Abstract
Endosymbiotic bacteria were identified in the parasitic ciliate Ichthyophthirius multifiliis, a common pathogen of freshwater fish. PCR amplification of DNA prepared from two isolates of I. multifiliis, using primers that bind conserved sequences in bacterial 16S rRNA genes, generated an ∼1,460-bp DNA product, which was cloned and sequenced. Sequence analysis demonstrated that 16S rRNA gene sequences from three classes of bacteria were present in the PCR product. These included Alphaproteobacteria (Rickettsiales), Sphingobacteria, and Flavobacterium columnare. DAPI (4′,6-diamidino-2-phenylindole) staining showed endosymbionts dispersed throughout the cytoplasm of trophonts and, in most, but not all theronts. Endosymbionts were observed by transmission electron microscopy in the cytoplasm, surrounded by a prominent, electron-translucent halo characteristic of Rickettsia. Fluorescence in situ hybridization demonstrated that bacteria from the Rickettsiales and Sphingobacteriales classes are endosymbionts of I. multifiliis, found in the cytoplasm, but not in the macronucleus or micronucleus. In contrast, F. columnare was not detected by fluorescence in situ hybridization. It likely adheres to I. multifiliis through association with cilia. The role that endosymbiotic bacteria play in the life history of I. multifiliis is not known.
The ciliate Ichthyophthirius multifiliis is an obligate parasite of freshwater fish that infects epithelia of the skin and gills. The life cycle of I. multifiliis consists of three stages: an infective theront, a parasitic trophont, and a reproductive tomont. Infection is initiated by invasion of the skin and gills by free-swimming, 40-μm-long, pyriform-shaped theronts that burrow several cell layers deep into epithelial tissue of the skin and gills and rapidly differentiate into trophonts. Trophonts feed on epithelial cells and grow into 500- to 800-μm-diameter cells, causing extensive damage to skin and gills, which in severe infections results in mortality (10-12). After feeding for 5 to 7 days, trophonts leave the host, form encysted tomonts, and undergo up to 10 cell divisions over 18 to 24 h, producing as many as 103 daughter cells, which exit the cyst as infective theronts to reinitiate the life cycle. I. multifiliis is ciliated at all stages (9).
DNA sequencing of the I. multifiliis genome at the J. Craig Venter Institute unexpectedly revealed that bacterial DNA sequences, including sequences with homology to Rickettsia, were present in the DNA preparations (R. S. Coyne, 2009 [http://www.jcvi.org/cms/research/projects/ich/overview]). The origin of these sequences was unclear, but they represented evidence for either horizontal gene transfer into the I. multifiliis genome (17, 27) or the presence of intracellular bacteria. No previous evidence suggested the presence of intracellular bacteria in I. multifiliis, even though the fine structure of I. multifiliis theronts and trophonts has been examined by transmission electron microscopy (10-12). Intracellular or endosymbiotic bacteria, however, are commonly found in protists, and about 200 ciliate species are known to harbor intracellular bacteria (13, 15). Sonneborn and Preer in their classic studies on endosymbionts in Paramecium characterized a number of different endosymbionts, including “killers,” named for their ability to kill uninfected strains of Paramecium. Cytoplasmic endosymbionts in Paramecium now include Caedibacter taeniospiralis (Gammaproteobacteria), and Pseudocaedibacter conjugates, Tectibacter vulgaris, and Lyticum flagellatum (Alphaproteobacteria). Macronuclear endosymbionts include the Alphaproteobacteria, Holospora caryophila, and Caedibacter caryophila, which can also infect the cytoplasm (4, 16, 22, 26). The roles these endosymbionts play in protists are not well understood.
The presence of sequences with homology to bacterial genomes prompted us to determine if I. multifiliis contained endosymbionts, or if these sequences represented evidence for horizontal gene transfer into the I. multifiliis genome. Our identification of the same two endosymbionts, in two different isolates of I. multifiliis, suggests that endosymbionts are common in I. multifiliis. However, the physiological relationships between I. multifiliis and its resident endosymbionts are unclear. It is not known if the endosymbionts contribute to the growth of I. multifiliis, if they contribute to the severity or pathogenicity of infection, or if they provide their host with any selective advantage, as occurs with Paramecium containing killer particles (4). It has not been determined if they influence the immune response of fish infected with I. multifiliis. It is possible that they may simply be parasites of this parasitic ciliate.
MATERIALS AND METHODS
I. multifiliis.
I. multifiliis isolates were maintained by serial passage on juvenile channel catfish (Ictalurus punctatus) as previously described (31). Strain G5 (serotype D) was isolated from an albino channel catfish obtained from a local aquarium store in 1995 and was subsequently continuously passaged on juvenile channel catfish in our laboratory (31). Strain G13 (uncharacterized serotype) was isolated in September 2008 from a longnose dace (Rhinichthys cataractae) collected in 2008 from Coopers Creek, GA. G13 was then passaged on juvenile channel catfish in our laboratory.
DNA isolation, PCR, cloning, and sequencing.
Tomonts and theronts were collected from infected channel catfish as previously described (31). Slow-swimming G5 tomonts were individually isolated by hand-pipetting. They were transferred in a volume of 1 to 2 μl of charcoal filtered tap water (CFW) per tomont from crystallization dishes (Fisher) into 25 ml of sterile CFW containing 100 μg/ml normocin (InvivoGen). They were washed twice by hand-pipetting into fresh, sterile CFW. Tomonts were collected by hand-pipetting into 25 ml of CFW containing 100 μg/ml normocin and held for 1 h at 4°C to control growth of extracellular bacteria. They were then hand-pipetted into fresh CFW and transferred to a 2-ml microcentrifuge tube for DNA isolation. Rapidly swimming G13 theronts were collected as previously described (31). They were washed twice in 100 ml of CFW by centrifugation at 500 × g for 2 min in 100 ml oil-testing centrifuge tubes (Fisher). Theronts were then transferred to 2-ml microcentrifuge tubes and collected by centrifugation at 500 × g for 2 min. DNA was prepared from tomonts and theronts by lysing cells in 0.5 M EDTA, 1% sodium dodecyl sulfate, and 10 mM Tris (pH 9.5) at 65°C for 20 min followed by incubation in 0.5 mg/ml pronase in a mixture of 0.5 M EDTA, 0.6% sodium dodecyl sulfate, and 10 mM Tris (pH 9.5) at 56°C for 18 h. DNA was isolated by phenol-chloroform extraction, precipitated with ethanol, and resuspended in 10 mM Tris-1 mM EDTA (pH 8.0) (8).
PCR amplification of bacterial 16S rRNA genes was performed using universal primers that target highly conserved regions of 16S rRNA genes in Bacteria (32). The sequence of the forward primer was 5′ GTTTGATYMTGGCTCAG 3′ (Escherichia coli 16S rRNA gene bases 11 to 27) (Y = C + T and M = A + C), and the sequence of the reverse primer was 5′ GGHTACCTTGTTACGACT 3′ (E. coli 16S rRNA bases 1492 to 1509) (H = A + T + C) (6, 32). Degeneracies in the primer sequences were introduced based on the sequences of PCR primers used to amplify 16S rRNA genes of other endosymbionts, including Rickettsia sp. and Caedibacter sp. (3, 5). PCRs were performed using Platinum Taq DNA polymerase (Invitrogen) in an MJ Research thermocycler in hot-start tubes in a final volume of 50 μl containing 0.5 μg DNA. The following conditions were used: 94°C for 2 min; 30 cycles of 94°C for 30 s, 46°C for 30 s, and 68°C for 1.5 min; and a final extension at 68°C for 5 min. Amplified DNA from each reaction was separated in 1% agarose gels, stained with ethidium bromide, and photographed using a GelDoc system (Bio-Rad). Amplified PCR products were purified from 1% agarose gels, ligated into the pCR8/GW/TOPO vector, and transformed into E. coli using the pCR8/GW/TOPO TA cloning kit following the manufacturer's instructions (Invitrogen). Individual colonies were picked and then grown overnight, and plasmid DNA was isolated. Cloned inserts were sequenced using an ABI3730 DNA sequencer at the University of Georgia's DNA sequencing facility.
Comparative sequence analysis.
Sequence similarity searches against public databases were performed using BLASTN, available at the National Center for Biotechnology Information website. Phylogenetic trees were constructed using MEGA4 (28). Data were examined by minimum evolution distance, neighbor-joining, and maximum parsimony algorithms using bootstrap analysis of 1,000 replicates.
Probes.
The EUB338 probe binds a highly conserved region within 16S rRNA of Bacteria and is specific for the domain Bacteria (1). Oligonucleotide probes specific for the 16S rRNA sequences of the Rickettsiales, Sphingobacteriales, and Flavobacterium columnare bacteria identified in this study were designed using Primer3 software and are listed in Table 1 (23). The probes were fluorescently labeled at their 5′ termini with Cy3 or Cy5 (Integrated DNA Technologies). The probes were designed to target regions of limited secondary structure in 16S rRNA, based on the structure of E. coli 16S rRNA (33). In each case, two complementary probes were synthesized: one complementary to the 16S rRNA sequence and a second with the same sequence as the transcribed 16S rRNA, which served as a negative control to detect nonspecific binding. The probes were used alone, or in combination with EUB338, which served as a positive control for fluorescence in situ hybridization (FISH) experiments.
TABLE 1.
Oligonucleotide probes used in this study
| Probe (position) | Sequence (5′→3′) | Specificity | Source or reference |
|---|---|---|---|
| EUB338 (337-354) | 5′ (Cy3)GCTGCCTCCCGTAGGAGT 3′ | Bacteria | 1 |
| EUBN (337-354) | 5′ (Cy3)ACTCCTACGGGAGGCAGC 3′ | Bacteria | This study |
| RICP (803-822) | 5′ (Cy5)TGCTTAATGCGTTAGCTGCG 3′ | Rickettsiales | This study |
| RICN (803-825) | 5′ (Cy5)CGCAGCTAACGCATTAAGCACTC 3′ | Rickettsiales | This study |
| BACP (1215-1235) | 5′ (Cy5)TGCTCCACATCGCTGTATTGC 3′ | Sphingobacteriales | This study |
| BACN (1215-1235) | 5′ (Cy5)GCAATACAGCGATGTGGAGCA 3′ | Sphingobacteriales | This study |
| FLAP (820-841) | 5′ (Cy5)TCACTTTCGCTTAGCCACTCAG 3′ | Flavobacterium | This study |
| FLAN (820-841) | 5′ (Cy5)CTGAGTGGCTAAGCGAAAGTGA 3′ | Flavobacterium | This study |
DAPI staining.
To stain cells with DAPI (4′,6-diamidino-2-phenylindole), theronts and tomonts were fixed in 3% paraformaldehyde in CFW at 4°C for 1 h. They were then washed in cold 50 mM HEPES (pH 7.5) by centrifugation at 500 × g for 30 s, pipetted onto microscope slides, air dried, incubated with 1 μg/ml DAPI in 50 mM HEPES for 4 min, and washed twice with 50 mM HEPES.
FISH.
Fluorescently labeled DNA probes were hybridized to bacterial 16S rRNAs using modifications of standard procedures (5). Approximately 4 × 103 I. multifiliis theronts in 100 μl of CFW were pipetted onto poly-l-lysine-treated slides, an equal volume of 8.0% aqueous paraformaldehyde was added, and theronts were fixed for 30 min at room temperature (RT). The theronts were washed twice with 10.1 mM NaH2PO4, 1.5 mM KH2PO4, 2.7 mM KCl, and 136 mM NaCl (phosphate-buffered saline [PBS]), pH 7.2, for 3 min; air dried for 1 to 2 h; and stored at −20°C. In preparation for FISH, samples were treated with 1.0% Triton X-100 in sterile distilled water for 3 min at RT, 100 μg/ml proteinase K in PBS for 60 min at 37°C, and 0.2% glycine in PBS for 3 min at RT; washed twice with PBS at RT for 5 min; dehydrated in a graded ethanol series (70%, 95%, and 100% for 3 min each); and air dried. The samples were blocked with 0.25 μg/ml yeast tRNA in 2× standard saline citrate (SSC), 0.3 M NaCl, and 0.03 M Na3C6H5O7 (pH 7.0) for 30 min at RT. Samples were incubated in hybridization solution consisting of 0.25 μg/ml yeast tRNA in 1.4× SSC, 30% formamide, and 5 ng/μl probe in the dark in a humidified chamber for 18 to 24 h at 48°C. Unbound probe was removed by washing slides in 2× SSC for 10 min at RT, 1× SSC for 10 min at 37°C, 0.3× SSC for 10 min at 48°C, and 0.3× SSC for 10 min at RT. Slides were air dried, covered with Vectashield mounting medium (Vector Laboratories), and stored in the dark. Theronts were counterstained with DAPI following FISH by covering with Vectashield mounting medium containing DAPI.
RNase treatment before FISH.
To treat theronts with RNase before hybridization of probes, they were processed for FISH through the proteinase K and wash steps. They were then incubated with 100 μg/ml RNase A and 25 U/ml RNase T1 in 300 mM NaCl, 10 mM Tris-Cl (pH 7.4) for 60 min at 37°C. Slides were then washed twice with 0.25 μg/ml yeast tRNA in 1.4× SSC, 30% formamide for 5 min at RT, dehydrated in ethanol, blocked with 0.25 μg/ml yeast tRNA in 2× SSC for 30 min at RT, and hybridized with EUB338 as before.
Photomicroscopy and image analysis.
For confocal microscopy, slides were examined using a Zeiss AxioImager M1 microscope fitted with an LSM510 Meta confocal scan head and a Plan Apo 100×/1.4 oil lens. DAPI was excited with a 30-mW Diode 405-nm laser, Cy3 with a 1.2-mW HeNe 543-nm laser, and Cy5 with a 5-mW HeNe 633-nm laser, and fluorescence signals were visualized with appropriate bandpass filters. Differential interference contrast (DIC) images were obtained using the 5-mW HeNe 633-nm laser. Images were collected with Laser Scanning Microscope LSM510 software (4.0 SP2) and processed with Zeiss LSM Image Browser software (Version 4.0.0.12). Zeiss LSM510 software was used to determine the colocalization of signals from EUB338 (Cy3) and either RICP or BACP (Cy5) in theronts treated with these probes, and Manders' weighted-colocalization coefficients were calculated (19). A value of zero indicates that no fluorescence in the Cy3 channel colocalizes with fluorescence from the Cy5 channel, whereas a value of 1 indicates that 100% of fluorescence in the Cy3 channel colocalizes with fluorescence from the Cy5 channel.
Digital photographs of RNase-treated theronts were taken with an Olympus Q Color-3 digital camera mounted on an Olympus BH-2 microscope using a Plan Apo 40×/0.85 lens.
Electron microscopy.
Theronts were collected by centrifugation at 500 × g for 2 min, resuspended in CFW, passed through a 40-μm-pore filter to remove extracellular debris, washed in CFW, and fixed in an equal volume of 4.0% glutaraldehyde, 4.0% paraformaldehyde, 0.4% picric acid, and 0.2 M cacodylate-HCl buffer (pH 7.2) at 4°C overnight. Theronts were washed in 0.1 M cacodylate-HCl buffer, embedded in 3% agar, postfixed in 1% OsO4 in 0.1 M cacodylate-HCl buffer (pH 7.25) for 1 h at RT, washed in distilled water, stained with 0.5% aqueous uranyl acetate for 1 h at RT, and washed and dehydrated in a graded ethanol series. Theronts were embedded in 1:1 propylene oxide-epon-araldite, cut into 60-nm sections, stained with 5% methanolic uranyl acetate and Reynold's lead citrate, and viewed in a JEM-1210 transmission electron microscope.
Nucleotide sequence accession numbers.
The sequences for the endosymbionts in this study have been deposited in GenBank under accession no. GQ870455 for Rickettsiales clone c1312 and GQ870456 for Sphingobacteria clone c134.
RESULTS
PCR, cloning, and sequencing.
Sequencing of the I. multifiliis strain G5 genome revealed the presence of DNA sequences of bacterial origin, including those with homology to Rickettsia (R. S. Coyne, 2009 [http://www.jcvi.org/cms/research/projects/ich/overview]). To determine if bacterial 16S rRNA gene sequences could be amplified from I. multifiliis DNA preparations, PCR primers targeting highly conserved regions of bacterial 16S rRNA genes were designed. PCR amplification of G5 DNA, isolated from tomonts, using these 16S rRNA gene primers, generated a DNA product of ∼1,460 bp, which agreed with the predicted size (data not shown). The 1,460-bp PCR product was recovered from agarose gels, ligated into the pCR8/GW/TOPO vector, and cloned. Plasmid DNA was isolated from six randomly selected clones and sequenced. BLASTN searches of the NCBI database with these sequences revealed that DNAs from three different classes of bacteria, including Alphaproteobacteria (Rickettsiales), Sphingobacteria (Emticicia), and Flavobacterium columnare, were present in the samples.
The G5 strain of I. multifiliis had been cultured in our laboratory since 1995. To determine if the bacteria associated with G5 were present in other strains of I. multifiliis, including I. multifiliis strains collected more recently from wild populations of fish, a second strain of I. multifiliis (G13) was isolated from a wild fish in 2008. G13 theronts were collected, and DNA was isolated. The DNA was PCR amplified as before and a 1,460-bp product was again generated. It was cloned, and plasmid DNA was isolated from 14 colonies. Sequencing of these plasmids demonstrated that 16S rRNA gene sequences from the same three bacterial classes found in G5 DNA preparations were also present in the G13 DNA preparation.
Phylogeny. (i) Alphaproteobacteria (Rickettsiales).
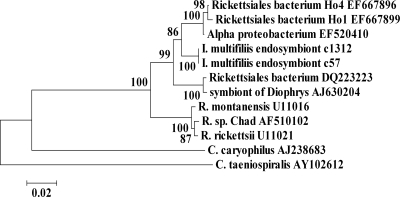
BLASTN searches showed that six clones (from G5, c56 and c57; from G13, c1311, c1312, c1314, and c1315) had high sequence similarity to 16S rRNA genes from Rickettsiales sp. Of these, c1312 and c1315, isolated from G13 theronts, had identical sequences and shared 99% sequence similarity with c57, isolated from G5 tomonts. These three clones had 96% sequence identity with 16S rRNA genes from two uncultured Rickettsiales bacterium clones, isolated from epithelial tissue of the cnidarian Hydra oligactis and an uncultured alphaproteobacterium obtained from a freshwater lake, and 95% similarity to 16S rRNA genes from an uncultured Rickettsiaceae endosymbiont of the marine ciliate Diophrys appendiculata (14, 21, 30). They had 93% to 94% similarity to type species of Rickettsia, such as R. rickettsia. Comparison by BLASTN to Paramecium endosymbionts demonstrated that they shared 86% sequence identity with the alphaproteobacterium C. caryophila, but only 78% sequence identity with the gammaproteobacterium C. taeniospiralis (5, 16, 22). Phylogenetic trees for clones c57 and c1312 were constructed using MEGA4 minimum evolution, neighbor-joining, and maximum parsimony algorithms (28). These methods all generated similar groupings of these clones and showed that clones c57 and c1312 cluster most closely with two uncultured Rickettsiales bacterium clones, isolated from epithelial tissue of the cnidarian H. oligactis and an uncultured alphaproteobacterium obtained from a freshwater lake, and less closely with the Paramecium endosymbionts (Fig. 1).
FIG. 1.
Phylogenetic relationships of I. multifiliis Rickettsiales endosymbionts c57 and c1312. Minimum evolutionary distance trees were calculated using 16S rRNA gene sequences. Relationships of I. multifiliis endosymbionts c57 and c1312 to representative Alphaproteobacteria, including uncultured bacteria isolated from Hydra (EF667896 and EF667899), an uncultured isolate of Alphaproteobacteria (EF520410) from a freshwater lake in the Adirondacks, Rickettsia sp., and two endosymbionts of Paramecium, C. caryophila (Alphaproteobacteria) and C. taeniospiralis (Gammaproteobacteria). Numbers at nodes are percentages of bootstrap values based on 1,000 samplings. The bar indicates percent sequence dissimilarity.
The second group of clones, including c56, c1311, and c1314, shared 95% to 99% sequence identity among themselves but only 90% to 93% similarity with c1312. These three clones were chimeras of Rickettsiales and F. columnare sequences. The sequences of these three clones and c1312 were 99% similar from bases 1 to 1025, but differed substantially over the last ∼450 bases. For c56, the sequence from base 996 to the 3′ terminus (base 1452) was 99% similar to that of bases 1018 to 1474 of the F. columnare 16S rRNA gene sequence determined in this study. Abridged Rickettsiales PCR products terminating around bases 996 to 1025 could potentially bind to their complementary sequence in F. columnare PCR products and serve as a primer in PCRs to generate the chimeric clones. Chimeric clones are commonly generated from DNA preparations containing multiple species (2).
(ii) Sphingobacteria.
BLASTN searches showed that four other clones had essentially identical sequences (from G5, c53; from G13, c134, c135, and c1313). They showed 94% identity with the 16S rRNA gene of uncultured Bacteroidetes Hv1.2 and 93% identity to uncultured Sphingobacteriales Hv1.25. The Hv1.25 and Hv1.2 bacteria were also isolated from the epithelia of Hydra, in this case, H. vulgaris (14). c134 also showed sequence identity with an uncultured Sphingobacteria isolate from a freshwater lake (21) and with three species of the little characterized genus Emticicia: 92% with E. oligotrophica (24), 89% with Emticicia sp. (7), and 89% with E. ginsengisoli (18).
The sequence of c134 was used to construct phylogenetic trees by minimum evolution, neighbor-joining, and maximum parsimony algorithms, all of which possessed similar topologies (28). All three algorithms grouped c134 most closely with the Bacteroidetes Hv1.2 and Sphingobacteriales Hv1.25 isolated from H. vulgaris, and the genus Emticicia (Fig. 2). Based on these data, these clones may represent a new species within the genus Emticicia and the class Sphingobacteria.
FIG. 2.
Phylogenetic relationships of I. multifiliis Sphingobacteria endosymbiont c134. Minimum evolutionary distance trees were calculated using 16S rRNA gene sequences. Relationships of I. multifiliis endosymbiont c134 with uncultured bacteria isolated from Hydra (EF667904 and EF667913), uncultured bacteria isolated from freshwater lakes (EF520595 and FJ801214), and Emticicia are shown. Numbers at nodes are percentages of bootstrap values based on 1,000 samplings. The bar indicates percent sequence dissimilarity.
(iii) Flavobacteria.
The other 10 clones had highly similar sequences and showed 97 to 99% identity with F. columnare 16S rRNA gene sequences (data not shown). F. columnare colonizes the skin of fish (29). The potential for contamination of I. multifiliis DNA preparations with F. columnare, or other bacteria from fish skin or aquarium water, was anticipated, but not to the extent found.
In situ identification.
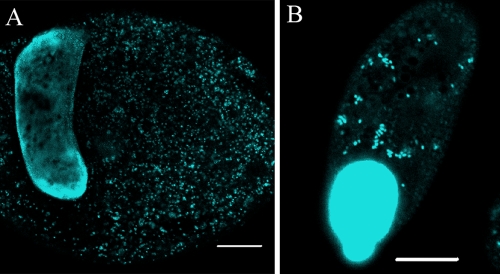
DAPI staining was used to determine if endosymbionts could be detected in I. multifiliis. Theronts and tomonts were stained by DAPI and examined by confocal microscopy, which revealed DAPI-positive endosymbionts in the cytoplasm, in addition to the brightly stained macronucleus and micronucleus. In theronts, DAPI-stained endosymbionts were concentrated in the middle and periphery of the theront, with fewer particles visible at the anterior or posterior ends. The number of endosymbionts detected by DAPI in theronts varied from a few to dense concentrations that prevented accurate counts of the number of endosymbionts. In tomonts, large numbers of DAPI-stained endosymbionts were distributed throughout the cytoplasm (Fig. 3).
FIG. 3.
Confocal images of an I. multifiliis G13 tomont and theront stained with DAPI. (A) DAPI-stained G13 tomont showing the macronucleus and endosymbionts (blue). Bar, 100 μm. (B) DAPI-stained G13 theront showing the macronucleus, micronucleus (merged with macronucleus), and endosymbionts (blue). Bar, 10 μm.
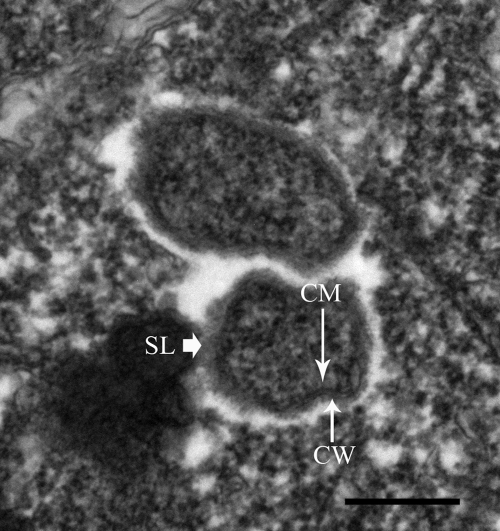
Transmission electron microscopy of sections of theronts showed oblong or rod-shaped endosymbionts distributed freely throughout the cytoplasm, singly or in groups of two or three (Fig. 4). Endosymbionts were not observed in vacuoles or nuclei and did not appear associated with cellular organelles. In our sections, the endosymbionts ranged from ∼0.35 to 0.43 by 0.53 to 0.99 μm in size, similar to Rickettsia (20, 25). They contained a granular cytoplasm, inner cytoplasmic membrane, outer envelope (cell wall), and a slime layer and were surrounded by a prominent, electron-translucent halo, characteristic of Rickettsia (20, 25).
FIG. 4.
Transmission electron micrograph of two endosymbionts in an I. multifiliis G13 theront. The inner cytoplasmic membrane (CM), outer envelope (CW), and slime layer (SL) are shown. Endosymbionts are surrounded by an electron-translucent halo. Bar, 300 nm.
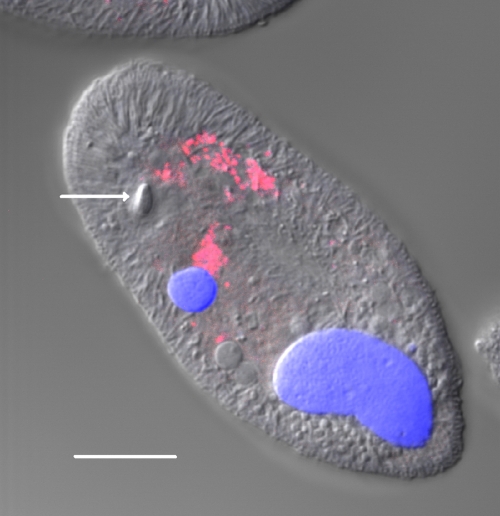
FISH was used to determine the distribution within theronts of the bacteria identified by sequencing. When I. multifiliis theronts were incubated with the EUB338 probe, which targets Bacteria, and poststained with DAPI, a hybridization pattern consistent with the DAPI staining was observed by confocal microscopy. EUB338-positive endosymbionts were primarily localized in the center of theronts and were less abundant near the anterior or posterior ends of cells. No intranuclear localization of the endosymbionts in either the micronucleus or the macronucleus of theronts was seen (Fig. 5). In contrast, no signal was detected in theronts when the EUBN probe, a noncomplementary, negative control probe that has the same sequence as 16S rRNA of Bacteria, was used for hybridization (not shown).
FIG. 5.
FISH analysis of an I. multifiliis G5 theront labeled with probe EUB338 and counterstained with DAPI. A confocal laser scanning image is shown. The merged DIC, DAPI, and FISH image shows endosymbionts labeled with EUB338 (red), DAPI-stained macronucleus and micronucleus (blue), and the organelle of Lieberkühn (arrow). Bar, 10 μm.
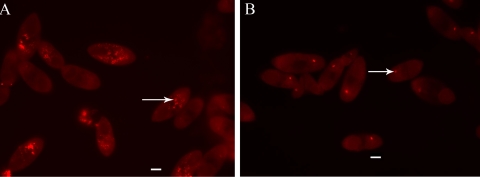
To confirm that the EUB338 signal resulted from hybridization of the probe with 16S rRNA, theronts were treated with 100 μg/ml RNase A and 25 U/ml RNase T1 prior to incubation with EUB338. This resulted in loss of the EUB338 signal, confirming that the EUB338 probe hybridized to 16S rRNA under these conditions, and demonstrated that these endosymbionts are transcribing 16S rRNA (Fig. 6). In some theronts, with or without RNase treatment, fluorescence was observed from the organelle of Lieberkühn, a photoreceptor found in some ciliates (10). This fluorescence appeared to result from autofluorescence or nonspecific interaction of probes with this organelle.
FIG. 6.
RNase treatment of I. multifiliis G5 theronts. (A) Endosymbionts labeled by FISH with probe EUB338 (red) are marked (arrow). (B) Theronts treated with RNase before hybridization with EUB338 do not show labeled endosymbionts. The organelle of Lieberkühn also showed fluorescence (arrow). Bar, 10 μm.
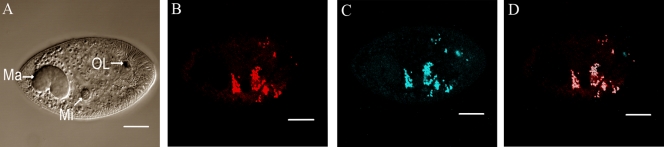
To identify the intracellular localization of the Rickettsia sp. in theronts, G13 theronts were incubated with both EUB338 and Rickettsiales-specific RICP, or the negative control RICN, and >50 theronts from two different experiments were examined by confocal microscopy. The pattern of hybridization with RICP was similar to that observed with EUB338. RICP-positive endosymbionts were found distributed throughout the central region of theronts and did not appear to cluster to specific cellular locations. No detectable staining of the nuclei was observed. When theronts were probed with both EUB338 and RICN, EUB338 hybridization to theronts was seen, but no hybridization with RICN was detected, confirming that the RICP signal resulted from specific hybridization of the RICP probe to Rickettsiales 16S rRNA. Merger of the EUB338 and RICP confocal images showed that a subset of the bacteria detected by EUB338 were also positive for RICP, in agreement with our sequencing results that multiple bacterial species are associated with I. multifiliis (Fig. 7). In these experiments, fluorescence from the organelle of Lieberkühn was also detected in the Cy5 channel.
FIG. 7.
FISH analysis of an I. multifiliis G13 theront labeled with probe EUB338 and the Rickettsiales-specific probe RICP. Confocal laser scanning images are shown. (A) DIC image of G13 theront. The macronucleus (Ma), micronucleus (Mi), and the organelle of Lieberkühn (OL) are indicated. (B) Median section of the same theront showing EUB338-labeled endosymbionts (red). (C) Median section of the same theront showing RICP-labeled endosymbionts (blue). (D) Merged image of panels B and C showing endosymbionts stained with both probes in white, bacteria labeled only with EUB338 in red, and autofluorescence from the organelle of Lieberkühn in blue. Bar, 10 μm.
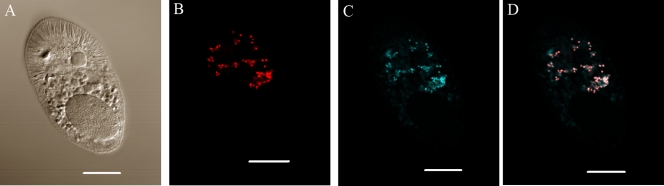
To identify the intracellular localization of Sphingobacteriales in theronts, G5 theronts were incubated with both EUB338 and the Sphingobacteriales-specific BACP or negative control BACN probe. BACP-positive endosymbionts in >50 theronts from two different experiments were detected by confocal microscopy. Endosymbionts were located in the central region of theronts, with fewer detected in the anterior or posterior regions of cells. No intranuclear staining was detected. When theronts were probed with both EUB338 and BACN, EUB338 hybridization was detected, but no BACN signal was observed. Merged images of EUB338- and BACP-stained theronts showed that a subset of the EUB338-positive bacteria were also positive for BACP (Fig. 8).
FIG. 8.
FISH analysis of an I. multifiliis G5 theront labeled with probe EUB338 and the Bacteroidetes-specific probe BACP. Confocal laser scanning images are shown. (A) DIC image of G5 theront. (B) Median section of the same theront showing EUB338-labeled endosymbionts (red); (C) median section of the same theront showing BACP-labeled endosymbionts (blue); (D) merged image of panels B and C showing endosymbionts stained with both probes in white, bacteria labeled only with EUB338 in red, and autofluorescence from the organelle of Lieberkühn in blue. Bar, 10 μm.
In contrast, no signal was detected when the FLAP or negative control FLAN probes, which were specific for F. columnare, were used for FISH. When theronts were incubated with EUB338 and FLAP, they were positive for EUB338, but no FLAP signal was detected. The lack of hybridization of FLAP to EUB338-positive theronts suggested that F. columnare was not an endosymbiont of I. multifiliis. EUB338 staining of extracellular bacteria associated with cilia was seen on some theronts, and DAPI-stained clumps of rod-shaped, extracellular bacteria associated with cilia and mucus on the surface of tomonts and theronts were also seen (not shown). F. columnare is found on the skin of fish. This data suggests that it is also associated with I. multifiliis cilia and was not removed by our washing protocols. These extracellular bacterial aggregates were probably the source of the F. columnare 16S rRNA gene that was amplified by PCR.
We used Manders' weighted colocalization coefficients to determine the extent of colocalization of the fluorescence signal from the Cy3-labeled EUB338 probe with that from the Cy5-labeled RICP or BACP probes (19). Single theronts were optically cross-sectioned in the z-dimension to determine the distribution of EUB338 and RICP or BACP signals in cross-sections of the entire theront. The mean weighted colocalization coefficients of the EUB338 signal were 0.39 ± 0.006 (mean ± standard deviation) with the RICP signal (n = 16) and 0.58 ± 0.09 with the BACP signal (n = 10). The combined values of the RICP and BACP signals accounted for all of the EUB338 signal in these cross-sectional analysis. We also determined the weighted colocalization coefficients from confocal images of single sections collected from theronts that had been selected for their strong RICP or BACP signals. In this case, the mean colocalization coefficients were 0.52 ± 0.24 for EUB338 with RICP (n = 6) and 0.9 ± 0.11 for EUB338 with BACP (n = 7). These samples from single sections overestimated the colocalization coefficients for either probe with the EUB338 signal compared to the cross-sectional analysis. The Manders' weighted colocalization coefficients determined from cross-sectional analysis of theronts demonstrated that Rickettsia accounted for 40% of the endosymbionts, while the Sphingobacteriales represented the other 60%, and confirmed that the Rickettsia and Sphingobacteriales are the only two endosymbionts found in I. multifiliis strain G5 or G13.
Not all I. multifiliis theronts have endosymbionts.
Not all theronts showed a positive EUB338 hybridization signal (Fig. 6). Theronts that lacked a EUB338 signal may have failed to hybridize with EUB338 or, alternatively, may harbor few or even no endosymbionts and thus showed no detectable signal following incubation with EUB338. To distinguish between these two possibilities, G5 and G13 theronts were stained with DAPI and examined by microscopy for DAPI-positive endosymbionts. A total of 336 G5 theronts were examined, of which 219 contained at least one DAPI-positive endosymbiont (65%). In G13, 362 theronts of 400 examined were DAPI positive (91%). These results demonstrated that endosymbionts were not found in all theronts.
DISCUSSION
There was no evidence to suggest the presence of endosymbionts in I. multifiliis until genome sequencing led to the identification of DNA sequences with homology to bacterial sequences (11, 12; R. S. Coyne, 2009 [http://www.jcvi.org/cms/research/projects/ich/overview]). These bacterial sequences could represent evidence for horizontal gene transfer into the I. multifiliis genome, or they could have originated from endosymbionts in I. multifiliis (17, 27). Our studies show that two different classes of bacteria, Alphaproteobacteria (Rickettsia) and Sphingobacteria, are found in the cytoplasm of I. multifiliis. Manders' weighted colocalization coefficients, calculated from z-section colocalization analysis of theronts, demonstrated that the signal from endosymbionts labeled with either the RICP or BACP probes colocalized with the signal from a subset of endosymbionts labeled with the EUB338 probe. The combined values for the weighted colocalization coefficients from the RICP and BACP probes account for all of the EUB338 signal. This confirmed that these Rickettsia and Sphingobacteriales are the only two bacterial endosymbionts found in I. multifiliis strains G5 and G13. In theronts, these endosymbionts appeared by FISH to have overlapping cytoplasmic distributions, primarily in the central region of the cell. DAPI staining of tomonts showed endosymbionts distributed throughout the cytoplasm. Endosymbionts were not detected in the macronucleus or micronucleus by FISH.
Six clones displayed sequence similarity to the Alphaproteobacteria and, more specifically, to the order Rickettsiales. Proteobacteria are endosymbionts of many different species of protozoa. The killer particles of Paramecium, first described by Sonneborn in 1938, and later extensively characterized by Preer and coworkers, include both Alphaproteobacteria and Gammaproteobacteria (4, 5, 16, 22, 26). Rickettsiales (Alphaproteobacteria) have been identified more recently in a wide variety of different protozoa, including acidophilic protists from acid mine drainages, a marine ciliate protozoan, D. appendiculata, and Acanthamoeba, a free-living amoeba (3, 16, 30). In addition, Rickettsiales are associated with the epithelia of the cnidarian Hydra (14). Minimum-evolutionary-distance phylogenetic trees and BLASTN searches indicated that the Rickettsiales endosymbionts in I. multifiliis showed the highest sequence similarity to Rickettsiales 16S rRNA genes isolated from two bacteria associated with epithelial tissue of H. oligactis.
Other clones had highest sequence similarity to the 16S rRNA genes of two members of the phylum Bacteroidetes (uncultured Bacteroidetes Hv1.2 and Sphingobacteriales Hv1.25), both of which were isolated from epithelial tissue of H. vulgaris (14). Thus, both I. multifiliis endosymbionts show the greatest 16S rRNA sequence similarity with bacteria associated with epithelial tissue of Hydra. The reason for this is unclear.
These Sphingobacteria clones also had ∼90% sequence similarity to the three described members of the genus Emticicia. The Emticicia species were isolated from freshwater ponds in India and Korea and a Korean ginseng field. They were not characterized as endosymbionts and are reported to grow on nutritionally poor media (7, 18, 24). This suggests that this I. multifiliis endosymbiont represents a novel and previously uncharacterized Sphingobacteria organism possibly related to the Emticicia.
The sequences of the other 10 clones were 97% to 99% similar to 16S rRNA genes of F. columnare. However, FISH analysis did not detect hybridization of the FLAP probe to EUB338-positive theronts, suggesting that F. columnare is not an intracellular endosymbiont of I. multifiliis. Calculation of Manders' weighted colocalization coefficients demonstrated that the combined value of the signal from the two endosymbionts labeled with the RICP and BACP probes accounted for all of the EUB338 signal. This confirmed that F. columnare is not an endosymbiont, but rather an extracellular contaminant.
F. columnare organisms are extracellular, gram-negative bacteria found on the skin and gills of fish. They are opportunistic pathogens that cause columnaris disease, a potentially fatal infection (29). Their potential presence, along with other bacteria commonly found in water samples used to collect I. multifiliis, was anticipated. Thus, the methods used for isolation of I. multifiliis theronts and tomonts were designed to minimize contamination of I. multifiliis with F. columnare and other environmental bacteria from the water used to collect tomonts and hatched theronts. I. multifiliis tomonts were individually collected and washed such that a minimal volume of water was transferred between washes. Theronts were collected by centrifugation and washed extensively in sterile water. Despite this, F. columnare clones represented 50% of the clones sequenced from tomont or theront DNA preparations. EUB338-positive bacteria were seen on cilia of theronts, and clumps of DAPI-positive extracellular bacteria were seen associated with cilia on DAPI-stained I. multifiliis theronts and tomonts. Thus, the association of F. columnare with I. multifiliis appears to be extracellular through interactions with cilia and enmeshment in mucus secreted by theronts and tomonts. Because of the close physical association of Flexibacter columnare with the surface of I. multifiliis, it is possible that the parasite serves as a carrier of bacteria to fish, including individuals that are susceptible to bacterial infection.
The DNA used to generate these clones was isolated from two different strains of I. multifiliis: G5, passaged in our laboratory since 1995, and G13, recently isolated from a wild fish and minimally passaged in our laboratory. However, DNA isolated from G5 tomonts and G13 theronts contained the same two endosymbionts in the same relative abundance. This suggests that these two endosymbionts are commonly found in I. multifiliis populations and are not lost even after long-term passage in the laboratory.
The physiological relationship between these endosymbionts and I. multifiliis is not understood. As all theronts do not contain detectable endosymbionts, the endosymbionts do not appear to play a critical role in supporting the growth of I. multifiliis, but their presence must also not be particularly detrimental to the growth of I. multifiliis. It is not known whether they play a role in the pathogenesis of I. multifiliis infections or if they affect the immune response of infected fish.
Acknowledgments
We thank W. Whitman for help with constructing phylogenetic trees, particularly Bacteroidetes, and M. Ard for help with electron microscopy.
This work was supported by USDA grant 2007-35600-18539 (D.H.-C., T.G.C., and R.S.C.), VMES grant 08-002 (R.C.F.), and a grant from the School of Life Sciences, Sun Yat-sen University (H.Y.S.).
Footnotes
Published ahead of print on 9 October 2009.
REFERENCES
- 1.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, B. J., P. Hugenholtz, S. C. Dawson, and J. F. Banfield. 2003. Extremely acidophilic protists from acid mine drainage host Rickettsiales-lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl. Environ. Microbiol. 69:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beale, G. H., and J. H. Preer, Jr. 2008. Paramecium genetics and epigentics. CRC Press, Boca Raton, FL.
- 5.Beier, C. L., M. Horn, R. Michel, M. Schweikert, H.-D. Goertz, and M. Wagner. 2002. The genus Caedibacter comprises endosymbionts of Paramecium spp. related to the Rickettsiales (Alphaproteobacteria) and to Francisella tularensis (Gammaproteobacteria). Appl. Environ. Microbiol. 68:6043-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius, J., M. L. Palmer, P. J. Kennedy, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, J. H., S. R. Jiang, J. C. Cho, J. Song, M. C. Lin, and W. M. Chen. 2008. Azonexus hydrophilus sp. nov., a nifH gene-harbouring bacterium isolated from freshwater. Int. J. Syst. Evol. Microbiol. 58:946-951. [DOI] [PubMed] [Google Scholar]
- 8.Clark, T. G., T. L. Lin, D. A. Jackwood, J. Sherrill, Y. Lin, and H. W. Dickerson. 1999. The gene for an abundant parasite coat protein predicts tandemly repetitive metal binding domains. Gene 229:91-100. [DOI] [PubMed] [Google Scholar]
- 9.Dickerson, H. W., and T. Clark. 1998. Ichthyophthirius multifiliis: a model of cutaneous infection and immunity in fishes. Immunol. Rev. 166:377-384. [DOI] [PubMed] [Google Scholar]
- 10.Ewing, M. S., and K. M. Kocan. 1992. Invasion and development strategies of Ichthyophthirius multifiliis, a parasitic ciliate of fish. Parasitol. Today 8:204-208. [DOI] [PubMed] [Google Scholar]
- 11.Ewing, M. S., K. M. Kocan, and S. A. Ewing. 1983. Ichthyophthirius multifiliis: morphology of the cyst wall. Trans. Am. Microsc. Soc. 102:122-128. [Google Scholar]
- 12.Ewing, M. S., K. M. Kocan, and S. A. Ewing. 1985. Ichthyophthirius multifiliis (Ciliophora) invasion of gill epithelium 1. J. Protozool. 32:305-310. [DOI] [PubMed] [Google Scholar]
- 13.Fokin, S. I. 2004. Bacterial endosymbionts of ciliophora and their interactions with the host cell. Int. Rev. Cytol. 236:181-249. [DOI] [PubMed] [Google Scholar]
- 14.Fraune, S., and T. C. G. Bosch. 2007. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. USA 104:13146-13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goertz, H. D. 2006. Symbiotic associations between ciliates and prokaryotes, p. 364-402. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. A handbook on the biology of bacteria: symbiotic associations, biotechnology, applied microbiology, 3rd ed. Springer, New York, NY.
- 16.Horn, M., T. R. Fritsche, R. K. Gautom, K. H. Schleifer, and M. Wagner. 1999. Novel bacterial endosymbionts of Acanthamoeba spp. related to the Paramecium caudatum symbiont Caedibacter caryophilus. Environ. Microbiol. 1:357-367. [DOI] [PubMed] [Google Scholar]
- 17.Huang, J., N. Mullapudib, T. Sicheritz-Pontenc, and J. C. Kissinger. 2004. A first glimpse into the pattern and scale of gene transfer in the Apicomplexa. Int. J. Parasitol. 34:265-274. [DOI] [PubMed] [Google Scholar]
- 18.Liu, Q. M., L. N. Ten, H. S. Yu, F. X. Jin, W. T. Im, and S. T. Lee. 2008. Emticicia ginsengisoli sp. nov., a species of the family ‘Flexibacteraceae’ isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 58:1100-1105. [DOI] [PubMed] [Google Scholar]
- 19.Manders, E. M. M., F. J. Verbeek, and J. A. Aten. 1993. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169:375-382. [DOI] [PubMed] [Google Scholar]
- 20.Munderloh, U. G., S. F. Hayes, J. Cummings, and T. J. Kurtti. 1998. Microscopy of spotted fever Rickettsia movement through tick cells. Microsc. Microanal. 4:115-121. [Google Scholar]
- 21.Percent, S. F., M. E. Frischer, P. A. Vescio, E. B. Duffy, V. Milano, M. McLellan, B. M. Stevens, C. W. Boylen, and S. A. Nierzwicki-Bauer. 2008. Bacterial community structure of acid-impacted lakes: what controls diversity? Appl. Environ. Microbiol. 74:1856-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Preer, J. R., Jr., and L. B. Preer. 1982. Revival of names of protozoan endosymbionts and proposal of Holospora caryophila nom. nov. Int. J. Syst. Bacteriol. 32:140-141. [Google Scholar]
- 23.Rosen, S., and H. J. Skaletsky. 2003. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 24.Saha, P., and T. Chakrabarti. 2006. Emticicia oligotrophica gen. nov., sp. nov., a new member of the family ‘Flexibacteraceae,’ phylum Bacteroidetes. Int. J. Syst. Evol. Microbiol. 56:991-995. [DOI] [PubMed] [Google Scholar]
- 25.Silverman, D. J., and C. L. Wisseman, Jr. 1978. Comparative ultrastructural study on the cell envelopes of Rickettsia prowazekii, Rickettsia rickettsia, and Rickettsia tsutsugamushi. Infect. Immun. 21:1020-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonnenborn, T. M. 1950. The cytoplasm in heredity. Heredity 4:11-36. [DOI] [PubMed] [Google Scholar]
- 27.Striepen, B., A. J. P. Pruijssers, J. Huang, C. Li, M. J. Gubbels, N. N. Umejiego, L. Hedstrom, and J. C. Kissinger. 2004. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc. Natl. Acad. Sci. USA 101:3154-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 29.Tripathi, N. K., K. S. Latimer, C. R. Gregory, B. W. Ritchie, R. E. Wooley, and R. L. Walker. 2005. Development and evaluation of an experimental model of cutaneous columnaris disease in koi (Cyprinus carpio). J. Vet. Diagn. Investig. 17:45-54. [DOI] [PubMed] [Google Scholar]
- 30.Vannini, C. G. Petroni, F. Verni, and G. Rosati. 2005. A bacterium belonging to the Rickettsiaceae family inhabits the cytoplasm of the marine ciliate Diophrys appendiculata (Ciliophora, Hypotrichia). Microb. Ecol. 49:434-442. [DOI] [PubMed] [Google Scholar]
- 31.Wang, X., T. G. Clark, J. Noe, and H. W. Dickerson. 2002. Immunisation of channel catfish, Ictalurus punctatus, with Ichthyophthirius multifiliis immobilisation antigens elicits serotype-specific protection. Fish Shellfish Immunol. 13:337-350. [DOI] [PubMed] [Google Scholar]
- 32.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yilmaz, L. Ş., H. E. Ökten, and D. R. Noguera. 2006. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl. Environ. Microbiol. 72:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]