Abstract
Environmental conditions can change dramatically over a crop season and among locations in an agricultural field and can increase denitrification and emissions of the potent greenhouse gas nitrous oxide. In a previous study, changes in the overall size of the denitrifier community in a potato crop field were relatively small and did not correlate with variations in environmental conditions or denitrification rates. However, denitrifying bacteria are taxonomically diverse, and different members of the community may respond differently to environmental changes. The objective of this research was to understand which portion of the nirK denitrifying community is active and contributes to denitrification under conditions in a potato crop field. Denaturing gradient gel electrophoresis (DGGE) of nirK genes in soil-extracted DNA showed changes in the composition of the nirK denitrifier community over the growing season and among spatial locations in the field. By contrast, the composition of the active nirK denitrifier community, as determined by DGGE analysis of nirK transcripts derived from soil-extracted mRNA, changed very little over time, although differences in the relative abundance of some specific transcripts were observed between locations. Our results indicate that the soil denitrifier populations bearing nirK genes are not all contributing to denitrification and that the denitrifying populations that are active are among the most abundant and ubiquitous nirK-bearing denitrifiers. Changes in the community composition of the total and active nirK denitrifiers were not strongly correlated with changes in environmental factors and denitrification activity.
Conditions in an agricultural field can vary dramatically over a crop growing season due to natural phenomena, such as cool, wet springs and hot, dry summers, and agronomic practices, such as application of fertilizers. Environmental changes have the potential to influence the metabolic activity of soil microbes that can lead to undesirable effects, including the emission of nitrous oxide (N2O), an important greenhouse gas and ozone-depleting substance, through microbial denitrification. Denitrification is an alternative respiratory process in which nitrogen oxides such as nitrate (NO3−) and nitrite (NO2−) are used as electron acceptors and reduced to N2O and dinitrogen (N2) gases under oxygen-limited conditions (37). The capacity to use nitrogen oxides as final electron acceptors is widespread among bacteria (43). Denitrification not only yields a potent greenhouse gas but is also responsible for nitrogen loss from agricultural soils (3, 7, 15).
Denitrification in soil is influenced by numerous interacting physical, chemical, and biological factors (26, 37). Soil aeration, temperature, and carbon and nitrate availability fluctuate over time and are major parameters regulating denitrification in agricultural soils (15, 30, 34, 35). Conditions known to be conducive to denitrification activity include low soil aeration (influenced by soil structure, texture, organic matter, and moisture content) and high soil nitrate concentrations (influenced mainly by nitrification and nitrogen fertilization). In potato fields, variations in denitrification activity and N2O emissions associated with environmental changes have been observed over the course of the potato growing season and among spatial locations in the field (6, 9, 29, 30). For example, the potato furrow shows higher denitrification rates (N2 production) but lower levels of N2O emissions compared to the potato hill. High N2O emissions in the hill are due to the presence of high nitrate content (nitrogen fertilizer is applied in bands close to the seeds at planting time) and aerated soil conditions. By contrast, high soil moisture and compaction and low nitrate concentration in the furrow favor the completion of the denitrification reaction in this location (6, 9).
Although the effects of changes in environmental conditions on denitrification rates and denitrification enzyme activity (DEA [i.e., potential activity]) have been extensively studied (6, 29, 30, 34, 35), the roles of the abundance and composition of the denitrifer community on denitrification rates are poorly understood. Hallin et al. (13) demonstrated that differences in the abundance of nosZ denitrifiers (estimated by quantitative PCR) rather than the composition of a nosZ denitrifier community were correlated to differences in potential denitrification rates in fertilized and unfertilized soils. These results are in accordance with those from the study by Patra et al. (25), which indicated changes in the abundance of denitrifers (estimated by the most probable number technique) and potential denitrification rates in grassland soil under grazing but no changes in the composition of nitrate reducers. On the other hand, Ma et al. (19) showed that changes in the nosZ denitrifier community composition and abundance between cultivated wetlands and uncultivated wetlands were not related to differences in N2O emissions.
Beyond assessing the diversity of the denitrifying population, very few studies have assessed the diversity of denitrifying gene transcripts to determine which members of the denitrifying community actively contribute to denitrification (23, 32) or have compared the diversity of the community with that of the active component (5, 33). Bacteria that use denitrification as an alternative respiratory process belong to different taxonomic groups and under identical conditions may exhibit different growth rates and levels of denitrification activity. These differences may be due to differences in sensitivity to O2 (16), in the response to NO3− (38), and in the ability to use carbon substrates (24) or other available terminal electron acceptors. Denitrification is an opportunistic growth mechanism and not necessarily a requirement for growth in soil. Thus, all denitrifiers present in an environment may not contribute to denitrification, and when environmental conditions vary, the composition and relative abundance of active denitrifiers may change even when the overall abundance or composition of the total denitrifying community does not. To better explore the relationships between diversity and function in denitrifying communities, it is therefore important to assess the diversity of the denitrifiers that are actively expressing denitrifying genes (27, 40).
A previous experiment in an agricultural field cropped to potato showed that although environmental conditions (i.e., temperature, soil moisture, and nitrogen and carbon availability) and denitrification rates fluctuated greatly over the growing season and among spatial locations, changes in the overall size of the denitrifier community, estimated by quantitative PCR targeting broad groups of denitrifiers, were relatively small (1.7- to 2.7-fold) (9). In addition, changes in denitrifier abundance were not correlated to the observed changes in denitrification. The first objective of this study was therefore to determine if the composition of the denitrifer community changes over the growing season and among spatial locations in the potato field. We hypothesized that some members of the community may respond differently to environmental changes occurring over time and between different field locations. Alternatively, if the overall denitrifier community composition doesn't change, perhaps the active component does. Our second objective was therefore to assess changes in the composition of the active members of the denitrifier community over the potato growing season and in different field locations. Finally, we investigated the relationships between changes in the composition of the community and/or the active component of the community and changes in environmental parameters and denitrification rates. For instance, we predicted that the composition of the active nirK denitrifiers in the hill, where nitrate levels are high and the soil is aerated, may differ from the composition of active populations in the furrow, where the level of nitrate is lower and soil moisture and compaction are high.
Denaturing gradient gel electrophoresis (DGGE) was used to analyze the nirK (coding for Cu-nitrite reductase) genes and transcripts in soil samples collected from the agricultural field cropped to potato at different times over a growing season (from April to August) and at different locations (hill, close to plant roots, and furrow). These dates and soil locations were chosen because environmental parameters (such as soil moisture, temperature, and carbon and nitrate concentrations) and denitrification rates, DEA, and N2O emissions changed during that period and among these locations in this field (9).
MATERIALS AND METHODS
Field site and soil sampling.
Soil samples were collected during 2006 from the potato year of a potato-spring wheat rotation at the Potato Research Centre, Agriculture and Agri-Food Canada, Fredericton, New Brunswick, Canada (45°52′N, 66°31′W) (for soil characteristics, see reference 9). Composite soil samples (0- to 15-cm layer) were collected from four plots, where each plot consisted of six potato rows 0.91 m apart and 8 m in length. The plots were planted on 15 May with potato cultivar Russet Burbank, with 0.51-m within-row spacing to allow bulk soil, with limited influence from potato root systems, to be collected between plants in the potato hill. Fertilizer was banded at ∼7.5 cm to each side and 5 cm below the potato seed pieces at planting, according to normal production practices, at 200 kg N ha−1 as NH4NO3, 150 kg P2O5 ha−1, and 150 kg K2O ha−1. On 10 July surface soil was removed at the base of the plants to form a hill in order to promote potato tuber development. Plots were not irrigated, and diseases, insects, and weeds were controlled using standard agricultural practices (2).
Soil samples were analyzed from each of the four plots on five dates from April to August (i.e., 20 April, 25 May, 20 June, 14 July, and 10 August). These dates over the growing season were chosen because denitrification activity levels varied in the field (from no activity to the maximum activity level) during this period (9). Soil samples were collected separately from three spatial locations over the crop growing season (i.e., from June to August): the bulk soil between plants in the potato hill (H), the soil in the hill close to the plant roots (Hp), and the bulk soil in the furrow (F). Prior to planting (i.e., in April and May), samples were also collected from the bulk soil and were designated as “H” because at this stage the hill and bulk soil both consist of the same topsoil layer. For the H and F locations, in each plot, six cores were randomly sampled and then mixed to form one composite sample. For Hp samples taken during June and July, the soil on the roots of three to four potato plants was collected by shaking it into a sample bag. Very little soil stayed on the roots of the plants pulled up later in the season (10 August); therefore, a sample of loose soil was collected from the area immediately adjacent to the position from which the plant was uprooted, where root density was greatest.
On each sampling date, the composite samples were split into subsamples to perform different analyses. For nucleic acid extraction, each soil subsample was transferred to a sterile plastic tube (15 ml) and immediately flash frozen in liquid nitrogen in the field in a cryogenic vapor shipper. Frozen soil samples were later stored in the laboratory at −80°C for further processing. Soil temperature, water-filled pore space (WFPS), levels of extractable organic carbon (EOC), NO3−, and NH4+, CO2 and N2O fluxes, denitrification rates (by measuring N2O accumulation over a 24-h period from soil cores by the acetylene blockage method), DEA (activity measured under nonlimiting conditions of nitrate and carbon substrates), and the abundance of the denitrifying community were quantified for all the soil samples and have been described by Dandie et al. (9).
PCR and reverse transcription-PCR (RT-PCR) targeting nirK genes and DGGE analysis.
Soil samples were prepared for nucleic acid extraction by freeze drying overnight to complete dryness and were stored at −80°C. DNA was extracted from 0.5-g soil samples as described by Dandie et al. (8). RNA was extracted from 1.5-g soil samples using the RNA PowerSoil total RNA isolation kit (MO BIO Laboratories, Inc., Carlsbad, CA).
Following extraction, RNA was further purified by treatment with DNase I (Invitrogen, Burlington, Ontario, Canada) followed by a phenol-chloroform extraction. Briefly, RNA samples were incubated for 30 min at 37°C with 20 units of DNase I, a 0.11 volume of DNase buffer (10 mM CaCl2, 10 mM MgCl2 in Tris-HCl, pH 7.5), and sterile H2O to bring the volume to 200 μl. Then, 200 μl of phenol-chloroform-isoamyl alcohol (50:49:1) was added and samples were incubated for 5 min. After centrifugation at 10,000 × g for 5 min, the aqueous phase was transferred to a new tube and a 0.1 volume of 5 M NaCl and 2 volumes of 100% ethanol were added. Tubes were incubated at −20°C for 30 min followed by centrifugation at 10,000 × g for 10 min. The supernatant was removed and the RNA pellet air dried and resuspended in 50 μl sterile diethyl pyrocarbonate-treated H2O.
DNA and purified RNA samples were subjected to PCR and RT-PCR targeting nirK (coding for Cu-nitrite reductase) using primers Copper 583F and Copper 909R (18) followed by DGGE. Reverse transcription of RNA (using 100 to 200 ng of RNA) to cDNA was performed with SuperScript III reverse transcriptase (Invitrogen, Burlington, Ontario, Canada) according to the manufacturer's recommendations using 0.3 μM Copper 909R primer. PCR (using primers Copper 583F and Copper 909R primers) and DGGE analysis of PCR products were performed as described previously on soil samples tested by Wertz et al. (40), except that 0.3 μM concentrations of primers were used for the PCR step. Control PCRs without reverse transcription were performed to check for DNA contamination of RNA samples.
Cloning and sequencing of partial nirK gene sequences.
Partial nirK sequences (350 bp) corresponding to six DGGE band positions were characterized. Clone libraries were generated from four nirK PCR products and four nirK RT-PCR products obtained from templates representing different soil locations and sampling dates. PCR products were cloned into the pGEM T-Easy vector and transferred to competent Escherichia coli JM109 cells (Promega, Madison, WI). Isolated clones were subjected to PCR, and the PCR products were screened by DGGE (as described above). Clones were grouped on the basis of their band position on the DGGE gels. For each band position, a minimum of eight clones obtained from at least two nirK PCR products and two nirK RT-PCR products were selected for sequencing (except for band 16, for which only two clones from two nirK PCR products could be obtained). Sequencing was performed at the Centre d'Innovation Génome Québec et Université McGill (Montreal, QC, Canada). Sequences showing ≥99% similarity were not considered different due to methodological uncertainties linked to the PCR and cloning/sequencing procedure (1).
All clone sequences were compared to sequences available in the GenBank (http://www.ncbi.nlm.nih.gov/) and the Functional Gene Pipeline/Repository (http://fungene.cme.msu.edu/) databases. Afterwards, clone sequences were aligned with the closest related sequences using Clustal X 1.81 (EMBL, Heidelberg, Germany). Phylogenetic trees were constructed using the neighbor-joining method (31), and tree topology was evaluated by bootstrap analysis (500 replicates).
Statistical analyses.
DGGE banding profiles from all soil samples were analyzed as follows. A data matrix consisting of the position and relative intensity of each band for all the samples was constructed using Phoretix software (Phoretix International, Newcastle-Upon-Tyne, United Kingdom). For each sample, ratios of the intensity of each band versus the total band intensity were calculated. To assess the dissimilarity in the DGGE banding patterns between soil samples, rank similarity matrices were calculated from the data matrix using PRIMER-E Ltd. software (Plymouth, United Kingdom) and represented by multidimensional scaling (MDS) (17). Distortions between the computed dissimilarities among banding patterns (rank similarity matrices) and the MDS representations of dissimilarities were assessed by calculating a stress value. Stress values from 0 to 0.2 indicate excellent to moderately good representation of the DGGE profile similarities by the MDS. One-way analysis of similarity (ANOSIM) was then performed to test, for each spatial location (H, F, and Hp) separately, whether the effect of sampling date on the genetic structure of nirK genes and transcripts was significant. ANOSIM was also performed to test the effect of spatial location across sampling dates on the genetic structure of nirK genes and transcripts; in this case, the ANOSIM considered only the dates during the crop growth period (i.e., from June to August), when all three different spatial locations were sampled. ANOSIM results in the computation of P values indicating the level of significance and R statistic values indicating the degree of discrimination between groups (i.e., sampling dates or spatial locations) and ranging from 0 to 1. R is 0 if the similarities between and within replicates of groups are the same, and R is 1 if all the replicates of a group are more similar to each other than to any replicates of the other group. Dissimilarity percentages were also computed to compare the similarities within replicates of DGGE profiles of nirK genes and transcripts.
The total number of bands in the DGGE profiles was determined to compare the diversity in terms of richness between samples. Two-way analysis of variance (ANOVA) was performed to test, for each spatial location (H, F, and Hp), the effects of the following factors on richness: gene versus transcript, sampling date, and gene versus transcript sampling date. Two-way ANOVA was also performed to test, across the sampling dates from June to August, the effects of the following factors on richness: gene versus transcript, spatial location, and gene versus transcript spatial location. In each case, mean values (averages across samples from replicate plots) were then compared using Tukey's test.
Correlations between changes in the community structure of the total and active nirK denitrifiers and changes in environmental parameters (levels of EOC, NO3−, and WFPS), actual denitrification, and DEA were performed. Spearman rank correlation coefficients (R) and the significance level of the correlation (P) were computed using Ginkgo software (http://biodiver.bio.ub.es/ginkgo/Ginkgo.htm).
Nucleotide sequence accession numbers.
The partial nirK gene sequences generated in this study were deposited in the GenBank database under accession numbers FJ664516 to FJ664544.
RESULTS
Diversity of nirK genes and transcripts in three spatial locations over a growing season in a potato field.
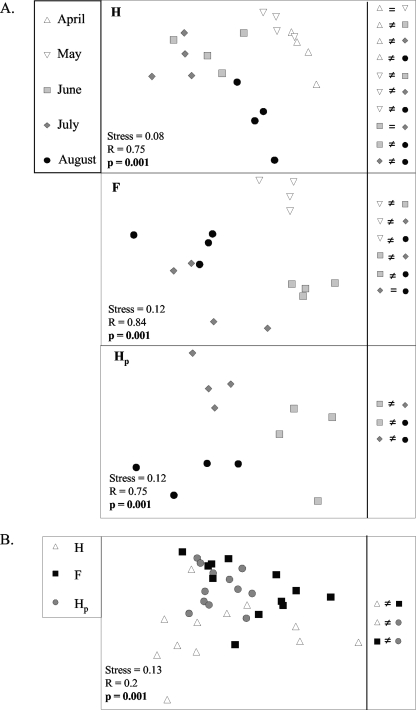
The structure and richness of the denitrifying community bearing nirK were characterized by DGGE analyses of nirK genes. DGGE analyses were also performed using cDNA derived from nirK transcripts to assess the diversity of active members of the nirK denitrifier community. In each spatial location (H, F, and Hp), changes in the nirK denitrifier community structure were observed between sampling dates (Fig. 1A; Table 1). Community structures were not different only between the dates of April and May and between June and July in the H location and between July and August in the F location. The nirK community structure also varied among the three spatial locations (H, F, and Hp) in the field but to a lesser extent (R = 0.2) than between the sampling dates (R = 0.75 to 0.84) (Fig. 1B; Tables 1 and 2).
FIG. 1.
MDS representation of the structure of the nirK denitrifier community (assessed by DGGE analysis of nirK genes) on different sampling dates for three spatial locations, H, Hp, and F, in soil under potato cultivation (A) and at different row locations across the sampling dates (from June to August) (B). The similarities or differences in community structures are indicated. See Tables 1 and 2 for results of ANOSIM.
TABLE 1.
One-way ANOSIM results for structure of total and active nirK denitrifier communities as determined by DGGE banding profiles from nirK genes and transcripts based on data for different sampling dates
| Comparison between dates | ANOSIM resultsa for structure of total or active nirK denitrifying communities |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total nirK |
Active nirK |
|||||||||||
| H |
F |
Hp |
H |
F |
Hp |
|||||||
| P | R | P | R | P | R | P | R | P | R | P | R | |
| April vs May | 0.11 | 0.24 | ND | ND | ND | ND | 0.54 | 0 | ND | ND | ND | ND |
| April vs June | 0.03 | 0.87 | ND | ND | ND | ND | 0.4 | 0.04 | ND | ND | ND | ND |
| April vs July | 0.03 | 1 | ND | ND | ND | ND | 0.03 | 0.55 | ND | ND | ND | ND |
| April vs Aug | 0.03 | 0.92 | ND | ND | ND | ND | 0.03 | 0.84 | ND | ND | ND | ND |
| May vs June | 0.03 | 0.75 | 0.03 | 1 | ND | ND | 0.6 | 0.03 | 0.06 | 0.39 | ND | ND |
| May vs July | 0.03 | 1 | 0.03 | 0.95 | ND | ND | 0.06 | 0.31 | 0.14 | 0.17 | ND | ND |
| May vs Aug | 0.03 | 0.94 | 0.03 | 0.98 | ND | ND | 0.17 | 0.15 | 0.06 | 0.48 | ND | ND |
| June vs July | 0.17 | 0.16 | 0.03 | 0.79 | 0.03 | 0.83 | 0.09 | 0.27 | 0.54 | 0.04 | 0.37 | 0.08 |
| June vs Aug | 0.03 | 0.63 | 0.03 | 0.97 | 0.03 | 0.71 | 0.03 | 0.31 | 0.09 | 0.41 | 0.43 | 0.07 |
| July vs Aug | 0.03 | 0.94 | 0.09 | 0.26 | 0.03 | 0.74 | 0.03 | 0.53 | 0.4 | 0.06 | 0.94 | 0.02 |
ANOSIM results are for analysis by sampling date at three locations, H, F, and Hp, in soil under potato cultivation. The R values indicate the degree of discrimination between two groups; P values indicate the significance level (values in bold indicate P ≤ 0.05). ND, not determined.
TABLE 2.
One-way ANOSIM results for the structure of the total and active nirK denitrifier communities as determined by DGGE banding profiles from nirK genes and transcripts based on data for different spatial locations
| Comparison between spatial locations | ANOSIM resultsa for structure of total or active nirK denitrifying communities |
|||
|---|---|---|---|---|
| Total nirK |
Active nirK |
|||
| P | R | P | R | |
| H vs F | 0.001 | 0.23 | 0.49 | 0.007 |
| H vs Hp | 0.008 | 0.21 | 0.32 | 0.02 |
| F vs Hp | 0.02 | 0.15 | 0.06 | 0.13 |
ANOSIM results are for analysis by row location across the sampling dates from June to August. The R values indicate the degree of discrimination between two groups; P values indicate the significance level (values in bold indicate P ≤ 0.05). ND, not determined.
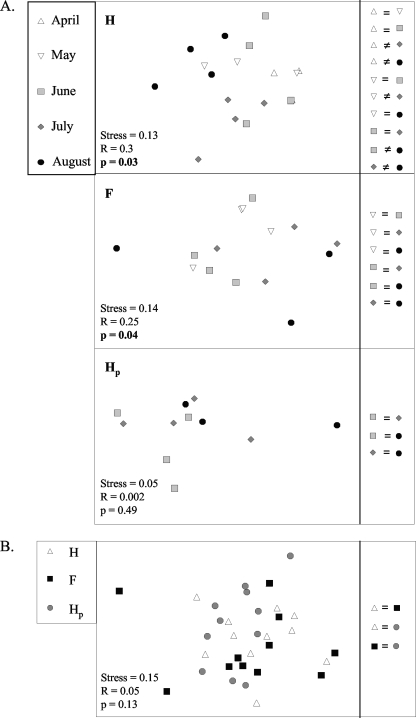
The community structure of active nirK denitrifiers did not change over time or among spatial locations, except in the H location (Fig. 2A and B; Tables 1 and 2). For this location, differences were observed between some dates. The community composition of nirK denitrifiers that were active early in the season (April) was different from the community composition of nirK denitrifiers active later in the season (July and August), and the active nirK community in August was also different from the active nirK community in June and July (Fig. 2A; Table 1). The variability between replicate plots (for a given date or spatial location) of DGGE profiles of nirK transcripts was high compared to the DGGE profiles of nirK genes (dissimilarities among replicates ranged from 39.2 to 84.8% and from 13.4 to 43.1%, respectively [data not shown]).
FIG. 2.
MDS representation of the structure of the active nirK denitrifier community (assessed by DGGE analysis of nirK transcripts) on different sampling dates for three spatial locations, H, Hp, and F, in soil under potato cultivation (A) and at different row locations across the sampling dates (from June to August) (B). The similarities or differences in community structures are indicated. See Tables 1 and 2 for results of ANOSIM.
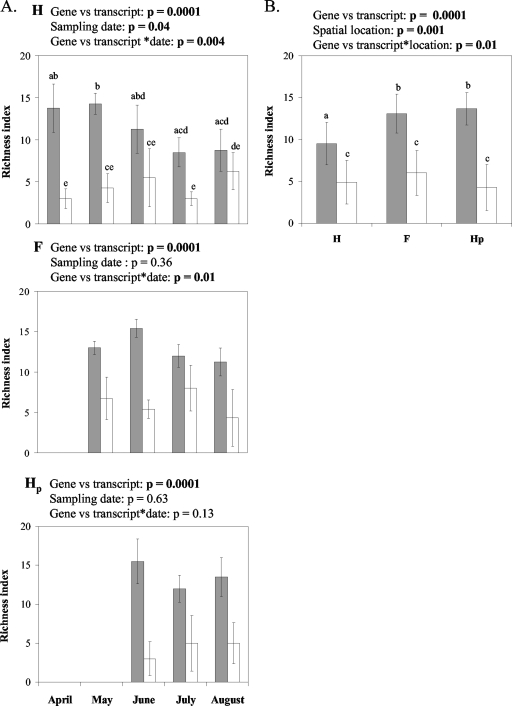
The richness of the nirK denitrifier community, as assessed by the number of bands in DGGE profiles of nirK genes, showed significant differences over time only in the H location, with a higher richness level in May compared to July and August (Fig. 3A). In addition, the richness of the nirK community across several sampling dates was lower in the H location than in the F and Hp locations (Fig. 3B). No differences in the richness of the active nirK denitrifier community were observed over time or among soil locations (Fig. 3A and B). The richness of nirK transcripts was lower than the richness of the nirK genes present in the soil, except for the sample for August in the H location (Fig. 3A).
FIG. 3.
Richness indices of nirK genes (averages of the total number of bands in DNA-derived DGGE profiles [shaded bars]) and nirK transcripts (averages of the total number of bands in cDNA-derived DGGE profiles [open bars]) on different sampling dates for three spatial locations, H, Hp, and F, in soil under potato cultivation (A) and at different row locations across the sampling dates from June to August (B). Means are represented along with standard errors. Results of two-way ANOVA testing are shown for gene versus transcript, sampling date, and gene versus transcript × sampling date (A) and for gene versus transcript, row location, and gene versus transcript × row location effects (B) on DGGE band numbers. P values indicate the significance level (P values of ≤0.05 are indicated in bold). Values with different letters differed significantly (P < 0.05).
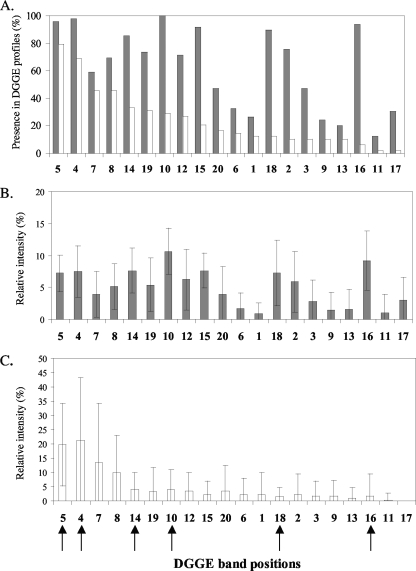
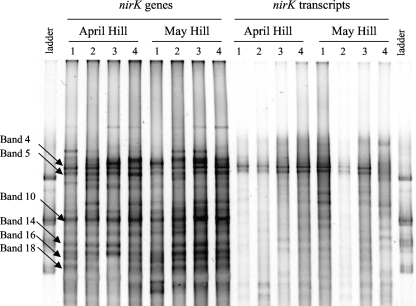
All of the different band positions (i.e., 20 in total) observed in the DGGE profiles of nirK genes were also found in the DGGE profiles of nirK transcripts (Fig. 4A). However, the relative intensities of the different bands varied between the DGGE profiles of nirK genes and transcripts (Fig. 4B and C). In the DGGE profiles of nirK genes, eight dominant bands (i.e., bands 4, 5, 10, 12, 14, 15, 16, and 18) represented 63.3% of the total intensity (Fig. 4B), while the DGGE profiles of nirK transcripts were characterized by only four dominant bands (i.e., bands 4, 5, 7, and 8, accounting for 64.7% of the total intensity) (Fig. 4C).
FIG. 4.
(A) The proportion of all of the DGGE profiles that contain each of the numbered band positions for nirK genes (shaded bars) and transcripts (open bars) (i.e., samples from all dates and spatial locations). (B and C) Relative intensity of each band position in all the DGGE profiles of nirK genes (B) and nirK transcripts (C). Band positions are shown in decreasing order of presence in DGGE profiles of nirK transcripts. Band positions chosen for sequencing are indicated by arrows.
Dominant bands in the DGGE profiles of nirK transcripts corresponded to some of the most dominant bands in the DGGE profiles of nirK genes (Fig. 4 and 5). Bands 4 and 5 were present in most of the DGGE profiles of nirK genes (98% and 96% for bands 4 and 5, respectively) (Fig. 4A) and were also present in the majority of the DGGE profiles of nirK transcripts (69% and 79% for bands 4 and 5, respectively) (Fig. 4A). These two bands were abundant in the DGGE profiles of nirK genes (representing 7.5% and 7.2% of the total intensity for bands 4 and 5, respectively) (Fig. 4B) and were also dominant in the DGGE profiles of nirK transcripts (representing 21.3% and 19.8% of the transcripts present for bands 4 and 5, respectively) (Fig. 4C). However, some dominant bands in the DGGE profiles of nirK genes were only observed either at moderate intensities (e.g., band positions 10 and 14) or at weak intensities (e.g., band positions 16 and 18) in the DGGE profiles of nirK transcripts (Fig. 4B and C). The relative intensities of the band positions 10 and 14 were 10.6% and 7.6%, respectively, in the DGGE profiles of nirK genes and were 4.1% and 3.9% in the DGGE profiles of nirK transcripts. The relative intensities of band positions 16 and 18 in the DGGE profiles of nirK genes were 9.2% and 7.3%, and they were 1.8% and 1.4% in the DGGE profiles of nirK transcripts. Band positions 10, 14, 16, and 18 were present in most of the DGGE profiles of nirK genes (90 to 100%); however, band positions 10 and 14 were moderately present (30% and 33%, respectively) in the DGGE profiles of nirK transcripts, and bands 16 and 18 were observed in only a few DGGE profiles of nirK transcripts (6% and 13%, respectively) (Fig. 4A).
FIG. 5.
Example of DGGE profiles of nirK genes and transcripts for hill soil samples in April and May. The ladder consisted of nirS sequences of cultured denitrifier strains. Arrows indicate bands that were sequenced.
The relative intensities of some cDNA bands differed among the DGGE profiles of the three spatial locations. Band 1 was significantly more abundant in the Hp location (relative intensity, 7.9%) than in the hill (0.4%) and the furrow (0.6%) (data not shown). The relative intensity of band 7 was significantly higher in the Hp location (25.6%) and the hill (14.6%) than in the furrow (2.9%) (data not shown). Band 13 was only observed in the cDNA DGGE profiles of the furrow. No significant differences in the intensities of other particular bands in the DGGE profiles of nirK transcripts were observed over time or among spatial locations.
Phylogenetic analyses of partial nirK sequences.
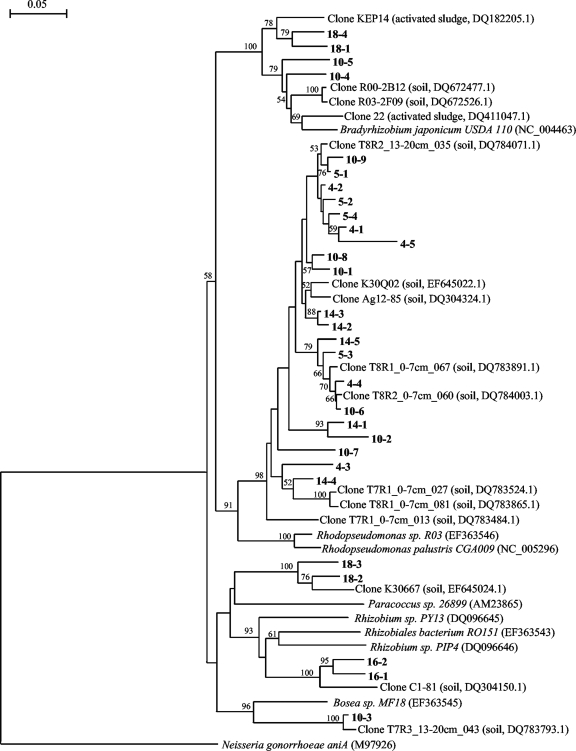
Partial nirK sequences corresponding to three couples of DGGE band positions with different relative intensities of nirK DNA and cDNA bands as described above (i.e., band positions 4 and 5, 10 and 14, and 16 and 18) were subjected to phylogenetic analyses. The partial nirK sequences of the clones representing the band positions were closer to nirK sequences of uncultured bacteria retrieved from the environment (similarities between 89% and 99%) than to nirK sequences of isolated strains (similarities between 79% to 87%) (Fig. 6). Sequences representing band positions 4 and 5, which were dominant in both nirK gene and transcript profiles, were clustered in the phylogenetic tree. The sequences of cultured strains that are closest to the sequences of these band positions were the nirK sequences of Rhodopseudomonas palustris CGA009 and Rhodopseudomonas sp. strain R03. Interestingly, clones representing band position 14 also clustered with this group. Although this band was dominant in the DGGE profiles of nirK genes, it was only moderately abundant in the cDNA DGGE profiles. Sequences representing band positions 16 and 18, which were dominant in gene profiles but only weakly abundant in transcript profiles, belonged to two clusters that are different from the one containing sequences of band positions 4, 5, and 14. The nirK sequences from bacteria from the genera Rhizobium, Bradyrhizobium, and Paracoccus were the cultured strain sequences most similar to the sequences of these band positions (Fig. 6). Sequences from band position 10 were spread in all the clusters of the phylogenetic tree (Fig. 6).
FIG. 6.
Neighbor-joining phylogenetic tree of partial nirK sequences of clones representative of some DGGE band positions in relation to nirK sequences of the closest uncultured and cultured relatives. Clone sequences from this study are labeled by numbers in bold. The first number corresponds to the band position, and the second number corresponds to clones with an identical DGGE band position but different sequences. The sources and accession numbers of the sequences are indicated in brackets. The scale bar indicates 5% nucleotide substitutions. Bootstrap values greater than 50% (500 replicates) are reported at the nodes. The sequence of aniA from Neisseria gonorrhoea served as an outgroup to root the phylogram.
Some clones showing identical migration in DGGE gels harbored different nirK sequences (Fig. 6). Clone sequences obtained from DNA and cDNA PCR products and from different sampling dates or spatial locations that represented the same band position on the DGGE gel were either different or identical. Similarly, clone sequences from a single PCR product representing the same DGGE band position were either different or identical. When a high number of clones were sequenced for a band position, identical sequences were found for the majority of the clones (e.g., for band position 5, 16 clones were sequenced and an identical sequence was obtained in 75% of the clones [data not shown]).
Correlation between community structure of total and active nirK denitrifiers and environmental factors and denitrification activity.
Environmental parameters such as nitrate concentration, EOC, and WFPS, and also denitrification rates in the field and DEA were monitored previously by Dandie et al. (9) for the soil samples analyzed in this study. Variations in these environmental parameters and denitrification activities have been observed over time and among spatial locations in the field. No strong correlations between changes in the community structure of the total and active nirK denitrifiers and changes in these environmental parameters and denitrification activities were observed. When all samples from the H, F, and Hp locations were included for the analysis of correlations, only a low correlation (i.e., low R value, significant P value) was observed between the nitrate concentration and the structure of the total nirK denitrifying community (Table 3). For samples taken from the H location, changes in the nitrate concentration, EOC, WFPS, and denitrification rate in the field were weakly correlated to changes in the structure of the total nirK denitrifying community (Table 3). Similarly, changes in the structure of the active component of the nirK denitrifying community were only weakly correlated with changes in the denitrification rate for this location (Table 3). In the F location, weak correlations were observed between changes in the structure of the active nirK denitrifying community and changes in the nitrate concentration and denitrification rate (Table 3). In the Hp location, the changes in environmental parameters and denitrification activities were not correlated to changes in the community structure of the total and active nirK denitrifiers (Table 3).
TABLE 3.
Correlations and significance for rank similarity matrices obtained for the community structures of the total and active nirK denitrifiers compared to environmental or microbial factors
| Community and factor or activity |
R coefficient (cP)a for samples from: |
|||
|---|---|---|---|---|
| All sites | H | F | Hp | |
| Total nirK denitrifiers | ||||
| Environmental factor | ||||
| Nitrate concn | 0.15 (0.003) | 0.18 (0.03) | 0.15 (0.09) | 0.04 (0.8) |
| EOC | 0.11 (0.14) | 0.2 (0.03) | 0.09 (0.41) | 0.03 (0.14) |
| WFPS | ND | 0.19 (0.03) | 0.07 (0.46) | ND |
| Microbial activity | ||||
| Denitrification | ND | 0.21 (0.01) | 0.12 (0.26) | ND |
| DEA | 0.02 (0.8) | 0.07 (0.56) | 0.04 (0.7) | 0.04 (0.7) |
| Active nirK denitrifiers | ||||
| Environmental factor | ||||
| Nitrate concn | 0.01 (0.83) | 0.07 (0.36) | 0.35 (0.002) | 0.02 (0.23) |
| EOC | 0.08 (0.29) | 0.03 (0.82) | 0.01 (0.93) | 0.02 (0.41) |
| WFPS | ND | 0.15 (0.16) | 0.12 (0.3) | ND |
| Microbial activity | ||||
| Denitrification | ND | 0.18 (0.03) | 0.39 (0.005) | ND |
| DEA | 0.01 (0.89) | 0.05 (0.7) | 0.02 (0.88) | 0.04 (0.81) |
ND, not determined. P values of ≤0.05 are indicated in bold.
DISCUSSION
Emission of the greenhouse gas N2O is a significant environmental problem and one that is greatly impacted by agricultural practices, especially nitrogen fertilizer application (15, 20, 29). Although denitrification and N2O emissions are known to increase in response to agronomic inputs and other environmental conditions, less is known about the dynamics of denitrifier communities in agricultural soils, for example, how diversity and abundance of communities change, which members of the community are major contributors to gas emissions, and what environmental factors are dominant contributors to denitrifier community changes. In this study we sought to determine if the composition of the nirK denitrifier community (assessed by DGGE analysis of nirK genes), and in particular the active component of the community (assessed by DGGE analysis of nirK transcripts), varied in response to changes in environmental conditions over a potato growing season and in different locations in the potato field and if this could at least partially explain the increases in denitrification observed in the potato field. While the community composition varied over time, and to a lesser extent among field locations, the active component was not significantly different among the samples. Rather, a few ubiquitous and abundant denitrifiers were responsible for most of the nirK gene expression.
Denitrifying bacteria bear either nirS or nirK genes encoding cytochrome cd1or copper-containing nitrite reductases, respectively. However, in the present study, we were not able to amplify nirS cDNA by RT-PCR using available primers suitable for subsequent DGGE analysis (36), although PCR amplification with a DNA template could be obtained (data not shown). We therefore focused our study on nirK denitrifiers. As noted in other studies, mRNA of genes involved in different steps of denitrification was not detected using RT-PCR, while amplification from DNA was successful (23, 32, 33). This may be due to the instability of mRNA or to a concentration of mRNA template that is below the detection limit by RT-PCR. In our study, nirK cDNA could be amplified by RT-PCR for all soil samples, even those for which no denitrification activity could be detected in situ (i.e., in April and August), although the amplification signals on agarose gels were generally weak in those cases.
Changes in the genetic structure of the nirK denitrifying community were observed over time during the potato growing season and to a lesser extent among different spatial locations in the field (i.e., in the hill, furrow, and hill close to plant roots). Differences in the community composition were greatest between the early and late seasons. For example, the nirK denitrifier represented by band 18, which is closely related to the nirK gene of culturable denitrifiers of the genera Rhizobium, Bradyrhizobium, and Paracoccus, was absent in April but increased in relative abundance as the season progressed. Wolsing and Priemé (42) also observed large seasonal variations in the structure of the nirK denitrifying community in an arable soil over an 8-month period. Likewise, temporal shifts in the structure of the nosZ denitrifying community of meadow and forest soils over a 4-year period have been reported (4). Other nirK denitrifiers may also have been present in the potato field but were not detected in our analysis because the sequences of some nirK genes may not have sufficient homology to the PCR primer sequences to generate amplification products for DGGE, sequences closely related to the primer sequences may have been preferentially amplified during PCR, some rare nirK sequences may not be detected in the samples, and PCR products may comigrate during DGGE.
Overall, there was no significant change in the genetic structure of the active component of the nirK community over time, except in the potato hill. In general, the denitrifying community in the hill that was active early in the season (April) was different from the denitrifying community that was active later in the season during mid- to late summer (July and August). Lack of detectable changes in other field locations over time or among spatial locations could be due to the high variability in the DGGE profiles among replicate plots (of a given sampling date or spatial location). This variability may be attributed to a high spatial heterogeneity in situ in the diversity of nirK mRNA and to the low concentration of extracted nirK mRNA template, which would limit reproducible amplification to the most abundant nirK transcripts (i.e., highly expressed nirK genes).
Only a portion of the total nirK denitrifier community that is present in a field location or on a particular date is active. A comparison of the DGGE profiles derived from DNA and cDNA of all the samples showed that the richness (assessed by the number of bands) of nirK genes expressed was lower overall than the richness of nirK genes present in the soil. The most active members of the nirK denitrifier community are the most abundant and also the most ubiquitous. A comparison of the dominant bands in the DGGE profiles derived from DNA and cDNA samples indicated that the most abundant nirK transcripts corresponded to some of the most abundant nirK genes present in the soil. In particular, the denitrifiers represented by bands 4 and 5 were present in all field locations and at all dates in more or less equal abundance. These bacteria may be responsible for most of the nirK expression throughout the growing season and in all locations, except in the area around plant roots, where the bacteria represented by band 7 were also dominant.
Analysis of partial nirK sequences corresponding to band positions 4 and 5, as well as others that were moderately and weakly represented in the profiles of nirK transcripts, confirmed that PCR specifically amplified nirK fragments for all the clones representing the different band positions. Some clone sequences from nirK PCR and RT-PCR products corresponding to the same DGGE band position were different. Migration of different sequences to the same position on DGGE gels has also been shown in the analysis of other genes (22, 41). In our study, differences in sequences found in a single DGGE band did not correspond to differences in the nature or source of the sample (i.e., DNA versus cDNA templates or different sampling dates or spatial locations). All of the clone sequences were more similar to other nirK sequences retrieved from uncultured soil bacteria than from known denitrifying isolates. Sharma et al. (32) investigated the diversity of nirK transcripts in the rhizosphere of three legumes and also showed that the rhizosphere nirK sequences that were expressed did not cluster with nirK sequences of cultured organisms. The nearest culturable relative in that study was Mesorhizobium sp. In our study, the nearest cultured strain sequences to the nirK sequences of the band positions 4 and 5 that were highly expressed in the potato field were the nirK sequences of Rhodopseudomonas palustris and Rhodopseudomonas sp. strain R03. Thus, bacteria belonging to the genus Rhodopseudomonas may be among the major denitrifiers contributing to denitrification in the potato field.
Other nirK denitrifiers are present in abundance and are ubiquitous but are not strong contributors to denitrification, as indicated by a low abundance of transcripts. These include the denitrifiers represented by bands 16 and 18, which are most closely related to the nirK sequences of the genera Rhizobium, Bradyrhizobium, and Paracoccus. Using a functional gene microarray, Bulow et al. (5) found that only three dominant groups of nirS denitrifiers along an estuarine gradient could be detected at the mRNA level, suggesting that the most abundant denitrifying groups are responsible for most of the nirS expression and are important contributors to the denitrification rate. In contrast, Sharma et al. (33) showed that, following a freeze-thaw event in soil microcosms, the most active nirS denitrifiers are not the most abundant ones. After freezing, the DGGE profiles derived from nirS cDNA were more complex in terms of the number of bands than the corresponding DNA-derived DGGE profiles, and the dominant bands in the cDNA-derived DGGE profiles were not detected in the corresponding DNA-derived DGGE profiles. Thus, the contribution to denitrification gene expression cannot be predicted on the basis of denitrifier abundance.
Expression of nirK by some denitrifiers differed among spatial locations in the potato field. Bacteria represented by band positions 1 and 7 were present in all field locations tested; however, they were more active in the area around plant roots. The expression of nirK in these bacteria may be influenced by plant roots or by the activity of other microbes in this typically densely colonized region. Plants release a variety of organic compounds from their roots that are carbon and energy sources for soil microbes (21). Metabolism of these compounds reduces oxygen levels through respiration, which can increase expression of nitrogen oxide reductases (43). Moreover, nitrate is generally removed more rapidly from the soil due to uptake by plants and microbes. Henry et al. (14) reported differences in the structure of the denitrifying community between microcosms amended or not with artificial root exudates, and DeAngelis et al. (10) recently showed that roots influence the composition of the microbial community and the abundance of some rhizosphere populations. At our study site, nirK denitrifiers were more abundant in the Hp location than in the other locations (9). Thus, genetic structure, abundance, and activity of the nirK denitrifying community may be influenced by plant and microbial activity in the rhizosphere.
A major goal of this study was to identify factors, especially agronomic factors, that contribute to changes in nirK gene expression in the denitrifier community. Temperature, soil aeration, and addition of carbon and nitrate, which vary in the potato field (9), may influence the abundance, composition, and/or activity of the denitrifying community. Previous studies were not able to identify strong correlations between increases in denitrifier abundance and changes in environmental conditions (9), and therefore we reasoned that perhaps only a portion of the targeted denitrifier community responds significantly to environmental changes. While increases in abundance are often not apparent when broad denitrifier groups are measured, changes in the size of targeted populations have been reported (9). Although correlations were relatively weak (low R; P < 0.05), nitrate, which influences N2O flux (9), has a stronger effect on denitrifier composition and activity than the other environmental factors measured (EOC and WFPS). Nitrate levels were high in May and June in the hill, where they are applied, and lower in the furrow and the areas around plant roots. Correlation analyses could not be used to assess the effect of temperature because only one temperature measurement was taken per sampling date (with no replicates within the plot). Nonetheless, temperature changes in the field over the course of the growing season may have induced changes in the genetic structure of nirK genes and transcripts. Although pH can affect the composition of denitrifying communities (11), in our study there was no correlation between soil pH and denitrifier diversity or activity (data not shown).
Variations in denitrification rates in the field and in DEA over time during the potato growing season and among spatial locations have been reported (9). Changes in denitrification rates were strongly influenced by variations in nitrate availability and WFPS. Denitrification rates in the potato hill increased from April to May, when nitrate concentration was the highest, and then decreased progressively until August. In the furrow, the denitrification rate was highest in May, June, and July and was low in August, when the soil moisture decreased. In addition, the cumulative denitrification value was higher in the soil of the furrow (less aerated) than in the hill (9). The composition of the nirK community was correlated with denitrification rates in the hill, albeit weakly. Changes in environmental conditions may more immediately modify the denitrification rate but may influence the diversity of nirK denitrifiers in the longer term (39), which may explain the weak correlation. The absence of a strong correlation also suggests that changes in the community composition of other denitrifier groups (e.g., nirS denitrifiers) might be more important in the denitrification response under our field conditions. Changes in the genetic structure of the active nirK community in the hill and the furrow were also weakly correlated with changes in denitrification rates. The active component of the nirK population in these locations remained stable during the period from May to July, when denitrification rates were highest, suggesting that these active nirK denitrifiers may be significant contributors to denitrification. In other studies that have assessed the relationship between diversity of denitrifiers and denitrification activity, conflicting results have been reported. For instance, differences in denitrifying community structure associated with differences in DEA between meadow and forest soils have been shown by Rich et al. (28). By contrast, Enwall et al. (12) observed that differences in the composition of the denitrifying community were not correlated with differences in denitrification activity, and both variables seemed to be influenced by different factors.
In conclusion, although the nirK denitrifier community composition changed over a potato growing season and among locations in a potato field, there is no evidence of a strong link with denitrification rates. Nor is the composition of the community strongly influenced by the environmental parameters measured, which correlate more strongly with denitrification rates. Differences between the composition of nirK denitrifiers that are present and those that are active highlight the importance of assessing the active members when relating denitrifier community structure with denitrification rates. The active portion of the community is relatively stable. That is, not all members of the nirK denitrifying community contribute to denitrification in the field, and some of the major contributors are highly abundant in all locations over the growing season. These populations are therefore likely not responsible for the sharp increases in denitrification and N2O emissions observed in the potato field over time. Rather, they are likely contributing to a basal level of activity. On the other hand, the nirK expression of other denitrifiers in the community does increase among field locations. Variations in denitrification rates may be explained by changes in the expression of a few specific denitrifiers, and future studies should develop PCR primers to specifically target these groups to assess their response to environmental parameters. The contribution of nirS denitrifiers also needs to be considered. Understanding which are the most active denitrifiers and the environmental factors to which they respond can help us develop appropriate agricultural management strategies to reduce greenhouse gas emissions.
Acknowledgments
This work was supported by a Natural Sciences and Engineering Research Council Strategic Grant to C.L.P., C.G., and J.T.T.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Acinas, S. G., R. Sarma-Rupavtarm, V. Klepac-Ceraj, and M. F. Polz. 2005. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71:8966-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernard, G., S. K. Asiedu, and P. Boswall (ed.). 1993. Atlantic Canada potato guide. Atlantic Provinces Agriculture Services Coordinating Committee publication 1300/93, Agdex 257/13. APASCC, Fredericton, New Brunswick, Canada.
- 3.Bockman, O. C., and H.-W. Olfs. 1998. Fertilizers, agronomy and N2O. Nutr. Cycl. Agroecosyst. 52:165-170. [Google Scholar]
- 4.Boyle, S. A., J. J. Rich, P. J. Bottomley, K. Cromack, Jr., and D. D. Myrold. 2006. Reciprocal transfer effects on denitrifying community composition and activity at forest and meadow sites in the Cascade Mountains of Oregon. Soil Biol. Biochem. 38:870-878. [Google Scholar]
- 5.Bulow, S. E., C. A. Francis, G. A. Jackson, and B. B. Ward. 2008. Sediment denitrifier community composition and nirS gene expression investigated with functional gene microarrays. Environ. Microbiol. 10:3057-3069. [DOI] [PubMed] [Google Scholar]
- 6.Burton, D. L., B. J. Zebarth, K. M. Gillam, and J. A. MacLeod. 2008. Effect of rate and time of fertilizer nitrogen application on N2O emissions from potato. Can. J. Soil Sci. 88:229-239. [Google Scholar]
- 7.Conrad, R. 1996. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60:609-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dandie, C. E., M. N. Miller, D. L. Burton., B. J. Zebarth, J. T. Trevors, and C. Goyer. 2007. Nitric oxide reductase-targeted real-time PCR quantification of denitrifier populations in soil. Appl. Environ. Microbiol. 73:4250-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dandie, C. E., D. L. Burton, B. J. Zebarth, S. L. Henderson, J. T. Trevors, and C. Goyer. 2008. Changes in bacterial denitrifier community abundance over time in an agricultural field and their relationship with denitrification activity. Appl. Environ. Microbiol. 74:5997-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeAngelis, K. M., E. L. Brodie, T. Z. DeSantis, G. L. Andersen, S. E. Lindow, and M. K. Firestone. 2009. Selective progressive response of soil microbial community to wild oat roots. ISME J. 3:168-178. [DOI] [PubMed] [Google Scholar]
- 11.Deiglmayr, K., L. Philippot, V. A. Hartwig, and E. Kandeler. 2004. Structure and activity of the nitrate-reducing community in the rhizosphere of Lolium perenne and Trifolium repens under long-term elevated atmospheric pCO2. FEMS Microbiol. Ecol. 49:445-454. [DOI] [PubMed] [Google Scholar]
- 12.Enwall, K., L. Philippot, and S. Hallin. 2005. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 71:8335-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallin, S., C. J. Jones, M. Schloter, and L. Philippot. 2009. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 3:597-605. [DOI] [PubMed] [Google Scholar]
- 14.Henry, S., S. Texier, S. Hallet, D. Bru, C. Dambreville, D. Cheneby, F. Bizouard, J. C. Germon, and L. Philippot. 2008. Disentangling the rhizosphere effect on nitrate reducers and denitrifiers: insight into the role of root exudates. Environ. Microbiol. 10:3082-3092. [DOI] [PubMed] [Google Scholar]
- 15.Hofstra, N., and A. F. Bouwman. 2005. Denitrification in agricultural soils: summarizing published data and estimating global annual rates. Nutr. Cycl. Agroecosyst. 72:267-278. [Google Scholar]
- 16.Ka, J.-O., J. Urbance, R. W. Ye, T.-Y. Ahn, and J. M. Tiedje. 1997. Diversity of oxygen and N-oxide regulation of nitrite reductases in denitrifying bacteria. FEMS Microbiol. Lett. 156:55-60. [DOI] [PubMed] [Google Scholar]
- 17.Kruskal, J. B., and M. Wish. 1978. Multidimensional scaling. Sage Publications, Beverley Hills, CA.
- 18.Liu, X., S. M. Tiquia, G. Holguin, L. Wu, S. C. Nold, A. H. Devol, K. Luo, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen-deficient zone off the Pacific coast of Mexico. Appl. Environ. Microbiol. 69:3549-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, K. W., A. Bedard-Haughn, S. D. Siciliano, and R. E. Farrell. 2008. Relationship between nitrifier and denitrifier community composition and abundance in predicting nitrous oxide emissions from ephemeral wetland soils. Soil Biol. Biochem. 40:1114-1123. [Google Scholar]
- 20.Mosier, A. R. 1998. Soil processes and global change. Biol. Fertil. Soils 27:221-229. [Google Scholar]
- 21.Neumann, G., and V. Römheld. 2001. The release of root exudates as affected by the plant's physiological status, p. 41-93. In R. Pinton, Z. Varanni, and P. Nanniperi (ed.), Rhizosphere: biochemistry and organic substances at the soil-plant interface. Marcel Dekker, New York, NY.
- 22.Nicolaisen, M. H., and N. B. Ramsing. 2002. Denaturing gradient gel electrophoresis (DGGE) approaches to study the diversity of ammonia-oxidizing bacteria. J. Microbiol. Methods 50:189-203. [DOI] [PubMed] [Google Scholar]
- 23.Nogales, B., K. N. Timmis, D. B. Nedwell, and A. M. Osborn. 2002. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otani, Y., K. Hasegawa, and K. Hanaki. 2004. Comparison of aerobic denitrifying activity among three cultural species with various carbon sources. Water Sci. Technol. 50:15-22. [PubMed] [Google Scholar]
- 25.Patra, A. K., L. Abbadie, A. Clays-Josserand, V. Degrange, S. J. Grayston, P. Loiseau, F. Louault, S. Mahmood, S. Nazaret, L. Philippot, F. Poly, J. I. Prosser, A. Richaume, and X. Le Roux. 2005. Effects of grazing on microbial functional groups involved in soil N dynamics. Ecol. Monogr. 75:65-80. [Google Scholar]
- 26.Payne, J. W. 1981. Denitrification. Wiley-Interscience Publications, New York, NY.
- 27.Philippot, L., and S. Hallin. 2005. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 8:234-239. [DOI] [PubMed] [Google Scholar]
- 28.Rich, J. J., R. S. Heichen, P. J. Bottomley, K. Cromack, Jr., and D. D. Myrold. 2003. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl. Environ. Microbiol. 69:5974-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruser, R., H. Flessa, R. Schilling, F. Beese, and J. C. Munch. 2001. Effect of crop-specific field management and N fertilization on N2O emissions from a fine-loamy soil. Nutr. Cycl. Agroecosyst. 59:177-191. [Google Scholar]
- 30.Ruser, R., H. Flessa, R. Russow, G. Schmidt, F. Buegger, and J. C. Munch. 2006. Emission of N2O, N2 and CO2 from soil fertilized with nitrate: effect of compaction, soil moisture and rewetting. Soil Biol. Biochem. 38:263-274. [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Sharma, S., M. K. Aneja, J. Mayer, J. C. Munch, and M. Schloter. 2005. Diversity of transcripts of nitrite reductase genes (nirK and nirS) in rhizospheres of grain legumes. Appl. Environ. Microbiol. 71:2001-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma, S., Z. Szele, R. Schilling, J. C. Munch, and M. Schloter. 2006. Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 72:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, K. A., I. P. McTaggart, K. E. Dobbie, and F. Conen. 1998. Emissions of N2O from Scottish agricultural soils, as a function of fertilizer N. Nutr. Cycl. Agroecosyst. 52:123-130. [Google Scholar]
- 35.Stres, B., T. Danevcic, L. Pal, M. M. Fuka, L. Resman, S. Leskovec, J. Hacin, D. Stopar, I. Mahne, and I. Mandic-Mulec. 2008. Influence of temperature and soil water content on bacterial, archaeal and denitrifying microbial communities in drained fen grassland soil microcosms. FEMS Microbiol. Ecol. 66:110-122. [DOI] [PubMed] [Google Scholar]
- 36.Thröback, I. N., K. Enwall, A. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 37.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley and Sons, Inc., New York, NY.
- 38.Tiedje, J. M. 1994. Denitrifiers, p. 245-267. In R. W. Weaver et al. (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. SSSA book series no. 5. Soil Science Society of America, Madison, WI.
- 39.Wallenstein, M. D., D. D. Myrold, M. Firestone, and M. Voytek. 2006. Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16:2143-2152. [DOI] [PubMed] [Google Scholar]
- 40.Wertz, S., V. Degrange, J. I. Prosser, F. Poly, C. Commeaux, T. Freitag, N. Guillaumaud, and X. Le Roux. 2006. Maintenance of soil functioning following erosion of microbial diversity. Environ. Microbiol. 8:2162-2169. [DOI] [PubMed] [Google Scholar]
- 41.Wertz, S., F. Poly, X. Le Roux, and V. Degrange. 2008. Development and application of a PCR-denaturing gradient gel electrophoresis tool to study the diversity of Nitrobacter-like nxrA sequences in soil. FEMS Microbiol. Ecol. 63:261-271. [DOI] [PubMed] [Google Scholar]
- 42.Wolsing, M., and A. Prieme. 2004. Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments. FEMS Microbiol. Ecol. 48:261-271. [DOI] [PubMed] [Google Scholar]
- 43.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]