Abstract
Seafood consumption-related diarrhea became prevalent in Chile when the pandemic strain of Vibrio parahaemolyticus serotype O3:K6 reached a region in the south of Chile (Region de los Lagos) where approximately 80% of the country's seafood is produced. In spite of the large outbreaks of clinical infection, the load of V. parahaemolyticus in shellfish of this region is relatively low. The pandemic strain constitutes a small but relatively stable group of a diverse V. parahaemolyticus population, composed of at least 28 genetic groups. Outbreaks in Region de los Lagos began in 2004 and reached a peak in 2005 with 3,725 clinical cases, all associated with the pandemic strain. After 2005, reported cases steadily decreased to a total of 477 cases in 2007. At that time, 40% of the clinical cases were associated with a pandemic strain of a different serotype (O3:K59), and 27% were related to V. parahaemolyticus isolates unrelated to the pandemic strain. In the results published here, we report that in the summer of 2008, when reported cases unexpectedly increased from 477 to 1,143, 98% of the clinical cases were associated with the pandemic strain serotype O3:K6, a change from 2007. Nevertheless, in 2009, when clinical cases decreased to 441, only 64% were related to the pandemic strain; the remaining cases were related to a nonpandemic tdh- and trh-negative strain first identified in shellfish in 2006. Overall, our observations indicate that the pandemic strain has become a relatively stable subpopulation and that when the number of diarrhea cases related to the pandemic strain is low, previously undetected V. parahaemolyticus pathogenic strains become evident.
Diarrhea associated with seafood consumption is caused primarily by pathogenic V. parahaemolyticus. This species includes marine bacterial strains, only a few of which are pathogenic in humans (13). The load of pathogenic strains in shellfish depends on physical environmental variables, such as temperature and salinity, and on biological variables including the presence of protozoan predators, competing nonpathogenic bacteria, and bacteriophages capable of killing V. parahaemolyticus (21). Therefore, diarrhea outbreaks caused by V. parahaemolyticus are mainly an environmental problem. Records of the Public Health Institute of Chile indicate that from 1992 to 1997 diarrhea cases related to seafood consumption were not widespread in Chile in spite of the large consumption of raw shellfish. Cases of seafood-related diarrhea increased greatly with the arrival of the pandemic strain O3:K6, originally observed in Southeast Asia (9). This strain corresponds to a clonal complex. The clonal nature of the V. parahaemolyticus pandemic isolates obtained worldwide has been ascertained by the high degree of similarity among their genomes. This comparison includes the presence of specific genetic markers and similarity of the restriction patterns of their genomes, demonstrated by genome restriction fragment length polymorphism-pulsed-field gel electrophoresis (22), direct genome restriction enzyme analysis (DGREA) (8), arbitrarily primed PCR (15, 18), and multilocus sequence typing (6, 10). Characteristics of isolates of the O3:K6 pandemic clone are the O3:K6 antigens, a distinctive toxRS sequence (toxRSnew) (15), orf8 (17) and tdh genes, and the absence of the trh gene found in some pathogenic strains. However, numerous serovariants have apparently emerged since 1996 (16). Genome sequencing of the RIMD 2210633 pandemic strain revealed two sets of gene clusters encoding a type III secretion system apparatus, one in each of its two chromosomes (14).
Since 2004, we have characterized the strains of V. parahaemolyticus in both clinical cases and shellfish in a southern region of Chile (Region de los Lagos) in an effort to understand the proliferation of the pathogenic strains in the environment (7, 8, 11). Region de los Lagos extends from 40°13′S to 44°3′S and produces approximately 80% of the seafood in Chile (Anuario 2008 Sernapesca [http://www.sernapesca.cl]). It is generally accepted that the seafood from this region causes most of the clinical cases of V. parahaemolyticus-associated diarrhea observed in the entire country. The large diarrhea outbreaks related to seafood consumption started in this region in 2004. In 2005, cases reported by the Ministry of Health reached a peak of 3,600 and 10,984 in Region de los Lagos and the whole country, respectively. Since then, the number of cases has oscillated between 450 and 1,100 cases annually in Region de los Lagos and between 1,500 and 3,500 in the country as a whole (19).
Until 2007, more than 95% of the cases were related to the classical pandemic V. parahaemolyticus strain O3:K6 (7, 8). Variants of the pandemic strain were recovered in the summer of 2007, when the outbreaks diminished to 477 reported cases in Region de los Lagos. That year, many cases were caused by a new serovar of the pandemic strain, O3:K59 (11). This same year, a larger percentage of cases analyzed (27%) were due to nonpandemic strains. Some of these last cases corresponded to a strain apparently generated by transference of the pathogenicity island containing the type III secretion island from the pandemic clone to an indigenous V. parahaemolyticus strain (11). Another example of interactions between the pandemic strain and native microflora is the finding of variants containing a 42-kb plasmid corresponding to a telomeric temperate phage (24). The observations in 2007 suggested that the changes in the epidemiology of seafood-related diarrhea represented an inflection point in outbreak trends and a decreased prevalence of the pandemic strain in clinical cases. We present here the results of the analysis of V. parahaemolyticus in clinical cases and shellfish samples obtained during the summer of 2008, when reported cases unexpectedly increased from 477 to 1,143, and the summer of 2009, when clinical cases decreased to 441 (http://epi.minsal.cl/epi/html/elvigia/elvigia.htm). The number of cases observed in 2009 was the lowest since the beginning of large outbreaks in 2004. Overall, our observations illustrate the dynamics of V. parahaemolyticus population in outbreaks of diarrhea. They show the following: (i) that the pandemic strain has become a relatively stable subpopulation of the V. parahaemolyticus population in shellfish, (ii) that pandemic strain variants have emerged, and (iii) that V. parahaemolyticus pathogenic strains unrelated to the pandemic strains become evident when the number of diarrhea cases due to the pandemic strain are low. These data will be helpful in the understanding of V. parahaemolyticus ecology and improving the risk analysis of seafood related diarrhea.
MATERIALS AND METHODS
Strains.
V. parahaemolyticus RIMD 2210633 (also called VpKX) was obtained from the Research Institute for Microbial Diseases, Osaka University, Osaka, Japan. Strains identified with the prefix PMC (see Table 1) correspond to isolates from clinical samples obtained from people seeking attention at the Hospital Regional de Puerto Montt. The last digit of the designation corresponds to the year of isolation. The environmental strains, identified by the prefix PMA (see Table 1), were obtained from shellfish samples taken during the season in which outbreaks occurred (December to March). Isolates from preceding years have been described previously (7, 8, 11).
TABLE 1.
Properties of V. parahaemolyticus clinical isolates collected during the summers of 2008 and 2009 in Puerto Montt, Chile
| V. parahaemolyticus isolate(s) by year and isolate typea | Genetic profileb |
Serotype | DGREA profile | 42-kb phageb | ||||
|---|---|---|---|---|---|---|---|---|
| tlh | tdh | trh | orf8 | toxRSnew | ||||
| Isolates from 2008 | ||||||||
| Classical pandemic isolates | ||||||||
| PMC3.8, 7.8, 12.8, 15.8, 17.8, 20.8, 24.8, 26.8, 30.8, 34.8, 41.8, 42.8, 45.8, 47.8, 2.8, 6.8, 8.8, 9.8, 11.8, 13.8, 16.8, 18.8, 19.8, 21.8,23.8, 29.8, 31.8, 32.8, 33.8, 35.8, 37.8, 43.8, 44.8,46.8 | + | + | − | + | + | O3:K6 | KX | − |
| Other pandemic isolates | ||||||||
| PMC1.8, 10.8, 22.8, 36.8, 38.8, 48.8, 49.8, 50.8 | + | + | − | + | + | O3:K6 | KX | + |
| PMC4.8, 14.8 | + | + | − | + | − | O3:K6 | KX | − |
| PMC5.8 | + | + | − | + | + | O3:KUT | KX | − |
| Nonpandemic isolate | ||||||||
| PMC39.8 | + | − | − | − | − | OUT:KUT | 39.8 | − |
| Isolates from 2009 | ||||||||
| Classical pandemic isolates | ||||||||
| PMC31.9, 33.9, 47.6, 64.9, 69.9, 70.9, 72.9 | + | + | − | + | + | O3:K6 | KX | − |
| Other pandemic isolates | ||||||||
| PMC29.9, 51.9 | + | + | − | + | + | O3:K6 | KX | + |
| Nonpandemic isolates | ||||||||
| PMC25.9, 34.9, 40.9, 41.9, 44.9 | + | − | − | − | − | O3:KUT | 34.6 | − |
Underlining, DGREA performed; italics, serotyping not performed.
+, present; −, absent.
Analysis.
Samples from clinical cases and shellfish were obtained and analyzed as described previously (8). Briefly, samples of shellfish soft tissue were enriched for V. parahaemolyticus in three-tube serial dilutions in alkaline peptone water for assessment of bacterial load by the most probable number (MPN) method; tubes with bacterial growth were tested for tlh, tdh, or trh by multiplex PCR (2). Total and pandemic V. parahaemolyticus loads were calculated according to the number of tubes positive for tlh and for tdh and trh, respectively. Positive enrichment tubes were plated on CHROMagar Vibrio (CHROMagar Microbiology, Paris, France), and bacterial colonies with the morphology and color expected for V. parahaemolyticus were purified. Isolates were characterized for different properties, as described previously: the O and K antigens of the V. parahaemolyticus strains were determined by slide agglutination with rabbit antiserum obtained from Seiken (Denka Seiken Co., Ltd., Tokyo, Japan), as described by the supplier; PCR assays were performed for tlh, tdh, and trh (2); orf8 (17); and toxRSnew (15). PCR was performed using approximately 10 ng of total bacterial DNA per reaction tube. DGREAs were performed as described previously (8). Each of the DGREA patterns found in 2008 and 2009 was compared to patterns described in previous years, and when similarities were observed, their identities were evaluated by comparing the patterns obtained during the same electrophoresis run. The presence of the 42-kb plasmid was examined by alkaline extraction and electrophoresis in agarose gel, as described previously (24).
RESULTS
V. parahaemolyticus associated with clinical cases in 2008 and 2009.
Isolates from diarrhea cases that occurred in Region de los Lagos during the summers of 2008 and 2009 were analyzed and grouped according to serotype, the presence of genetic markers (orf8, toxRSnew, tlh, tdh, and trh), and the distinctiveness of their DGREA patterns. Forty-six cases were analyzed in 2008, and 14 were analyzed in 2009. One isolate from each patient was characterized (Table 1). In 2008, isolates from 45 cases (98%) corresponded to the pandemic clonal group according to the genetic markers and their DGREA patterns. That year 11 isolates from this group could be differentiated from the classical pandemic strain: eight contained a 42-kb plasmid consisting of a previously described telomeric prophage (24), two failed to PCR amplify toxRSnew, and one did not contain the classical K6 antigen but instead showed a K antigen that did not react with any of the antisera provided by Denka Seiken. The single nonpandemic isolate obtained in 2008 from a clinical case was negative for the genes associated with pathogenicity, tdh and trh. In contrast to the observations in 2008, only 64% of the 14 cases observed in 2009 were associated with the pandemic strain. The nonpandemic isolates obtained from the other five cases lacked the pathogenesis-related genes tdh and trh, and all corresponded to the same DGREA group 34.6. This group, observed in relative abundance in shellfish since 2006, had not been previously observed in clinical cases of infection. Two of the nine pandemic isolates contained the 42-kb telomeric prophage.
V. parahaemolyticus associated with shellfish.
Twenty-seven and 17 shellfish samples were analyzed in 2008 and 2009, respectively. V. parahaemolyticus enrichment parallel serial dilutions of the soft meat in alkaline peptone water were tested for MPN estimation. The presence of tlh, which is specific to the species, and tdh and trh, which are associated with pathogenic strains, was tested by multiplex PCR (2). In 2008, V. parahaemolyticus (tlh+) was detected in every sample; only eight contained tdh. None of the samples contained trh. The MPN of tdh-positive bacteria in the shellfish fluctuated from undetectable (<0.3 g−1) to 24 g−1. On the other hand, total V. parahaemolyticus (tlh+) ranged from 1.5 g−1 to >110 g−1, generally 10 to 100 times higher than pandemic isolates (tdh+). In 2009, V. parahaemolyticus was detected in only 14 of the 17 samples, and tdh was found in only 4. None of the samples contained trh. The load of tdh-positive bacteria fluctuated from undetectable to 1.1 g−1, while total V. parahaemolyticus ranged from <0.3 g−1 to 24 g−1. In general, samples from 2009 contained a smaller load of V. parahaemolyticus than those from 2008.
Single colonies were obtained from the enrichment cultures and characterized after purification; their properties are described in Table 2. Isolates positive for tdh were obtained from only four of the eight tdh-positive samples observed in 2008 and from none of four tdh-positive samples observed in 2009. Failure to obtain tdh-positive isolates from tdh-positive enrichments was probably due to their low frequency among total V. parahaemolyticus bacteria. Only 4 to 10 colonies were tested for tdh from each sample, and among these the probability of finding isolates present in a proportion lower than 1:10 was very low. Twenty and 12 pandemic and nonpandemic isolates obtained from shellfish in 2008 and 2009, respectively, were characterized in detail for the presence of genetic markers (orf8, toxRSnew, tlh, tdh, and trh) and the distinctiveness of their DGREA patterns. Serotyping was performed only for selected isolates (Table 2). The three tdh-positive isolates obtained in 2008 showed the characteristic genetic markers and DGREA pattern of the pandemic strain. None of these isolates contained the 42-kb plasmid.
TABLE 2.
Properties of V. parahaemolyticus isolates collected from shellfish during the summers of 2008 and 2009 in Región de Los Lagos, Chile
| V. parahaemolyticus isolate by year of collection and DGREA groupa | Genetic profileb |
Serotype | DGREA group | ||||
|---|---|---|---|---|---|---|---|
| tlh | tdh | trh | orf8 | toxRSnew | |||
| Isolates from 2008 | |||||||
| PMA18.8, 23.8,24.8 | + | + | − | + | + | O3:K6 | KX |
| PMA6.8, 13.8, 16.8, 20.8, 21.8, 26.8 | + | − | − | NDc | ND | ND | 118 |
| PMA1.8, 2.8, 4.8, 5.8 | + | − | − | ND | ND | ND | 1.8 |
| PMA3.8, 8.8, 10.8, 11.8, 14.8, 15.8, 17.8, 18.8 | + | − | − | ND | ND | ND | 3.8 |
| PMA7.8 | + | − | − | ND | ND | ND | 7.8 |
| PMA9.8 | + | − | − | ND | ND | ND | 9.8 |
| PMA25.8 | + | − | − | ND | ND | O3:KUT | 25.8 |
| Isolates from 2009 | |||||||
| PMA 10.9, 26.9, 31.9, 33.9 | + | − | − | − | − | ND | 118 |
| PMA6.9 | + | − | − | − | − | ND | 128 |
| PMA21.9 | + | − | − | − | − | ND | 187 |
| PMA11.9 | + | − | − | − | − | ND | 34.6 |
| PMA29.9 | + | − | − | − | − | ND | 40.6 |
| PMA4.9 | + | − | − | − | − | ND | 21.7 |
| PMA2.9 | + | − | − | − | − | ND | 2.9 |
| PMA18.9 | + | − | − | − | − | ND | 18.9 |
Boldface, strain corresponds to the prototype strain of each DGREA group.
+, present; −, absent.
ND, not determined.
V. parahaemolyticus population diversity in shellfish.
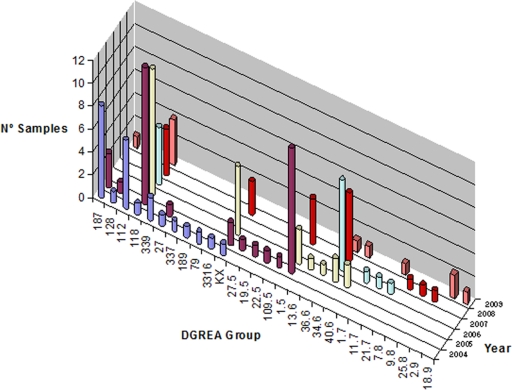
The V. parahaemolyticus population isolated from shellfish in Region de los Lagos comprises a large number of strains belonging to different DGREA groups that change considerably each summer (7, 8, 11). Isolates belonging to 23 DGREA groups have been isolated from 2004 to 2007. Sixteen nonpandemic isolates characterized in 2008 were differentiated into seven DGREA groups. Four groups had been observed in previous years. The 12 isolates obtained from shellfish in 2009 were differentiated into eight DGREA groups, six of which were previously observed. The new groups observed in 2008 and 2009 increase the number of DGREA groups isolated to date to 28, including the group KX corresponding to the pandemic strain. Figure 1 shows the number of samples containing isolates from the different DGREA groups since 2004 using the data reported here and previously (7, 8, 11).
FIG. 1.
Histogram showing the number of shellfish samples containing V. parahaemolyticus corresponding to the different DGREA groups observed each summer since 2004.
DISCUSSION
Seafood-related diarrhea outbreaks reached a peak in 2005, with 3,600 and 10,984 cases reported by the Ministry of Health in Region de los Lagos and the whole country, respectively. Levels subsequently declined to reach their lowest numbers in 2007 with 477 and 1,008 cases in Region de los Lagos and the whole country, respectively. In 2007, only 73% of the V. parahaemolyticus infections were caused by the pandemic strain, in contrast to almost 100% of these infections observed in previous years (11). Furthermore, 40% of the pandemic isolates analyzed in 2007 had the serotype O3:K59 instead of O3:K6. These results seemed to indicate a shift in the epidemiology of outbreaks caused by V. parahaemolyticus, which was expected to continue in the following years. However, in 2008 the reported cases unexpectedly increased from 477 to 1,143. At the same time, the epidemiology returned to the pattern observed in years prior to 2007 (Table 3). In 2007, in addition to the new O3:K59 serotype observed, a strain apparently generated by transference of the pathogenicity island from the pandemic clone to an indigenous V. parahaemolyticus strain (11) was found in 13% of clinical cases. However, in 2008 none of the changes observed in 2007 were maintained. Instead, 98% of clinical cases were related to the canonical pandemic strain, as observed prior to 2007, and neither the new pathogenic strains nor the serovar O3:K59 observed in 2007 was detected. In 2009 only 441 cases were reported, in contrast to the more than 1,000 cases reported in other years (with the exception of 2007). Also, similar to the observations in 2007, only 64% of the clinical cases (9 of 14 cases analyzed) were related to the pandemic strain. The other five cases corresponded to a single DGREA group and lacked the pathogenicity-associated genes tdh and trh. Since the same isolates were found in the five clinical cases, it is unlikely that they correspond to nonvirulent strains. This is an important observation for risk analysis since these two genes are considered markers of pathogenic strains and are used to estimate the load of pathogenic strains in seafood. The pandemic strain serovar O3:K59 and the other nonpandemic strains observed in clinical cases of 2007 were not detected in 2009. These overall results suggest the existence of a background level of diarrhea cases related to a diverse group of nonpandemic strains of pathogenic V. parahaemolyticus, which become evident when there are relatively few cases produced by the pandemic strain. This assertion is supported by observations in Japan and Taiwan in 1996, when the large diversity of V. parahaemolyticus serotypes observed in clinical cases was surpassed by the predominance of serotype O3:K6 (4, 23). The load of V. parahaemolyticus in shellfish and the number of pandemic isolates recovered from the enrichments seemed to parallel the extent of the outbreaks, but the sample number analyzed was too low to confirm a definite relationship.
TABLE 3.
V. parahaemolyticus isolates in clinical and shellfish samples in Región de los Lagos, Chile, from 2004 to 2009a
| Yr | Clinical case data |
Shellfish data |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of cases | No. of cases analyzed | No. of pandemic strain-positive cases (%) | Pandemic isolate characteristic (no. of isolates [%])b |
Sample data (no. [%]) |
V. parahaemolyticus loadc | Pandemic strain loadc | No. of pandemic strain isolates | ||||||
| O3:K6 negative | orf8 negative | toxRSnew negative | 42-kb phage positive | Total analyzed | V. parahaemolyticus positive | Pandemic strain positive | |||||||
| 2004 | 1,500 | 24 | 23 (96) | * | * | * | 0/9 (0) | NI | NI | NI | NI | NI | 1 |
| 2005 | 3,725 | 40 | 40 (100) | 0 | 0 | 0 | 1/9 (11) | NI | NI | NI | NI | NI | 2 |
| 2006 | 1,083 | 19 | 19 (100) | 0 | 0 | 0 | 6/11 (54) | 20 | 17 (85) | 10 (50) | 255 | 9.3 | 6 |
| 2007 | 477 | 37 | 27 (73) | 13 | 0 | 0 | 0/27 (0) | 20 | 16 (80) | 4 (20) | 22.6 | 2.5 | 0 |
| 2008 | 1,153 | 46 | 45 (98) | 1 | 0 | 2 | 2/8 (25) | 27 | 27 (100) | 8 (30) | 5.0 | 2.2 | 3 |
| 2009 | 441 | 14 | 9 (64) | 0 | 0 | 0 | 2/13 (15) | 17 | 14 (82) | 4 (24) | 3.3 | 0.5 | 0 |
Data for 2004 to 2007 was extracted or calculated from data from previous publication of our group (8, 9, 11). NI, data were not incorporated because the analysis was performed by a different and less sensitive method than that used since 2006.
Asterisk, 5 of 24 clinical strains were reported as variants of the pandemic strain (9), but further examination of these isolates showed that they corresponded to a pandemic strain contaminated with other V. parahaemolyticus strains. The data reported for the 42-kb phage represent the number of isolates positive/total number of pandemic strain isolates.
Total and pandemic V. parahaemolyticus loads were calculated according to the number of enrichment tubes positive for tlh and for tdh and trh, respectively; numbers correspond to the average geometric mean of bacteria/g in positive samples.
The arrival and massive proliferation of the V. parahaemolyticus pandemic strain in Region de los Lagos offered us an exceptional opportunity to study the evolution of a clonal strain in its natural environment. As stated by Achtman and Wagner (1), these genetically monomorphic organisms may reveal evolutionary mechanisms undetectable in bacterial populations with greater sequence diversity, in which millions of years of evolutionary history have blurred genomic signals of phylogenetic history through recombination or have eliminated them through genomic reduction. Substantiating this claim, we found that two of the pandemic strains from clinical patients lacked the toxRSnew marker, one had serotype O3:KUT, and four contained a 42-kb linear prophage plasmid observed in pandemic strains since 2005 (24). However, except for the lysogenized variants, most of the variants seem to persist for short times and are observed only sporadically (Table 3). The emergence and disappearance of serotype variants have been reported previously (5).
Taking into account the five additional new DGREA groups found in 2008 and 2009, a total of 28 groups have now been detected in shellfish. According to the number of isolates observed at a single time and those observed more than once, the richness of V. parahaemolyticus DGREA groups in shellfish (mainly mussels) may be estimated by Chao1 analysis to be 60 (12). However, the number of different V. parahaemolyticus strains in shellfish is probably much larger since most of the samples analyzed correspond to a single shellfish species extracted from an area of less than 1 km2. Using the Chao1 method, Thompson et al. (20) estimated that 1,287 Vibrio splendidus genotypes, as determined by pulsed-field gel electrophoresis patterns, occur in the water column.
The systematic follow-up of the V. parahaemolyticus strains in shellfish and clinical cases (summarized in Table 3) has contributed to a better description of the ecology of this species in seafood. The results presented here complement previous findings (7, 8, 11) and offer new observations that increase our knowledge of V. parahaemolyticus outbreaks in southern Chile. Overall, they show that the pandemic strain has become a relatively stable bacterial subpopulation of the diverse V. parahaemolyticus population present in shellfish in Chile. However, the pandemic strain is evolving by serotype changes and interactions with phages (24) and other V. parahaemolyticus subpopulations (11). The load of pandemic strain in shellfish seems to fluctuate yearly, and this oscillation could be related to the fluctuation of clinical cases. The results reported here together with those obtained from the summer of 2007 (11) indicate that there are other V. parahaemolyticus pathogenic strains that become evident when the prevalence of diarrhea cases related to the pandemic strain declines. Some of these pathogenic strains may consist of strains that are negative for tdh and trh. The isolation of the same strains—considering their grouping in the same DGREA group—from five clinical cases suggests that these isolates are not nonvirulent strains that proliferate during infection with a virulent strain, as has been suggested when isolates negative for tdh and trh are obtained (3). This is an important observation for risk analysis since these two genes are considered markers of pathogenic strains and are used to estimate the load of pathogenic strains in seafood.
Acknowledgments
We thank Gastón Higuera, Roberto Bastías, and Beatriz Zabala for their help with sample collection, isolation, and characterization of bacterial strains. We thank B. Suarez for access to his laboratory for collection of shellfish samples in 2008.
This work was supported in part by grant 1070658 from FONDECYT, Chile.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Achtman, M., and M. Wagner. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6:431-440. [DOI] [PubMed] [Google Scholar]
- 2.Bej, A. K., D. P. Patterson, C. W. Brasher, M. C. Vickery, D. D. Jones, and C. A. Kaysner. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36:215-225. [DOI] [PubMed] [Google Scholar]
- 3.Bhoopong, P., P. Palittapongarnpim, R. Pomwised, A. Kiatkittipong, M. Kamruzzaman, Y. Nakaguchi, M. Nishibuchi, M. Ishibashi, and V. Vuddhakul. 2007. Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J. Clin. Microbiol. 45:1544-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou, C. S., S. Y. Hsu, S. I. Chiu, T. K. Wang, and C. S. Chao. 2000. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J. Clin. Microbiol. 38:4621-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chowdhury, A., M. Ishibashi, V. D. Thiem, D. T. N. Tuyet, T. Van Tung, B. T. Chien, L. von Seidlein, D. G. Canh, J. Clemens, D. D. Trach, and M. Nishibuchi. 2004. Emergence and serovar transition of Vibrio parahaemolyticus pandemic strains isolated during a diarrhea outbreak in Vietnam between 1997 and 1999. Microbiol. Immunol. 48:319-327. [DOI] [PubMed] [Google Scholar]
- 6.Chowdhury, N. R., O. C. Stine, J. G. Morris, and G. B. Nair. 2004. Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J. Clin. Microbiol. 42:1280-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuenzalida, L., L. Armijo, B. Zabala, C. Hernandez, M. L. Rioseco, C. Riquelme, and R. T. Espejo. 2007. Vibrio parahaemolyticus strains isolated during investigation of the summer 2006 seafood related diarrhea outbreaks in two regions of Chile. Int. J. Food Microbiol. 117:270-275. [DOI] [PubMed] [Google Scholar]
- 8.Fuenzalida, L., C. Hernandez, J. Toro, M. L. Rioseco, J. Romero, and R. T. Espejo. 2006. Vibrio parahaemolyticus in shellfish and clinical samples during two large epidemics of diarrhoea in southern Chile. Environ. Microbiol. 8:675-683. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez-Escalona, N., V. Cachicas, C. Acevedo, M. L. Rioseco, J. A. Vergara, F. Cabello, J. Romero, and R. T. Espejo. 2005. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg. Infect. Dis. 11:129-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Escalona, N., J. Martinez-Urtaza, J. Romero, R. T. Espejo, L. A. Jaykus, and A. Depaola. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harth, E., L. Matsuda, C. Hernandez, M. L. Rioseco, J. Romero, N. Gonzalez-Escalona, J. Martinez-Urtaza, and R. T. Espejo. 2009. Epidemiology of Vibrio parahaemolyticus outbreaks, southern Chile. Emerg. Infect. Dis. 15:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iida, T., K. S. Park, and T. Honda. 2006. Vibrio parahaemolyticus, p. 340-348. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 14.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto, C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Rammamurthy, H. C. Wong, A. Depaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair, G. B., T. Ramamurthy, S. K. Bhattacharya, B. Dutta, Y. Takeda, and D. A. Sack. 2007. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K. S. Park, K. Yokoyama, K. Makino, H. Shinagawa, and T. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okuda, J., M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olea, A. M., C. González, M. Chiu, C. Vallebuona, M. Labraña, and F. Martiniello. 2005. Brote de gastroenteritis por Vibrio parahaemolyticus en Chile. Revista Chilena Salud Pública 9:51-53. [Google Scholar]
- 20.Thompson, J. R., S. Pacocha, C. Pharino, V. Klepac-Ceraj, D. E. Hunt, J. Benoit, R. Sarma-Rupavtarm, D. L. Distel, and M. F. Polz. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311-1313. [DOI] [PubMed] [Google Scholar]
- 21.Urakawa, H., and I. N. Rivera. 2006. Aquatic environment, p. 175-189. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 22.Wong, H. C., S. H. Liu, T. K. Wang, C. L. Lee, C. S. Chiou, D. P. Liu, M. Nishibuchi, and B. K. Lee. 2000. Characteristics of Vibrio parahaemolyticus O3:K6 from Asia. Appl. Environ. Microbiol. 66:3981-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. 1999. Vibrio parahaemolyticus, Japan, 1996-1998. Wkly. Epidemiol. Rec. 74:361-363. [PubMed] [Google Scholar]
- 24.Zabala, B., K. Garcia, and R. T. Espejo. 2009. Enhancement of UV light sensitivity of a Vibrio parahaemolyticus O3:K6 pandemic strain due to natural lysogenization by a telomeric phage. Appl. Environ. Microbiol. 75:1697-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]