Abstract
mothur aims to be a comprehensive software package that allows users to use a single piece of software to analyze community sequence data. It builds upon previous tools to provide a flexible and powerful software package for analyzing sequencing data. As a case study, we used mothur to trim, screen, and align sequences; calculate distances; assign sequences to operational taxonomic units; and describe the α and β diversity of eight marine samples previously characterized by pyrosequencing of 16S rRNA gene fragments. This analysis of more than 222,000 sequences was completed in less than 2 h with a laptop computer.
Since Pace and colleagues (18) outlined the culture-independent framework for sequencing 16S rRNA gene sequences in 1985, microbial ecologists have experienced an exponential improvement in the ability to sequence not only this primary phylogenetic marker but also numerous functional genes from diverse environments. Twenty-five years later, there are over 106 rRNA gene sequences deposited in public repositories such as GenBank and the number of sequences continues to double every 15 to 18 months (http://www.arb-silva.de/news/view/2009/03/27/editorial/). The development of pyrosequencing technologies has enabled the Human Microbiome Project (29), the International Census of Marine Microbes (ICoMM; http://icomm.mbl.edu), and individual investigators to collectively amass over 109 16S rRNA gene sequence tags since 2006. Because of this development in sequencing technology, individual studies have shifted from sequencing 101 to 102 sequences from multiple samples (e.g., references 2 and 16) to sequencing 104 to 105 sequences from multiple samples (e.g., references 27 and 28). These impressive statistics are indicative of the excitement that the field enjoys over relating changes in microbial community structure with changes in ecosystem performance.
Advances in computational tools have improved our ability to address ecologically relevant questions. Because of the development of tools including ARB (13), DOTUR (22), SONS (23), LIBSHUFF (25, 26), UniFrac (11, 12), AMOVA and HOMOVA (15, 21), TreeClimber (24), and rRNA-specific databases (3, 4, 20), microbial ecology has progressed from being a descriptive to an experimental endeavor. Although these tools have been widely successful, a number of limitations will affect their use as sequencing capacity increases and studies become more complex. First, for ease of use many of the rRNA-specific databases have online tools including aligners, classifiers, and analysis pipelines; however, these tools allow a limited set of generic analyses, and we must begin to question whether transferring gigantic data sets across the Internet for analysis is a sustainable practice. Second, much of the existing software was developed for analyzing 102 to 104 sequences. As the number of sequences expands, it is essential that existing software be refactored to use more efficient algorithms. In addition, although the use of scripting languages such as Perl and Python has been useful for the online analysis of small data sets, they are relatively slow compared to code written in C and C++. Finally, the boutique nature of the existing tools has limited their integration and further development. One consequence of this is that the generation of field-wide analysis standards has not been developed, making it difficult to perform meta-analyses. As sequencing capacity increases and our research questions become more sophisticated, it is critical that the software be flexible and easily maintained.
Introducing mothur.
To overcome these limitations, we have developed a single software platform, mothur (Table 1). mothur implements the algorithms implemented in previous tools including DOTUR, SONS, TreeClimber, LIBSHUFF, ∫-LIBSHUFF, and UniFrac. Beyond the implementation of these approaches, we have incorporated additional features including (i) over 25 calculators for quantifying key ecological parameters for measuring α and β diversity; (ii) visualization tools including Venn diagrams, heat maps, and dendrograms; (iii) functions for screening sequence collections based on quality; (iv) a NAST-based sequence aligner (5); (v) a pairwise sequence distance calculator; and (vi) the ability to call individual commands either from within mothur, using files with lists of commands (i.e., batch files), or directly from the command line, providing for greater flexibility in setting up analysis pipelines.
TABLE 1.
Features from preexisting software that have been integrated into mothura
| Existing tool | Description | Implementation in mothur | Reference(s) |
|---|---|---|---|
| Pyrosequencing pipeline (RDP) | Online tool that trims and deconvolutes sequences using user-supplied data | Stand-alone implementation; increased speed; greater flexibility; additional screening options | 3 |
| NAST, SINA, and RDP aligners | Online tools that align user-supplied sequences with specific databases | Stand-alone implementation; can utilize multiple processors; increased speed; greater flexibility; open source | 3-5, 20 |
| DNADIST | Calculates pairwise distances between sequences (does not penalize for gaps) | Can utilize multiple processors; more efficient use of RAM; various ways to penalize gaps | 6 |
| DOTUR and CD-HIT | Assigns sequences to OTUs, constructs sampling curves, and estimates richness and diversity | More efficient clustering; requires less memory; additional calculators; greater flexibility | 10, 22 |
| SONS | Calculates estimates of the fraction and richness of OTUs shared between communities | Generates dendrograms, heat maps, and Venn diagrams; additional calculators; greater flexibility | 23 |
| ∫-LIBSHUFF | Uses the Cramer-von Mises statistic to test whether two communities have the same structure | Eliminates the need for a sorted distance matrix; can specify pairwise comparisons | 25, 26 |
| TreeClimber | Uses a parsimony-based test to determine whether two or more communities have the same structure | Greater flexibility; can specify pairwise comparisons | 14, 15,24 |
| UniFrac | Compares the phylogenetic distance between communities to detect differences in community structure | Stand-alone implementation; greater flexibility; can input bootstrap trees | 12 |
In all cases, modifications have been made to the mothur implementation of the algorithms for greater flexibility, speed, and resource utilization.
Object oriented, responsive, free, and platform independent.
mothur is written in C++ using modern object-oriented programming strategies (17, 19). Design patterns are used extensively to improve the maintenance and flexibility of the software (7). Since releasing the first version of mothur in February 2009, we have made use of an iterative release design model. This means that instead of releasing mothur once a year with many modifications, we release smaller updates to mothur throughout the year. The advantage to this approach is the ability to more quickly address bugs, incorporate user suggestions, and get new features to users. By making mothur an open-source software package under the GNU General Public License (http://www.gnu.org/licenses/gpl.html), we have ensured that the software is free and open to modification by other investigators developing their own analysis methods. mothur is available from the project website (http://www.mothur.org) as a Windows-compatible executable or as source code for compilation in Unix/Linux or Mac OS X environments.
Open documentation and support.
Extensive community-supported documentation and support are available through a MediaWiki-based wiki (http://www.mediawiki.org/) and a phpBB-based discussion forum (http://www.phpbb.com). The wiki format serves two important functions. First, it is a source of documentation that users are free to read, edit, and expand to help themselves and others understand the theory and implementation behind the commands provided in mothur. For example, the wiki page describing each calculator includes manual calculations. Numerous undergraduate and graduate courses have used these example calculations to improve their students' numeracy. Second, users are encouraged to create pages describing how they used the software to analyze a set of data as a medium for teaching others the diverse ways that one can design experiments and analyze their data. These “example workflows” include the original data, commands, and commentary from unpublished and published studies (e.g., references 1, 8, and 9). The discussion forum allows users to ask questions that anyone can answer, and the forum allows users to suggest improvements to the software.
Example workflow: the ocean's rare biosphere.
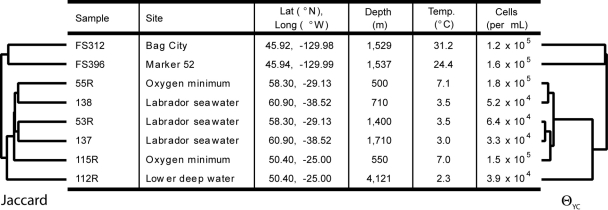
Although mothur is fully capable of analyzing traditional clone-based sequences, here we demonstrate the ability of mothur to efficiently analyze a pyrosequencing data set. Sogin and colleagues, in a seminal 2006 study that outlined the use of pyrosequencing in microbial ecology studies, obtained 216,243 high-quality sequence reads from the V6 region of the 16S rRNA gene from eight samples (27). They obtained six-paired samples from the meso- and bathypelagic realms from three sites in the North Atlantic Deep Water loop and two samples from diffuse hydrothermal vent fluids near the site of an eruption in the Axial Seamount in the northeast Pacific Ocean (Fig. 1). Their analysis primarily considered their inability to exhaustively sample the biodiversity of sites in spite of record sequencing depths. The sequence data were obtained from http://jbpc.mbl.edu/research_supplements/g454/20060412-private/, and we used the 2 February 2008 version of the data set. These data differ from those described in the original publication because the data processing algorithms internal to the GS20 machine were updated; therefore, it is not possible to make a direct comparison to the findings of the original analysis. Although these data were already trimmed and sorted into individual files for each sample, mothur has the capacity to generate these files from the FASTA-formatted sequence file generated by a sequencer. Furthermore, mothur has a number of functions for performing hypothesis tests, but here we will focus on operational taxonomic unit (OTU)-based methods of describing and comparing communities.
FIG. 1.
Description and comparison of the eight samples analyzed by Sogin et al. (27). The dendrogram to the left represents the similarity of the samples based on the membership-based Jaccard coefficient calculated using Chao1 estimated richness values. The dendrogram on the right represents the similarity of the samples based on the structure-based θYC coefficient. The distance from the tip of the dendrogram to the root is 0.50 for both trees.
mothur makes several improvements that allow users with modest computing resources to analyze large data sets. Most significant are the ability to analyze only the unique sequences in a data set but retain information about the number of times that each sequence was observed and the use of sparse matrices that represent only distances smaller than a user-specified cutoff. Using a PHYLIP-based approach would have required approximately 145 GB to represent 2.3 × 1010 distances. Our improvements resulted in an 18.9-MB file containing 5.2 × 105 pairwise distances that were smaller than 0.10. The only mothur-imposed limit is the number of distances that can be processed, which is 264. The more likely limitation will be the amount of random-access memory (RAM) available on the user's computer. With the reduced memory requirement also comes significantly improved processing speed. Considering that most computers have multiple processors, users can obtain further increases in speed by utilizing the parallelization features provided in the alignment and distance calculation commands.
mothur can cluster sequences using the furthest neighbor, nearest neighbor, or UPGMA (unweighted-pair group method using average linkages) algorithms (22). The ability to let the data speak for themselves in determining OTUs is advantageous compared to database-based approaches that can form clusters, in which sequences are similar to the same database sequences but not to each other. Furthermore, mothur uses the approach employed in DOTUR where OTUs are defined for multiple cutoffs up to the distance threshold so that alternative OTU definitions can be compared. For example, using the furthest neighbor algorithm, we clustered sequences into OTUs up to a distance threshold of 0.10 and observed 13,202, 11,317, and 7,971 OTUs at cutoffs of 0.03, 0.05, and 0.10 distance units, respectively. A similar type of analysis using the approach used in programs such as CD-HIT would limit the user to a nearest neighbor-based approach, and the users would need to run the program for each distance level in which they were interested (10).
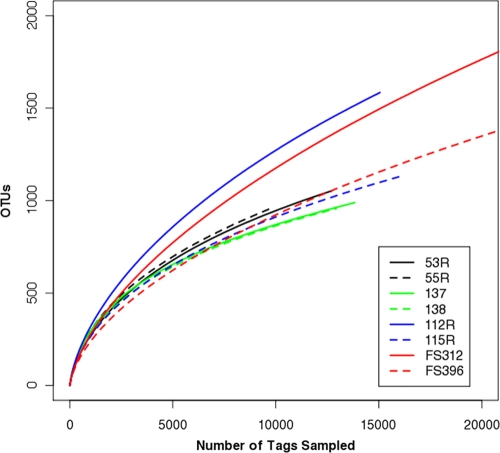
By inputting a file that maps each sequence to a sample identifier, the clusters could be parsed to perform α-diversity analyses. First, we calculated the richness and diversity of the eight samples at OTU cutoffs of 0.03, 0.05, and 0.10 distance units by using the number of observed OTUs, Chao1 estimated minimum number of OTUs, and a nonparametric Shannon diversity index (Table 2). Second, we calculated rarefaction curves for the eight samples for a 0.10 distance cutoff (Fig. 2); the original Sogin analysis built rarefaction curves using frequencies acquired from a database-based OTU assignment analysis. Interestingly, mothur calculated the coverage of these samples to be between 0.94 and 0.98, and yet the rarefaction curves continued to climb with increasing sequencing effort. These types of analysis were the extent of the α-diversity measurements performed in the original Sogin analysis, and each sample required up to 4 days to complete on a Quad Opteron 875 2.2-GHz series Dual Core machine with 28 GB of RAM (S. Huse, personal communication). The analysis described in this paper—from aligning of sequences through β-diversity analyses—required less than 2 h with use of a MacBook Pro laptop with 2 GB RAM and with only one of the 2.0-GHz dual processors.
TABLE 2.
Measures of α diversity for the samples characterized by Sogin et al. (27) for three OTU definitionsa
| Sample | No. of reads | 0.03 |
0.05 |
0.10 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OTU | Chao | H′ | OTU | Chao | H′ | OTU | Chao | H′ | ||
| 53R | 12,725 | 1,599 | 3,222 | 5.29 | 1,420 | 2,622 | 5.19 | 1,053 | 1,733 | 4.81 |
| 55R | 9,848 | 1,469 | 2,994 | 5.54 | 1,302 | 2,496 | 5.43 | 962 | 1,741 | 5.03 |
| 112R | 15,057 | 2,258 | 5,189 | 5.91 | 2,032 | 4,282 | 5.79 | 1,584 | 2,992 | 5.44 |
| 115R | 16,181 | 1,749 | 3,600 | 5.31 | 1,552 | 3,088 | 5.21 | 1,135 | 1,919 | 4.83 |
| 137 | 13,831 | 1,425 | 2,687 | 5.44 | 1,295 | 2,430 | 5.36 | 989 | 1,645 | 5.07 |
| 138 | 12,938 | 1,425 | 2,542 | 5.24 | 1,253 | 2,131 | 5.14 | 957 | 1,479 | 4.81 |
| FS312 | 54,894 | 4,371 | 10,691 | 5.23 | 3,948 | 9,259 | 5.16 | 3,095 | 6,409 | 4.94 |
| FS396 | 80,769 | 4,359 | 10,208 | 4.67 | 3,806 | 8,609 | 4.60 | 2,804 | 5,437 | 4.42 |
0.03, 0.05, and 0.10 are the OTU cutoffs in distance units. OTU signifies the number of OTUs observed, Chao signifies the Chao1 estimated minimum number of OTUs, and H′ signifies the nonparametric Shannon diversity index.
FIG. 2.
Rarefaction curves describing the dependence of discovering novel OTUs as a function of sampling effort for OTUs defined at a 0.10 distance cutoff. The curves for FS312 and FS396 climb to 3,095 and 2,804 OTUs after sampling of 54,894 and 80,769 sequences, respectively.
Due to software limitations, it was not possible to assess the β diversity of the samples in the original Sogin analysis. With the software improvements implemented in mothur, we were able to transform the original OTU information into heat maps, Venn diagrams, and dendrograms (Fig. 1) to describe the similarities in membership and structure of the eight samples. Several interesting observations can be made from this analysis. First, although the dendrograms generated using the Jaccard coefficient and the θYC community structure similarity coefficient have similar topologies, the terminal branch lengths of the Jaccard coefficient dendrogram are considerably longer for samples 53R, 55R, 115R, and 137. This is interesting because it indicates that while these samples have considerably different memberships (Jaccard), the relative abundances of the shared OTUs are similar. Thus, the differences between the communities are likely found in the rarer OTUs. Second, the two diffuse hydrothermal flow samples clearly cluster away from the others. This is intuitive because of the considerable differences in temperature and chemistry. Third, the only available piece of metadata that explains the clustering of the seawater samples is extreme depth; the deepest sample, 112R, clearly clusters away from the other seawater samples and was taken 2,411 m deeper than was any of the other samples. Considering that this was the only sample taken at such an extreme depth, additional sampling is required in order to have confidence in such a correlation.
Looking forward.
The development of computational tools to describe and analyze microbial communities is in a “Red Queen”-type race where advances in computational power are met with expansions in sequencing capacity and vice versa. As the length and number of reads multiply, data analysis resources must meet the challenge. Although mothur goes a long way toward making data analysis efficient, flexible, and simple, the analyses are by no means trivial, and researchers must take care to ensure that their experiments are well designed and thought out and that their results are biologically plausible. The field of microbial ecology is experiencing an amazing revolution where we can now design experiments with sophisticated experimental designs. Tools such as mothur open new possibilities so that the primary limitation is our imagination.
Acknowledgments
Funding for mothur has been provided by the College of Natural Resources and the Environment at the University of Massachusetts, a grant from the Sloan Foundation, a grant from the National Science Foundation (award 0743432), and the Austrian GEN-AU project BIN.
We appreciate the input and support of the more than 900 users who registered their use of DOTUR, SONS, ∫-LIBSHUFF, or TreeClimber over the past 5 years.
P.D.S. conceived, designed, and prepared the manuscript; P.D.S., S.L.W., T.R., and G.G.T. generated source code; and P.D.S., S.L.W., T.R., J.R.H., M.H., E.B.H., R.A.L., B.B.O., D.H.P., C.J.R., J.W.S., B.S., D.J.V., and C.F.W. provided documentation. All authors helped in the final editing of the manuscript.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Antonopoulos, D. A., S. M. Huse, H. G. Morrison, T. M. Schmidt, M. L. Sogin, and V. B. Young. 2009. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77:2367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borneman, J. 1999. Culture-independent identification of microorganisms that respond to specified stimuli. Appl. Environ. Microbiol. 65:3398-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, et al. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeSantis, T. Z., Jr., P. Hugenholtz, K. Keller, E. L. Brodie, N. Larsen, et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394-W939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package. Cladistics 5:164-166. [Google Scholar]
- 7.Gamma, E., R. Helm, R. Johnson, and J. M. Vlissides. 1995. Design patterns: elements of reusable object-oriented software. Addison-Wesley, Reading, MA.
- 8.Hall, J. R., K. R. Mitchell, O. Jackson-Weaver, A. S. Kooser, B. R. Cron, L. J. Crossey, and C. D. Takacs-Vesbach. 2008. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl. Environ. Microbiol. 74:4910-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann, M., and F. Widmer. 2006. Community structure analyses are more sensitive to differences in soil bacterial communities than anonymous diversity indices. Appl. Environ. Microbiol. 72:7804-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, W., and A. Godzik. 2006. CD-HIT: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658-1659. [DOI] [PubMed] [Google Scholar]
- 11.Lozupone, C., M. Hamady, and R. Knight. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maddison, W. P., and M. Slatkin. 1991. Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution 45:1184-1197. [DOI] [PubMed] [Google Scholar]
- 15.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McConnell, S. 2004. Code complete, 2nd ed. Microsoft Press, Redmond, WA.
- 18.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1985. Analyzing natural microbial populations by rRNA sequences. ASM News 51:4-12. [Google Scholar]
- 19.Pilone, D., and R. Miles. 2008. Head first software development. O'Reilly, Sebastopol, CA.
- 20.Pruesse, E., C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schloss, P. D. 2008. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2:265-275. [DOI] [PubMed] [Google Scholar]
- 22.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool that compares the membership of microbial communities. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss, P. D., and J. Handelsman. 2006. Introducing TreeClimber, a test to compare microbial community structure. Appl. Environ. Microbiol. 72:2379-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. USA 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbaugh, P. J., M. Hamady, T. Yatsunenko, B. L. Cantarel, A. Duncan, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, et al. 2007. The human microbiome project. Nature 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]