Abstract
Genome annotation of the chlorinated ethene-respiring “Dehalococcoides ethenogenes” strain 195 indicated the presence of a complete nitrogenase operon. Here, results from long-term growth experiments, gene expression, and 15N2-isotope measurements confirm that strain 195 is capable of fixing atmospheric dinitrogen when a defined fixed-nitrogen source such as ammonium is unavailable.
“Dehalococcoides ethenogenes” strain 195 is the first isolated bacterium that is capable of reductively dechlorinating tetrachloroethene and trichloroethene (TCE) to vinyl chloride (VC) and ethene (22). Annotation of the 1.5-Mbp genome of strain 195 has identified 17 intact reductive dehalogenase (RDase) genes (25). The variety of RDases has essentially defined the metabolic capabilities of strain 195 and other Dehalococcoides strains for respiration of chlorinated ethenes (8, 9, 15, 23, 27) and other chlorinated compounds (1, 2, 6, 21), making them important participants in bioremediation processes (19). Expression of different putative RDase genes has been examined previously in pure culture (6) and in Dehalococcoides-containing enrichment cultures (3, 4, 13, 17, 24, 28).
Genome annotation of strain 195 has revealed the presence of a nitrogenase-encoding operon (nif) (DET1151-58) typical of those found in anaerobes (25). According to the published genome annotations of four strains of Dehalococcoides, strain 195 is the only one that contains a nif operon (16, 25; Joint Genome Institute, 2009, Integrated Microbial Genomes system [www.jgi.doe.gov]). A nif operon closely related to that in strain 195 has also been identified in a mixed Dehalococcoides-containing community (29); thus, the nitrogen-fixing function might be present in other unsequenced strains of Dehalococcoides.
Phylogenetically, the nitrogenase structural genes of strain 195 are clustered with diverse anaerobic Bacteria, including the molybdenum (Mo)-nitrogenase in Clostridium pasteurianum, as well as Archaea, including the Mo-nitrogenase in Methanosarcina barkeri (25, 30). In the genome of strain 195, the presence of an ABC transporter for molybdenum (DET1159-61) and a nifV gene (DET1614), which encodes homocitrate synthetase used in nitrogenase FeMo-cofactor biosynthesis, suggests that the nitrogenase is of the typical molybdenum-iron type (25). While strain 195 is the only sequenced Dehalococcoides isolate that contains a nif operon, Ju et al. (14) previously identified functional nifH genes in dechlorinating organisms from diverse genera such as Sulfurospirillum multivorans, Desulfovibrio dechloracetivorans, and Desulfomonile tiedjei.
Aquifers containing groundwater contaminated with chlorinated ethenes can potentially be limited in nutrients. For example, at the Wurtsmith Air Force Base, the chlorinated ethene-contaminated groundwater was found to contain less than 0.09 mM of ammonia, prompting ammonium amendment (26). Little is currently known about the potential effects of nitrogen limitation on reductive dechlorination in the environment, and the demonstration of nitrogen fixation in strain 195 was previously hindered by the use of an undefined medium (21). Here, we present results demonstrating that strain 195 is capable of fixing atmospheric dinitrogen and the physiological implications of the stress caused by nitrogen limitation.
Establishment of nitrogen-fixing D. ethenogenes strain 195.
To induce nitrogen fixation, strain 195 was grown in 100 ml liquid medium as described by He et al. (7), except with the following modifications: (i) NH4Cl and TES buffer [N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] were omitted, (ii) 0.2-μm-pore-filtered ultra-high-purity nitrogen gas was overpressurized into the bottle headspace, creating an H2/CO2/N2 ratio of 44:11:45 under 12 lb/in2 gauge pressure, and (iii) anoxic Na2MoO4·2H2O was added to increase the Mo concentration to 2.7 μM. After inoculation with 2 ml of cells that had previously been grown in ammonium-containing medium (5.6 mM), the first subcultures took ∼12 weeks to completely reduce the 40 μmol of TCE that was provided, coupling it with growth. Most of the incubation represented a lag period prior to growth. Subsequent subcultures (0.5% [vol/vol]) took 4 to 5 weeks for growth, indicating that cells had adapted to fixed-nitrogen limitation. Cultures of nitrogen-fixing strain 195 have now been growing in our laboratory for more than 3.5 years.
The purity of the nitrogen-fixing strain 195 cultures was assessed by microscopy and by sequencing the PCR products amplified with universal bacterial and archaeal primers (5, 10). In all cases, the archaeal primers returned no 16S rRNA band and the PCR products resulting from the bacterial primers generated sequences identical to the 16S rRNA gene of strain 195, demonstrating the purity of the culture.
15N2 incorporation.
The classic acetylene reduction assay (20) was not an applicable method to demonstrate nitrogen fixation in this study, as the diagnostic product ethene is a by-product of the respiratory activity of strain 195. Instead, the incorporation of labeled 15N2 into biomass was measured (n = 3) using a GVI IsoPrime mass spectrometer along with a Eurovector elemental analyzer (EuroEA3028-HT). After completely reducing 40 μmol of TCE, the N-isotope ratio of the biomass that was incubated with a 16.4 ± 0.70 atom% 15N headspace without ammonium amendment was 5.1 ± 0.69 atom% 15N, compared to 0.36 ± 0.0001 atom% 15N, close to natural abundance, for the biomass of control cultures grown with ammonium (5.6 mM). The biomass N-isotope ratio could be further enriched to 12.5 ± 0.67 atom% 15N when reducing agent cysteine (0.2 mM) was eliminated from the growth medium. A mass balance of the 15N in the headspace, medium, and biomass indicates that fixed atmospheric dinitrogen contributed approximately 29% and 75% of the nitrogen in the biomass pool in the presence and absence of cysteine, respectively. The significant difference in 15N enrichment in the biomass compared with ammonium controls confirms that strain 195 can grow by fixing atmospheric dinitrogen and the extent of nitrogen fixation is governed by the availability of fixed-nitrogen sources including cysteine.
Growth and dechlorination activity while fixing nitrogen.
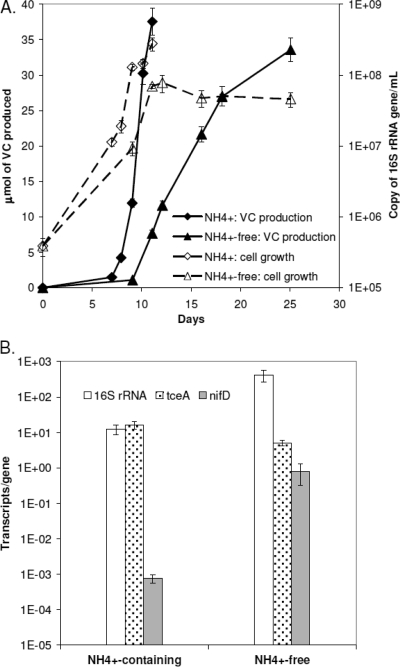
After achieving stable growth of nitrogen-fixing strain 195 cultures, experiments were carried out to characterize the effects of nitrogen fixation on growth and activity. Figure 1A shows the growth and dechlorination profile of strain 195 with and without 5.6 mM ammonium amendment. For the cells challenged to fix nitrogen, two distinct growth phases were observed, an exponential phase (days 0 to 11) followed by a long stationary phase (days 11 to 25). Transition into stationary phase after day 11 when less than 20% of the TCE had been reduced to VC was accompanied by continued dechlorination, exhibiting decoupling between growth and TCE-dechlorinating activity and generating limited energy for cell maintenance. These growth and dechlorination characteristics were consistently observed in repetitions of this experiment. In contrast, when ammonium was present as a fixed-nitrogen source, exponential growth continued until almost all of the TCE had been dechlorinated to VC on day 11.
FIG. 1.
(A) Dechlorination activity plotted as VC production and cell growth for strain 195 in medium with and without ammonium amendment. Cell growth data are averages of triplicate RT-qPCR analyses from each biological duplicate, and error bars represent 1 standard deviation. VC measurements are averages of biological duplicates, and error bars represent 5% uncertainty. The RT-qPCR method for 16S rRNA gene measurement has previously been described (11), and the chlorinated ethene concentrations were determined by gas chromatography-flame ionization detection (17). (B) Late-exponential-phase 16S rRNA, tceA, and nifD gene expression for cells growing with (day 10) and without (day 11) ammonium amendment. The results are averages of triplicate RT-qPCRs from each biological duplicate, and error bars represent 1 standard deviation.
The cell density normalized rate of VC production and cell yield during exponential phase were higher in the presence of ammonium. The VC production rates during exponential phase in the presence and absence of ammonium were 10.0 ± 1.2 μmol VC/day/1010 cells and 6.7 ± 1.2 μmol VC/day/1010 cells, respectively, and yields were (4.7 ± 0.5) × 108 cells/μmol Cl− and (2.8 ± 0.8) × 108 cells/μmol Cl−. Furthermore, a sixfold difference in final cell density between the ammonium-free culture [(4.6 ± 1.0) × 107 copies of gene/ml] and the ammonium-amended culture [(2.8 ± 0.6) × 108 copies of gene/ml] was measured.
Expression of nifD, tceA, and 16S rRNA genes.
In order to evaluate the regulatory impact of fixed-nitrogen limitation on strain 195, expression of the nifD gene as well as the 16S rRNA and tceA genes was measured in the presence and absence of ammonium amendment by multiplex reverse transcription-quantitative PCR (RT-qPCR) as previously described (12). Total RNA was isolated from frozen cell pellets using the RNeasy mini kit (Qiagen, CA). RNA standards were synthesized previously (12, 18) or according to previously outlined methods (18). The nifD gene (DET1155) encodes the nitrogenase molybdenum-iron protein α-chain and the primer and probe sequences are as follows: forward primer, 5′-AACTGGCCCAGGCTATAGAAGA; reverse primer, 5′-CCCACCGGACAGGTAGCAT; and probe 5′-6-carboxyfluorescein-ACCAGCTTTTCAAGCCCAAGACCATAGTTGT-6-carboxytetramethylrhodamine. Other primers and probe sequences have been published previously (11, 12).
Comparison of transcript quantities per gene during late exponential phase for the cultures shown in Fig. 1A shows that the nifD gene was upregulated 1,000-fold when ammonium was not provided (Fig. 1B), indicating that expression of the nif operon increases in response to fixed-nitrogen limitation and suggesting that cells were fixing nitrogen prior to stationary phase. In contrast, expression of the tceA gene was similar between the two conditions, while expression of 16S rRNA per gene copy was higher for cells that were fixing nitrogen, suggesting a high level of metabolic activity for these cells prior to stationary phase.
Effect of ammonium on cell growth, activity, and expression.
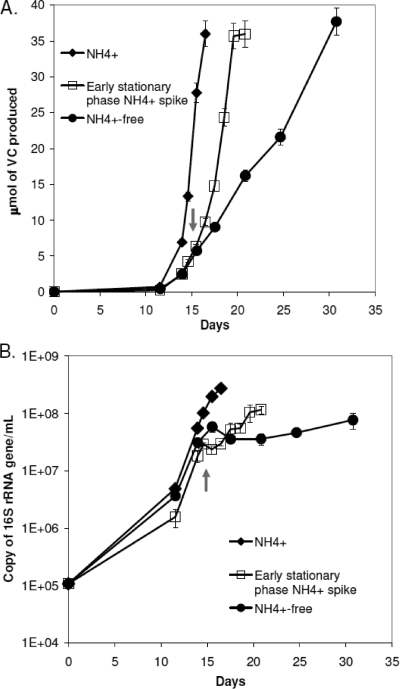
Following observations that ammonium deprivation consistently caused reduced growth and activity, a study was carried out to determine the cellular responses of adding ammonium to cultures that were actively growing while fixing nitrogen (Fig. 2). As shown in Fig. 2, ammonium addition after nitrogen-fixing cells had just entered stationary phase (day 15) had an immediate stimulatory effect on both VC production activity (Fig. 2A) and cell growth (Fig. 2B). For VC production, rather than requiring 15 days to reduce the remainder of the TCE to VC after transitioning into stationary phase, only 5 days were required after ammonium was spiked into the nitrogen-fixing cultures (Fig. 2A). An equivalent experiment with ammonium addition on day 18, after cells were further into stationary phase, showed similar activity with increases in both VC production and cell growth.
FIG. 2.
Effects of ammonium addition on dechlorination activity (A) and cell growth (B) in ammonium-free cultures. The arrow indicates the time (day 15) when ammonium was added to the ammonium-free cultures.
The increase in dechlorination activity was accompanied by a return to exponential cellular growth (Fig. 2B), with cell densities increasing fivefold from (2.3 ± 0.4) × 107 copies of gene/ml to (1.2 ± 0.2) × 108 copies of gene/ml within 5 days of ammonium amendment. An equivalent cellular increase to (1.4 ± 0.07) × 108 copies of gene/ml was observed for the day 18 amendment. The yield of (1.1 ± 0.2) × 108 cells/μmol Cl− after ammonium addition on day 15 was slightly lower than the yield of (1.6 ± 0.3) × 108 cells/μmol Cl− during the first exponential phase, but nevertheless, demonstrated a return to active growth. As a comparison, in the culture that was not amended with ammonium, cell density changed by a factor of only 1.3 after day 15. The increases in growth after ammonium amendment are likely the result of cells diverting energy and electrons previously used for nitrogen fixation to other metabolic processes.
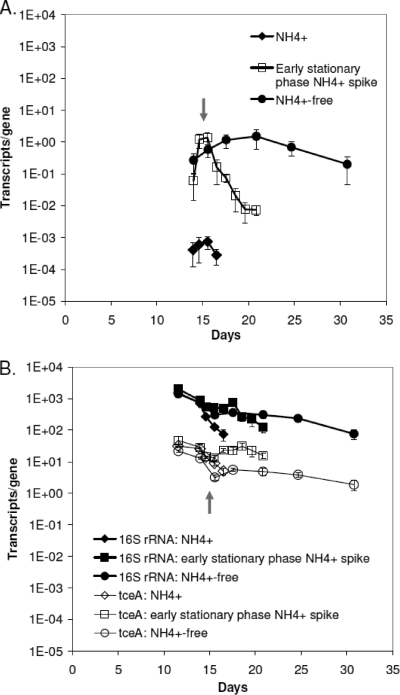
Ammonium amendment also resulted in changes in nifD gene expression (Fig. 3A). In the ammonium-free cultures, cells expressed nifD at quantities that were 3 orders of magnitude greater than cultures amended with ammonium, and nifD expression was relatively constant throughout the stationary phase. In contrast, cells that were grown initially ammonium free and amended with ammonium on day 15 exhibited a marked decrease in nifD expression in response to the amendment. A similar expression profile was exhibited by the day 18-amended cells. In the 24 h following the ammonium amendment, the nifD expression decreased by factors of 6 and 8 in the two ammonium-free cultures, with eventual downregulation reaching 2 orders of magnitude, suggesting that the nif operon is regulated according to the availability of ammonium as a fixed-nitrogen source. Conversely, the tceA and 16S rRNA genes were expressed at similar levels regardless of whether cells were provided with ammonium as a fixed-nitrogen source, with transcript number per gene declining as cells transitioned from exponential into stationary phase (Fig. 3B). Overall, no order-of-magnitude changes in gene expression were measured for tceA and the 16S rRNA genes.
FIG. 3.
nifD expression (A) and 16S rRNA gene and tceA expression (B) when cells were grown with or without ammonium amendment and the response to abrupt ammonium addition. The arrow indicates when the nitrogen-fixing cultures were amended with ammonium (day 15). No time zero point sample was collected for any transcription analysis, and nifD transcripts were unquantifiable at day 11.5 in all cultures. The analytical detection limit of nifD transcripts is 10−4 transcripts/gene as determined using the described RT-qPCR method and serial dilution of the quantified RNA standards.
Implications.
The subsurface matrix at many contaminated aquifer sites is oligotrophic with low levels of carbon and nutrients. Here we have shown that while D. ethenogenes strain 195 can fix nitrogen to sustain growth, this activity is accompanied by decreased cell density and dechlorination activity that may adversely affect the performance of bioremediation processes. Hence, the concentrations of fixed nitrogen and expression of genes related to nitrogen metabolism could serve as useful biomarkers for effective bioremediation.
Acknowledgments
We thank Wenbo Yang and Donald Herman of the University of California, Berkeley, for their expertise in N-isotope analysis.
This research was supported by the NIEHS Superfund Basic Research Project ES04705-19 and SERDP grant ER-1587.
Footnotes
Published ahead of print on 9 October 2009.
REFERENCES
- 1.Adrian, L., S. K. Hansen, J. M. Fung, H. Görisch, and S. H. Zinder. 2007. Growth of Dehalococcoides strains with chlorophenols as electron acceptors. Environ. Sci. Technol. 41:2318-2323. [DOI] [PubMed] [Google Scholar]
- 2.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Amos, B. K., K. M. Ritalahti, C. Cruz-Garcia, E. Padilla-Crespo, and F. E. Löffler. 2008. Oxygen effect on Dehalococcoides viability and biomarker quantification. Environ. Sci. Technol. 42:5718-5726. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S., M. F. Azizian, P. J. McMurdie, A. Sabalowsky, M. E. Dolan, L. Semprini, and A. M. Spormann. 2008. Monitoring abundance and expression of “Dehalococcoides” species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Appl. Environ. Microbiol. 74:5695-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodie, E. L., T. Z. DeSantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fung, J. M., R. M. Morris, L. Adrian, and S. H. Zinder. 2007. Expression of reductive dehalogenase genes in Dehalococcoides ethenogenes strain 195 growing on tetrachloroethene, trichloroethene, or 2,3-dichlorophenol. Appl. Environ. Microbiol. 73:4439-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He, J., V. F. Holmes, P. K. H. Lee, and L. Alvarez-Cohen. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73:2847-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He, J., K. M. Ritalahti, K. L. Yang, S. S. Koenigsberg, and F. E. Löffler. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62-65. [DOI] [PubMed] [Google Scholar]
- 9.He, J., Y. Sung, R. Krajmalnik-Brown, K. M. Ritalahti, and F. E. Löffler. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442-1450. [DOI] [PubMed] [Google Scholar]
- 10.Hershberger, K. L., S. M. Barns, A. L. Reysenbach, S. C. Dawson, and N. R. Pace. 1996. Wide diversity of Crenarchaeota. Nature 384:420. [DOI] [PubMed] [Google Scholar]
- 11.Holmes, V. F., J. He, P. K. H. Lee, and L. Alvarez-Cohen. 2006. Discrimination of multiple Dehalococcoides strains in a trichloroethene enrichment by quantification of their reductive dehalogenase genes. Appl. Environ. Microbiol. 72:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, D. R., P. K. H. Lee, V. F. Holmes, and L. Alvarez-Cohen. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 71:3866-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, D. R., P. K. H. Lee, V. F. Holmes, A. C. Fortin, and L. Alvarez-Cohen. 2005. Transcriptional expression of the tceA gene in a Dehalococcoides-containing microbial enrichment. Appl. Environ. Microbiol. 71:7145-7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju, X. F., L. P. Zhao, and B. L. Sun. 2007. Nitrogen fixation by reductively dechlorinating bacteria. Environ. Microbiol. 9:1078-1083. [DOI] [PubMed] [Google Scholar]
- 15.Krajmalnik-Brown, R., T. Hölscher, I. N. Thomson, F. M. Saunders, K. M. Ritalahti, and F. E. Löffler. 2004. Genetic identification of a putative vinyl chloride reductase in Dehalococcoides sp. strain BAV1. Appl. Environ. Microbiol. 70:6347-6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kube, M., A. Beck, S. H. Zinder, H. Kuhl, R. Reinhardt, and L. Adrian. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269-1273. [DOI] [PubMed] [Google Scholar]
- 17.Lee, P. K. H., D. R. Johnson, V. F. Holmes, J. He, and L. Alvarez-Cohen. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl. Environ. Microbiol. 72:6161-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, P. K. H., T. W. Macbeth, K. S. Sorenson, Jr., R. A. Deeb, and L. Alvarez-Cohen. 2008. Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol. 74:2728-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löffler, F. E., and E. A. Edwards. 2006. Harnessing microbial activities for environmental cleanup. Curr. Opin. Biotechnol. 17:274-284. [DOI] [PubMed] [Google Scholar]
- 20.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Brock biology of microorganisms, 9th ed. Prentice-Hall, Inc., Upper Saddle River, NJ.
- 21.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maymó-Gatell, X., Y. T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 23.Müller, J. A., B. M. Rosner, G. von Abendroth, G. Meshulam-Simon, P. L. McCarty, and A. M. Spormann. 2004. Molecular identification of the catabolic vinyl chloride reductase from Dehalococcoides sp. strain VS and its environmental distribution. Appl. Environ. Microbiol. 70:4880-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahm, B. G., R. M. Morris, and R. E. Richardson. 2006. Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes. Appl. Environ. Microbiol. 72:5486-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seshadri, R., L. Adrian, D. E. Fouts, J. A. Eisen, A. M. Phillippy, B. A. Methe, N. L. Ward, W. C. Nelson, R. T. Deboy, H. M. Khouri, J. F. Kolonay, R. J. Dodson, S. C. Daugherty, L. M. Brinkac, S. A. Sullivan, R. Madupu, K. E. Nelson, K. H. Kang, M. Impraim, K. Tran, J. M. Robinson, H. A. Forberger, C. M. Fraser, S. H. Zinder, and J. F. Heidelberg. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105-108. [DOI] [PubMed] [Google Scholar]
- 26.Skubal, K. L., M. J. Barcelona, and P. Adriaens. 2001. An assessment of natural biotransformation of petroleum hydrocarbons and chlorinated solvents at an aquifer plume transect. J. Contam. Hydrol. 49:151-169. [DOI] [PubMed] [Google Scholar]
- 27.Sung, Y., K. M. Ritalahti, R. P. Apkarian, and F. E. Löffler. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl. Environ. Microbiol. 72:1980-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waller, A. S., R. Krajmalnik-Brown, F. E. Löffler, and E. A. Edwards. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl. Environ. Microbiol. 71:8257-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.West, K. A., D. R. Johnson, P. Hu, T. Z. DeSantis, E. L. Brodie, P. K. H. Lee, H. Feil, G. L. Andersen, S. H. Zinder, and L. Alvarez-Cohen. 2008. Comparative genomics of “Dehalococcoides ethenogenes” 195 and an enrichment culture containing unsequenced “Dehalococcoides” strains. Appl. Environ. Microbiol. 74:3533-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]