amoA genes were amplified using previously published primers (12). Each 20-μl reaction mixture contained 10 μl iQ Supermix (Bio-Rad), a 0.5 μM concentration of each primer, and 1 μl of a 1:10 dilution of DNA (approximately 2 to 10 ng). Reactions were carried out using the following amplification cycle: 95°C for 5 min, followed by 35 cycles of 95°C for 20 s, 54°C for 20 s, and 72°C for 45 s, with a final elongation at 72°C for 5 min. All reactions were performed on an iQ iCycler (Bio-Rad). PCR products were cloned into the pSC vector using the StrataClone PCR cloning kit (Stratagene, Santa Clara, CA) according to the manufacturer's instructions. Transformants were randomly selected and inoculated into 100 μl LB broth with 100 μg ampicillin ml−1 in 96-well microtiter plates. All plates were incubated overnight at 37°C. Inserts were amplified from selected clones using the vector-specific primers T3 and T7. PCR products from clones containing the correctly sized insert were sequenced using the T3 primer. All sequencing was performed by High Throughput Sequencing Solutions (Seattle, WA).
Sequence analysis.
Sequences were compared to published sequences in GenBank using the Basic Local Alignment Search Tool (BLASTn) to identify related sequences and aligned using the sequence editor and Fast Align in ARB (20). All alignments were checked manually, and regions of ambiguous alignments were excluded from the analysis. All phylogenetic analyses were done with PAUP version 4.0 (36). Phylogenetic relationships were analyzed by using the neighbor-joining and parsimony algorithms. Parsimony analysis was performed using a full heuristic search with random addition sequence. Confidence in tree topology was assessed by 100 bootstrap replicates for both neighbor-joining and parsimony analyses. Sequences were checked for chimeras by comparing phylogenetic placement in trees constructed with the 5′ and the 3′ ends of the sequence. Pair-wise sequence comparisons were calculated in ARB, and operational taxonomic units (OTUs) were defined as sequences sharing ≥95% nucleotide sequence identity.
Real-time PCR of amoA genes.
Betaproteobacterial amoA genes were quantified as described by Bernhard et al. (4). Archaeal amoA genes were quantified using a modified version of CrenAmoAQ-F (21) and Arch-amoAR (12) (Table 1). The forward primer was modified to target new archaeal amoA genes recovered from estuarine and salt marsh samples that had two to three mismatches with the published CrenAmoAQ-F primer. Specificity of the archaeal amoA primers used in this study was confirmed by sequence analysis of clones generated with the same primers. All sequences were archaeal amoA genes (data not shown). PCR conditions were the same as above except we used 10 μl of iQ SYBR green I mix instead of iQ Supermix and we ran 50 cycles followed by melt curve analysis (95°C for 1 min, 55°C for 1 min, and then 0.5°C increase every 10 s, with fluorescence read continuously) to monitor product specificity. All samples were run in at least three separate experimental runs and compared to standard curves generated in each experimental run using five standards ranging in DNA concentration from 0.1 fg μl−1 to 1 pg μl−1, which is equivalent to 2.2 × 101 to 2.2 × 105 gene copies μl−1. Standards were purified plasmid DNAs from clones generated from archaeal amoA genes recovered previously from salt marsh sediments (A. Bernhard, unpublished data). Average PCR efficiencies for archaeal and bacterial amoA genes were 81 and 85%, respectively. We tested for inhibitory effects by running each sample at different dilutions (ranging from 1:5 to 1:15) and calculating the slope of the lines. Dilutions ranging from 1:8 to 1:12 gave similar slopes (coefficient of variation, 11.6%), so we used 1:10 dilutions of each sample for final analysis. Additionally, slopes were not significantly different among samples from different sites (P = 0.46). Data presented are the means of at least three separate analyses for each sample. Coefficients of variation among runs were 3.5 and 4.0% for β-AOB and AOA amoA genes, respectively.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Target | Reference |
|---|---|---|---|
| amoA-1F | GGGGTTTCTACTGGTGGT | β-AOB amoA | 31 |
| amoA-2R-TC | CCCCTCTGCAAAGCCTTCTTC | β-AOB amoA | 25 |
| ArchAmoAF | STAATGGTCTGGCTTAGACG | Archaeal amoA | 12 |
| ArchAmoAR | GCGGCCATCCATCTGTATGT | Archaeal amoA | 12 |
| CrenAmoAQModF | CARGTHGGNAARTTCTAYAAa | Archaeal amoA | This study |
| GAOB16S-F | GCGTGGGAATCTGGCCTCTA | γ-AOB 16S rRNA | This study |
| GAOB16S-R | CATCGCTGCTTGGCCACCT | γ-AOB 16S rRNA | This study |
| CGI.1b-270F | TGGATTGGACTGCGKCCGAT | CGI.1b 16S rRNA | 27 |
| CGI.1b-750R | GTCGAGCGCRTTCTGGMAAG | CGI.1b 16S rRNA | 27 |
| pSL12_750F | GGTCCRCCAGAACGCGC | pSL12 16S rRNA | 21 |
| pSL12_876R | GTACTCCCCAGGCGGCAA | pSL12 16S rRNA | 21 |
Bases in bold indicate modifications from those reported by Mincer et al. (21).
Real-time PCR of 16S rRNA genes.
16S rRNA genes affiliated with Crenarchaeota group I.1b were amplified using primers published by Park et al. (27) (Table 1) with the following cycle conditions: 95°C for 10 min, then 50 cycles of 95°C for 15 s, 64°C for 20 s, and 72°C for 30 s. Fluorescence was measured at 86°C to eliminate signals from nonspecific products with lower melting temperatures. Melt curve analysis was conducted to monitor product specificity (95°C for 1 min, 60°C for 1 min, and then 0.5°C increase every 10 s, with fluorescence read continuously). Specificity of primers was tested using DNA isolated from plasmids containing archaeal 16S rRNA inserts from a previous study (23) that represented Archaea from a variety of archaeal groups, including the CGI.1b and CGI.1a groups. The CGI.1b primers amplified all the sequences related to the CGI.1b group and no sequences that were not in this group. Archaeal 16S rRNA genes affiliated with pSL12 were amplified as reported by Mincer et al. (21). We also tested for gammaproteobacterial AOB 16SrRNA genes using primers in Table 1. Abundance of archaeal 16S rRNA genes related to Nitrosopumilus maritimus were reported previously (23).
Potential nitrification rates.
Potential nitrification rate experiments were set up within 4 to 6 h after samples were collected. Two-gram samples of sediment (wet weight) from the 0- to 2-cm horizon were transferred to 50-ml tubes containing 10 ml of artificial seawater (30 ppt) amended with 250 μM ammonium (as NH4Cl) and 60 μM phosphate (as KH2PO4). All samples were incubated at 15°C with shaking to keep oxygen conditions nonlimiting. One subsample from each replicate core was harvested at 0, 24, 48, and 72 h. Samples were centrifuged, filtered through GF/F filters (Whatman), and immediately frozen for nitrate analysis. Nitrate (plus nitrite) was measured as described above. Nitrification rates were calculated based on the change in nitrate (plus nitrite) concentration per gram of dry sediment over time.
Statistical analyses.
Multiple comparisons and correlations among quantitative variables were performed with Instat 3.0b (GraphPad Software, Inc.). Potential rate and amoA abundance data were log transformed to relieve heteroscedasticity. In cases where normality criteria were not met, data were analyzed by nonparametric tests.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in GenBank under accession numbers EU925166 to EU925374.
RESULTS AND DISCUSSION
β-AOB diversity.
We analyzed a total of 83 β-AOB amoA sequences to compare diversity in salt marsh sediments dominated by different grasses. Eighty-two of the 83 sequences were related to uncultured Nitrosospira-like amoA sequences recovered from other estuarine and marine environments (1, 3, 39) (Fig. 1). One sequence from SAS was related to Nitrosospira tenuis and Nitrospira briensis. β-AOB richness was low, with 3 to 4 OTUs detected at each site and only 5 OTUs detected overall (using a 5% cutoff at the nucleotide level) (Fig. 2A).
FIG. 1.
Phylogenetic relationships among deduced betaproteobacterial AmoA protein sequences. The unrooted neighbor-joining tree was inferred from an alignment of protein sequences with 134 amino acid residues. Bootstrap values (≥50) based on neighbor-joining and parsimony analyses are indicated above and below the nodes, respectively. Sequences with SAS, SAT, or SP as the prefix are from this study.
FIG. 2.
Rarefaction analyses of betaproteobacterial and archaeal amoA genes at the three sites. OTUs were defined as those with ≥95% nucleotide sequence identity.
The low β-AOB richness observed in the salt marsh is similar to β-AOB richness reported from other estuarine sediment environments (1, 3). Additionally, the dominance of Nitrosospira-like amoA sequences is consistent with previous studies of β-AOB amoA in other estuarine environments (3, 11, 13, 39). Unfortunately, there are still no cultured representatives of the dominant β-AOB found in most estuarine and marine systems, so their actual physiological tolerances remain speculative at best.
AOA diversity.
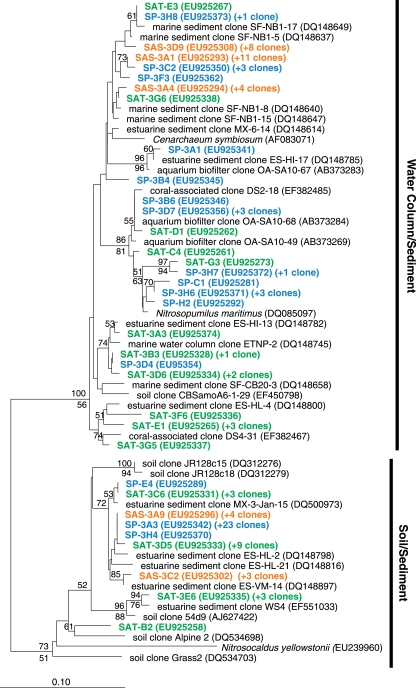
All but one of the AOA amoA sequences were related to sequences recovered from other marine or estuarine environments (1, 22, 33), with nine sequences most closely related to the amoA gene from the ammonia-oxidizing archaeon N. maritimus (17) (Fig. 3). One sequence, SAT-B2, was most closely related to a sequence recovered from soil. Topologies of trees constructed from alignments of deduced AmoA protein and amoA nucleic acid sequences were highly congruent (data not shown). Additionally, we detected a total of 20 AOA amoA OTUs, with 13 OTUs at SAT and 12 at SP but only 4 OTUs at SAS (Fig. 2).
FIG. 3.
Phylogenetic relationships among deduced archaeal AmoA protein sequences recovered from the three sites. The unrooted neighbor-joining tree was inferred from an alignment of protein sequences with 191 amino acid residues. Bootstrap support (≥50) based on neighbor-joining and parsimony analyses are indicated above and below the nodes, respectively.
We found approximately equal numbers of AOA amoA sequences affiliated with either the water column/sediment or soil/sediment clades at SAT and SP, but at SAS over 70% of the sequences fell within the water column/sediment clade. The recovery of AOA amoA genes within both the water column and soil clades is similar to results from other coastal marine or estuarine sites (1, 12, 22, 27). Additionally, some investigators have recovered AOA amoA sequences from soil environments that are affiliated with the water column/sediment clade (12, 37).
The richness of AOA amoA genes exceeded that of β-AOB amoA genes at two of the three sites (Fig. 2). Beman and Francis (1) found 42 AOA amoA OTUs in a subtropical estuary, but only 9 OTUs for β-AOB amoA genes, when using a 95% cutoff. Using the same 95% cutoff, Mosier and Francis (22) found 67 and 41 AOA and β-AOB OTUs respectively, in the San Francisco Bay. Although the numbers are considerably higher, the pattern is similar. In a subterranean estuary, only 2 β-AOB amoA OTUs were recovered, but 52 AOA amoA OTUs were found (33). We also found similar differences in richness of AOA and β-AOB amoA along a salinity gradient in Plum Island Sound (Bernhard, unpublished). These data suggest a consistent pattern of high AOA amoA diversity relative to β-AOB amoA diversity in estuarine systems.
AOA and β-AOB amoA abundance.
Abundance of β-AOB amoA genes ranged from 2.1 × 104 to 8.2 × 107 copies per g of dry sediment (or 3.6 × 103 to 2.6 ×107 copies per g of wet sediment) (Fig. 4). β-AOB amoA abundance was always lowest at SAS and was highest at SAT in April, June, and July. When data from all sampling dates were combined, β-AOB amoA abundance at SP and SAT was significantly greater than at SAS (Kruskal-Wallis, P < 0.0001).
FIG. 4.
Abundance of β-AOB and AOA amoA genes and 16S rRNA genes affiliated with N. maritimus, Crenarchaeota group I.1b, and pSL12 at the three study sites: SAS, SAT, and SP. Data are the means of triplicate core samples (except in April, when only duplicate cores were collected). Error bars represent the standard errors (in some cases the error bars are smaller than the symbols). pSL12 was not detected at SAT in October, at SP in June, July, or October, or in any samples at SAS. Additionally, N. maritimus and CGI.1b 16S rRNA genes were not detected at SAS in April.
β-AOB amoA abundance at our sites was similar to abundance measured by real-time PCR in other marine or estuarine environments (4, 22, 27, 30, 33) but about an order of magnitude higher than in a Georgia salt marsh (9). β-AOB amoA abundance in the Georgia salt marsh was measured by competitive PCR, which may not be as sensitive as real-time PCR. However, similar to our study, Dollhopf et al. (9) also found a significantly higher abundance of β-AOB amoA at sites dominated by the tall form compared to sites dominated by the short form of S. alterniflora.
AOA amoA gene abundance ranged from 1.6 × 106 to 5.8 × 109 copies per g of dry sediment (or 2.7 × 105 to 1.8 × 109 copies per g of wet sediment), was generally highest at the SP site and, similar to β-AOB, was always lowest at the SAS site (Fig. 4). As we found with β-AOB, AOA amoA abundance at SP and SAT sites was significantly greater than at the SAS site when all sampling dates were combined (Kruskal-Wallis, P < 0.0001). AOA amoA abundance at SAS site was similar to AOA abundance reported in other marine and estuarine sediments (22, 27, 33) using similar methods. However, AOA amoA abundance at SAT and SP was, on average, at least 10 times higher than levels reported in other studies, and at some sites about 100 times higher, based on comparisons of gene copies per g of wet sediment.
Our AOA amoA abundance data are corroborated by previous measurements of 16S rRNA genes related to the ammonia-oxidizing archaeon N. maritimus, for which Nelson et al. (23) found numbers as high as 109 copies per g of dry sediment at the same sites. Abundance of AOA amoA genes and N. maritimus-like 16S rRNA genes were highly correlated at SAT and SP (r = 0.88 and 0.91, respectively), but less so at SAS (r = 0.66).
Ratios of AOA amoA genes to N. maritimus-like 16S rRNA genes were similar at SAT (2.4 ± 0.3) and SP (2.9 ± 0.5) sites, but the average ratio at SAS was 41.7 ± 11.3. Ratios of amoA and 16S rRNA genes between 2 and 3 have been reported in other systems (2, 21). Although the genome of the only cultivated AOA, N. maritimus, has only one copy of both the 16S rRNA and amoA gene, some cultured β-AOB have as many as three copies of amoA (26). Additionally, the primers used by Nelson et al. (23) were designed to target all archaeal 16S rRNA genes related to N. maritimus recovered from the marsh, but it is likely that some 16S rRNA genes that represent Archaea with amoA genes were not targeted by these primers, which would skew our amoA/16S rRNA gene ratios.
The unexpectedly high ratios at the SAS site, however, suggest that other Archaea at this site harbor the amoA gene. To further investigate these ratios, we quantified members of the Archaea belonging to the CGI.1b (soil) group, some of which are known to have amoA (38), and the pSL12 group, which were previously implicated as potential amoA-harboring Archaea (21). No pSL12 16S rRNA genes were detected in any samples from SAS, but CGI.1b 16S rRNA genes were sometimes more abundant than N. maritimus-like genes (Fig. 4). When these two additional archaeal groups were included in the ratios, there was a slight decrease in ratios at SAT and SP sites, but the average ratio at the SAS site decreased about threefold (Table 2) due to the inclusion of CGI.1b genes. However, the ratios of amoA to 16S rRNA genes at SAS were still quite high compared to other reported values, suggesting that Archaea not targeted by the suite of 16S rRNA primers used in this study may contribute to the amoA abundance at SAS. The majority of archaeal 16S rRNA genes recovered from the SAS site belong to the group I.3b Crenarchaeota (23), for which amoA genes have not been reported.
TABLE 2.
Pearson correlation coefficients and ratios between AOA amoA and 16S rRNA gene abundance levels at the three sites
| Site |
r (P) |
Ratio |
||
|---|---|---|---|---|
| N. maritimus-like 16S rRNA only | All three rRNA genesa | N. maritimus- like 16S rRNA only | All three rRNA genesa | |
| SAS | 0.29 (0.33) | 0.73 (0.0012)b | 41.7 ± 11.3 | 11.9 ± 2.7b |
| SAT | 0.89 (<0.0001) | 0.95 (<0.0001) | 2.4 ± 0.3 | 1.9 ± 0.4 |
| SP | 0.75 (0.0006) | 0.92 (<0.0001) | 2.9 ± 0.5 | 2.6 ± 0.4 |
Data for the N. maritimus-like 16S rRNA, Crenarchaeota group I.1b 16S rRNA, and pSL12 16S rRNA genes.
No pSL12 16S rRNA genes were detected at SAS.
In a previous study of archaeal 16S rRNA gene diversity in the same salt marsh, Nelson et al. (23) recovered only one sequence affiliated with the group I.1b Crenarchaeota, but about half of our AOA amoA genes were affiliated with the soil/sediment cluster designated by Francis et al. (12). Although congruence between the phylogenies of archaeal 16S rRNA and amoA genes has been reported in other studies (24, 29, 32), we did not find this to be the case in our samples. The incongruencies we observed, however, may be due to primer biases or, more likely, differences in the AOA communities of the samples. Archaeal 16S rRNA genes reported in Nelson et al. (23) were generated from samples collected in October 2005, but amoA gene libraries reported in this study were generated from samples collected in March 2006 due to the high nitrification potentials compared to October. Additionally, the CGI.1b quantitative data indicate fairly high variability, so it is possible that the archaeal communities were quite different in October compared to March.
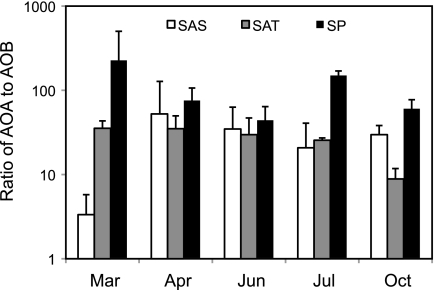
Similar to other recent studies, AOA amoA genes were significantly more abundant than β-AOB amoA genes at all three sites (Mann-Whitney, P < 0.0001), and the ratio of AOA to β-AOB amoA was highly variable (Fig. 5). At the SAS site, AOA amoA genes were only about 3 times more abundant than β-AOB in March, but on all other sampling dates at this site AOA amoA genes were 30 to 40 times more abundant. At SAT and SP, AOA amoA genes were approximately 9 to 215 times more abundant than β-AOB. When all sampling dates were combined, ratios of AOA to β-AOB amoA were significantly higher at SP compared to SAS or SAT (analysis of variance, P < 0.0001).
FIG. 5.
Ratios of AOA and β-AOB based on abundance of the amoA genes in the Barn Island salt marsh. Values represent the means of triplicate sediment cores (except in April, where n = 2), and error bars represent 1 standard deviation.
In other marine or estuarine sediments, ratios of AOA to β-AOB amoA gene abundance range from less than 1 to 80 (6, 22, 27, 33). In open ocean systems, AOA amoA genes are often 2 to 3 orders of magnitude greater than β-AOB levels (2, 21, 42). Conversely, β-AOB gene abundance was greater than AOA amoA genes in high-salinity areas of a subterranean estuary (33) and San Francisco Bay (22). Caffrey et al. (6) also reported that β-AOB amoA genes outnumbered AOA amoA genes at two highly sulfidic sites in an Alabama estuary. Such variable results in different systems suggest that the factors regulating marine and estuarine β-AOB and AOA amoA gene abundance may be quite complex. Identifying the factors that determine whether AOA or β-AOB dominate numerically in coastal systems should provide valuable insights into the physiological tolerances and ecological niches of the different nitrifying populations.
Abundances of AOA and β-AOB showed similar patterns and were significantly positively correlated (Table 3). However, the correlation of AOA and β-AOB appears to be driven by the strong relationships at SAT and SP sites, since the correlation at the SAS site was not significant when analyzed independently.
TABLE 3.
Pearson correlation coefficients between potential nitrification rates and AOA amoA and β-AOB amoA gene abundance levels for all data combined, by site and by sampling date
| Comparison and site or date |
r (P) fora: |
||
|---|---|---|---|
| Rates vs AOA | Rates vs β-AOB | AOA vs β-AOB | |
| All samples | 0.08 (0.71) | 0.06 (0.79) | 0.89 (<0.0001) |
| By site | |||
| SAS | −0.38 (0.24) | −0.27 (0.42) | 0.52 (0.10) |
| SAT | 0.60 (0.05) | 0.60 (0.05) | 0.95 (<0.0001) |
| SP | 0.12 (0.72) | 0.27 (0.43) | 0.93 (<0.0001) |
| By mo | |||
| March | 0.77 (0.01) | ||
| April | −0.38 (0.24) | −0.27 (0.42) | 0.52 (0.10) |
| June | 0.18 (0.63) | 0.14 (0.71) | 0.84 (0.005) |
| July | −0.51 (0.16) | −0.50 (0.17) | 0.72 (0.03) |
| October | 0.74 (0.02) | 0.79 (0.01) | 0.94 (0.0002) |
Values reported are correlation coefficients (r) followed by the P value in parentheses (correlations that were considered significant are indicated in bold) for the indicated comparison. No nitrification rates (“rates”) were measured in March.
It is clear from our results that the diversity and abundance of nitrifiers are consistently lower at SAS relative to SAT and SP sites. Differences in edaphic conditions among the three sites in this study likely contributed to the differences observed. In a previous study of a Georgia salt marsh, Dollhopf et al. (9) also found a lower abundance of β-AOB at SAS sites compared to SAT sites and attributed the differences to enhanced nitrification due to higher concentrations of Fe(III) and macrofauna burrowing activity at the SAT sites. Although we did not measure macrofauna activity or Fe(III) in our study, salt marsh vegetation patterns are highly predictable based on degree of tidal flooding and redox chemistry of the sediments (5, 14), so that the presence of dominant grasses can be used as proxies for prevailing sediment conditions. We think it is likely that the low abundance and diversity of nitrifiers at our SAS site may be a reflection of low redox or high sulfide conditions, which have been reported previously for this site (41). Joye and Hollibaugh (16) reported that sulfide may inhibit nitrification, and this might help explain the higher abundance and richness observed at SP relative to SAS. Unfortunately, redox and sulfide data for SAT are unavailable. However, since the SAT site is along a creek bank, it experiences greater tidal flushing, has a greater range of salinity, and has higher ammonium concentrations than the SP and SAS sites, which are higher on the marsh (Table 4). How these factors directly impact ammonia-oxidizing communities, however, is not clear. Additionally, the reason for the greater abundance of AOA and β-AOB amoA genes at the SP site also remains unclear but may be related to differences in plant root exudates between S. alterniflora and S. patens. Further research that is focused on the impact of plant roots is necessary to address these questions.
TABLE 4.
Ranges of pore water salinity, pH, and ammonium and nitrate (plus nitrite) concentrations for sediment samples collected at the three sites from March to October 2006a
| Site | Salinity (ppt) | pH | NH4+ (μM) | NO3− (μM) |
|---|---|---|---|---|
| SAS | 23.8-32.7 | 5.3-6.4 | 4.0-95.1 | 6.0-15.2 |
| SAT | 12.0-31.3 | 6.2-6.5 | 25.1-258.0 | 1.5-14.3 |
| SP | 24.0-30.0 | 5.3-6.3 | 12.5-111.0 | 4.6-13.8 |
Salinity, pH, and ammonium data are from Nelson et al. (23).
Nitrification potentials.
Potential nitrification rates showed a strong seasonal pattern at all three sites, with rates highest during April and decreasing to very low levels by October (Fig. 6). Average rates were highest overall at the SAT site (29.6 nmol NO3−/g [dry weight]/day) compared to 10.8 and 8.6 nmol NO3−/g (dry weight)/day at SAS and SP sites, respectively, but varied by sampling date. The seasonal patterns of nitrification potentials we report here are similar to those reported for other estuaries (4, 7), suggesting a common mechanism regulating nitrification rates in estuarine systems. One hypothesis for higher rates in April may be a reduced competition for ammonia (30), since it would still be early in the season for algal and plant growth. However, changes in other factors, such as salinity or oxygen, cannot be ruled out. Additionally, significantly lower ammonium concentrations were reported at all three sites in our study in October (23) and may have contributed to the extremely low rates we measured. However, potential nitrification rates and ammonium concentrations were not significantly correlated in this study.
FIG. 6.
Potential nitrification rates at the three sites over the growing season. Values represent the means of triplicate core samples (except in April, when n = 2), and error bars represent 1 standard deviation. No rates were measured in March.
Potential nitrification rates were significantly correlated with AOA and β-AOB abundance only at the SAT site (Table 3), suggesting that the resident AOA and β-AOB at this site are active ammonia oxidizers. When rates and nitrifier abundance were analyzed by sampling date, AOA and β-AOB were significantly positively correlated with rates only in October. In April and July, the correlations were actually negative (but not significant), and in July we did not detect any nitrification activity at the SP site despite the high numbers of AOA and β-AOB amoA genes. It is unclear to us why we were unable to detect potential rates at SP in July. Other studies have shown a significant decrease in nitrification potentials during late summer (4, 7). Although rates were still relatively high at SAS and SAT, conditions not conducive to nitrification at SP may have occurred already. We also tested for the presence of AOB belonging to the Gammaproteobacteria (γ-AOB), but we were unable to detect γ-AOB 16S rRNA genes in our samples (Bernhard, unpublished), so it is unlikely that potential rates could be attributed to γ-AOB activity, as was recently reported in a pelagic system (18).
Positive correlations between potential rates and β-AOB have been reported for marine sediment microcosms (30), salt marsh sediments (9), and estuarine sediments (4). Others, however, have reported no relationship between potential nitrification rates and β-AOB (6). Additionally, Caffrey et al. (6) reported positive correlations between AOA abundance and potential rates, but the relationship was significant at only two of six sites. Also, a recent study of nitrifiers in agricultural soils reported nitrification activity attributed to AOB and not AOA, despite high numbers of AOA amoA genes (15), suggesting that amoA gene abundance may not be an appropriate marker for nitrifying potential. It is important to consider that potential nitrification rates do not represent in situ rates and thus may not accurately reflect the nitrifying populations present. It may be that the conditions (such as oxygen, ammonium, or salinity) during the potential rate experiments may not be optimal for all resident nitrifiers. In future studies, it may be helpful to measure gene expression, in addition to abundance, to better quantify the active populations.
Site characteristics and correlation with nitrifying communities.
Similar to the results for N. maritimus 16S rRNA genes reported by Nelson et al. (23), the largest differences in AOA and β-AOB amoA gene abundance were found between samples at SAS and SP, yet there were few differences in pore water chemistry between these sites (Table 4). Differences in gene abundance at these sites are likely due to other environmental variables, such as redox or sulfide, which were previously reported to be significantly different at these sites (41). As has been reported in other estuarine environments (4, 22), salinity was significantly negatively correlated with AOA (r = −0.47, P = 0.02) and β-AOB (r = −0.52, P = 0.008) abundance. However, whether the effect on nitrifier abundance is due to a physiological response to salt or to some other factor that covaries with salinity has not been determined. The negative correlation between AOA abundance and pH (r = −0.46, P = 0.02) in our study also corroborates the results reported in soil samples (24). No significant correlations were detected between ammonium or nitrate concentrations and rates, AOA amoA abundance, or β-AOB amoA abundance.
Conclusions.
We report surprisingly high numbers of AOA amoA genes in some salt marsh sediments, suggesting a potentially important role of these Archaea in the ecology of the marsh. Additionally, AOA always outnumbered β-AOB at all sites and were considerably more diverse at two of the three sites. Our results also suggest that salinity and pH may be important environmental factors that regulate AOA abundance, as has been suggested by others (22, 24). Interestingly, differences in pore water nitrogen concentrations appeared to have little effect on nitrifier abundance in our study. Although the lack of consistent correlation of rates with nitrifier abundance may be a reflection of methodological limitations, it may also suggest that the relationship among AOA, β-AOB, and potential rates is highly complex and warrants further exploration. Measuring levels of amoA gene transcripts or AmoA protein levels in situ may help elucidate this relationship under different environmental conditions.
Acknowledgments
This work was supported in part by the National Science Foundation (MCB-0457183 and DEB-0814586 to A.E.B.), the George and Carol Milne Endowment at Connecticut College, the Long Island Sound License Plate Funds administered by the Connecticut Department of Environmental Protection, and the Keck Undergraduate Science Program through fellowships to K.A.N. and N.S.M.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Beman, J. M., and C. A. Francis. 2006. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahi'a del To'bari, Mexico. Appl. Environ. Microbiol. 72:7767-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beman, J. M., B. N. Popp, and C. A. Francis. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429-441. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard, A. E., T. Donn, A. E. Giblin, and D. A. Stahl. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289-1297. [DOI] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., J. Tucker, A. E. Giblin, and D. A. Stahl. 2007. Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ. Microbiol. 9:1439-1447. [DOI] [PubMed] [Google Scholar]
- 5.Bertness, M. D., and A. M. Ellison. 1987. Determinants of pattern in a New England salt marsh plant community. Ecol. Monogr. 57:129-147. [Google Scholar]
- 6.Caffrey, J. M., N. Bano, K. Kalanetra, and J. T. Hollibaugh. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660-662. [DOI] [PubMed] [Google Scholar]
- 7.Caffrey, J. M., N. Harrington, I. Solem, and B. B. Ward. 2003. Biogeochemical processes in a small California estuary. 2. Nitrification activity, community structure and role in nitrogen budgets. Mar. Ecol. Prog. Ser. 248:27-40. [Google Scholar]
- 8.Campbell, W. H., T. Kinnunen-Skidmore, M. J. Brodeur-Campbell, and E. R. Campbell. 2004. New and improved nitrate reductase for enzymatic nitrate analysis. Am. Lab. 22:12. [Google Scholar]
- 9.Dollhopf, S. L., J.-H. Hyun, A. C. Smith, H. J. Adams, S. O'Brien, and J. E. Kostka. 2005. Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl. Environ. Microbiol. 71:240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis, C. A., J. M. Beman, and M. M. M. Kuypers. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 1:19-27. [DOI] [PubMed] [Google Scholar]
- 11.Francis, C. A., G. D. O'Mullan, and B. B. Ward. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 12.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freitag, T. E., L. Chang, and J. I. Prosser. 2006. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ. Microbiol. 8:684-696. [DOI] [PubMed] [Google Scholar]
- 14.Howes, B. L., R. W. Howarth, J. M. Teal, and I. Valiela. 1981. Oxidation-reduction potentials in a salt marsh: spatial patterns and interactions with primary production. Limnol. Oceanogr. 26:350-360. [Google Scholar]
- 15.Jia, Z., and R. Conrad. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658-1671. [DOI] [PubMed] [Google Scholar]
- 16.Joye, S. B., and J. T. Hollibaugh. 1995. Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270:623-625. [Google Scholar]
- 17.Koenneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 18.Lam, P., M. M. Jensen, G. Lavik, D. F. McGinnis, B. Muller, C. J. Schubert, R. Amann, B. Thamdrup, and M. M. M. Kuypers. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. USA 104:7104-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leininger, S., T. Urich, M. Schloter, L. Schwark, J. Qi, G. W. Nicol, J. I. Prosser, S. C. Schuster, and C. Schleper. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806-809. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mincer, T. J., M. J. Church, L. T. Taylor, C. Preston, D. M. Karl, and E. F. DeLong. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific subtropical gyre. Environ. Microbiol. 9:1162-1175. [DOI] [PubMed] [Google Scholar]
- 22.Mosier, A. C., and C. A. Francis. 2008. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environ. Microbiol. 10:3002-3016. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, K. A., N. S. Moin, and A. E. Bernhard. 2009. Archaeal diversity and the prevalence of Crenarchaeota in salt marsh sediments. Appl. Environ. Microbiol. 75:4211-4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicol, G. W., S. Leininger, C. Schleper, and J. I. Prosser. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxiizing archaea and bacteria. Environ. Microbiol. 10:2966-2978. [DOI] [PubMed] [Google Scholar]
- 25.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approached to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 26.Norton, J. M., J. M. Low, and M. G. Klotz. 1996. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV. FEMS Microbiol. Lett. 139:181-188. [DOI] [PubMed] [Google Scholar]
- 27.Park, S. J., B. J. Park, and S. K. Rhee. 2008. Comparative analysis of archaeal 16S rRNA and amoA genes to estimate the abundance and diversity of ammonia-oxidizing archaea in marine sediments. Extremophiles 2008:1-11. [DOI] [PubMed] [Google Scholar]
- 28.Pennings, S. C., and M. D. Bertness. 2001. Salt marsh communities, p. 530. In M. D. Bertness, S. D. Gaines, and M. E. Hay (ed.), Marine community ecology. Sinauer Associates, Inc., Sunderland, MA.
- 29.Prosser, J. I., and G. W. Nicol. 2008. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10:2931-2941. [DOI] [PubMed] [Google Scholar]
- 30.Risgaard-Petersen, N., M. H. Nicolaisen, N. P. Revsbech, and B. A. Lomstein. 2004. Competition between ammonia-oxidizing bacteria and benthic microalgae. Appl. Environ. Microbiol. 70:5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sahan, E., and G. Muyzer. 2008. Diversity and spatio-temporal distribution of ammonia-oxidizing Archaea and Bacteria in sediments of the Westerschelde estuary. FEMS Microbiol. Ecol. 64:175-186. [DOI] [PubMed] [Google Scholar]
- 33.Santoro, A. E., C. A. Francis, N. R. De Sieyes, and A. B. Boehm. 2008. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10:1068-1079. [DOI] [PubMed] [Google Scholar]
- 34.Shea, M. L., R. S. Warren, et al. 1975. Biochemical and transplantation studies of growth form of Spartina alterniflora on Connecticut salt marshes. Ecology 56:461-466. [Google Scholar]
- 35.Strickland, J. D. H., and T. R. Parsons. 1972. A practical handbook of seawater analysis. Fisheries Research Board, Ottawa, Canada.
- 36.Swofford, D. L. 1991. PAUP: Phylogenetic Analysis Using Parsimony, version 4.0. Sinauer Associates, Inc., Sunderland, MA.
- 37.Tourna, M., T. E. Freitag, G. W. Nicol, and J. I. Prosser. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357-1364. [DOI] [PubMed] [Google Scholar]
- 38.Treusch, A. H., S. Leininger, A. Kietzin, S. C. Schuster, H. P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 39.Ward, B. B., D. Eveillard, J. D. Kirshtein, J. D. Nelson, M. A. Voytek, and G. A. Jackson. 2007. Ammonia-oxidizing bacterial community composition in estuarine and oceanic environments assessed using a functional gene microarray. Environ. Microbiol. 9:2522-2538. [DOI] [PubMed] [Google Scholar]
- 40.Warren, R. S., P. E. Fell, R. Rozsa, A. H. Brawley, A. C. Orsted, E. T. Olson, V. Swamy, and W. A. Niering. 2002. Salt marsh restoration in Connecticut: 20 years of science and management. Restor. Ecol. 10:497-513. [Google Scholar]
- 41.Warren, R. S., and W. A. Niering. 1993. Vegetation changes on a northeast tidal marsh: interaction of sea-level rise and accretion. Ecology 74:96-103. [Google Scholar]
- 42.Wuchter, C., B. Abbas, M. J. L. Coolen, L. Herfort, J. Van Bleijswijk, P. Timmers, M. Strous, E. Teira, G. J. Herndl, J. J. Middelburg, S. Schouten, and J. S. S. Damste. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. USA 103:12317-12322. [DOI] [PMC free article] [PubMed] [Google Scholar]