Abstract
We have been developing the cellulases of Thermobifida fusca as a model to explore the conversion from a free cellulase system to the cellulosomal mode. Three of the six T. fusca cellulases (endoglucanase Cel6A and exoglucanases Cel6B and Cel48A) have been converted in previous work by replacing their cellulose-binding modules (CBMs) with a dockerin, and the resultant recombinant “cellulosomized” enzymes were incorporated into chimeric scaffolding proteins that contained cohesin(s) together with a CBM. The activities of the resultant designer cellulosomes were compared with an equivalent mixture of wild-type enzymes. In the present work, a fourth T. fusca cellulase, Cel5A, was equipped with a dockerin and intervening linker segments of different lengths to assess their contribution to the overall activity of simple one- and two-enzyme designer cellulosome complexes. The results demonstrated that cellulose binding played a major role in the degradation of crystalline cellulosic substrates. The combination of the converted Cel5A endoglucanase with the converted Cel48A exoglucanase also exhibited a measurable proximity effect for the most recalcitrant cellulosic substrate (Avicel). The length of the linker between the catalytic module and the dockerin had little, if any, effect on the activity. However, positioning of the dockerin on the opposite (C-terminal) side of the enzyme, consistent with the usual position of dockerins on most cellulosomal enzymes, resulted in an enhanced synergistic response. These results promote the development of more complex multienzyme designer cellulosomes, which may eventually be applied for improved degradation of plant cell wall biomass.
In nature, some anaerobic cellulolytic bacteria produce cellulosomes, which are organized by the action of scaffoldin subunits that usually contain a single carbohydrate-binding module (CBM) and multiple cohesin modules (2, 7, 13, 14, 28, 36). This arrangement allows the integration of several dockerin-containing enzymes into a complex, which is then targeted to the cellulosic substrate by the common CBM. The cellulosomal enzymes then exhibit enhanced synergistic activity, presumably due to their spatial proximity and coordinated interaction. In contrast, the enzyme systems of aerobic bacteria and fungi comprise free (uncomplexed) enzymes, which differ from cellulosomal systems in that many of them contain their own CBM that delivers the individual catalytic module to the surface of the substrate (39, 41, 42).
In previous work, we used the designer cellulosome concept (5) to construct unique minicellulosomes of defined content (16, 32, 33). In order to construct designer cellulosomes, chimeric scaffoldins have been prepared which contained two or more cohesins that matched the dockerins of the enzymes (native cellulosomal or dockerin-fused chimeras). Enzymes that contain dockerins that match the specificity of a scaffoldin-borne cohesin can then be selectively integrated into the designer cellulosome at a specified site. Cellulosomal enzymes containing either a native dockerin or a divergent dockerin can be inserted on different sites of a chimeric scaffoldin. Alternatively, a free, noncellulosomal enzyme can be included in designer cellulosomes by replacing its native CBM with a dockerin of choice. In some cases, designer cellulosomes displayed enhanced synergistic activity over the parallel free-enzyme system (15, 17). This increased activity was shown to be a function of both a substrate-targeting effect (contributed by the CBM on the chimeric scaffoldin) and the enzyme proximity effect, thus supporting the initial hypothesis.
In recent studies, we have investigated the free-cellulase system of Thermobifida fusca for use in designer cellulosome systems. This aerobic thermophilic cellulolytic bacterium contains a limited set of six free cellulases, each composed of a catalytic module and a crystalline-cellulose binding family 2 CBM (CBM2) module on either the N or C terminus of the protein. T. fusca contains three endoglucanases (Cel5A, Cel6A, and Cel9B), two exocellulases (Cel6B and Cel48A), and one processive endoglucanase (Cel9A). Previously, we converted both family 6 cellulases and the family 48 exoglucanase from the free to the cellulosomal mode of action by replacing their native CBM2s with a dockerin module (11, 12). All three chimeric enzymes exhibited cellulose-degrading activity on both soluble and crystalline substrates. The results indicated that the family 48 exoglucanase appeared to be well adapted to the cellulosomal mode of action, whereas the family 6 exoglucanase is less appropriate for inclusion into cellulosomes. Indeed, family 48 cellulases have been found to be a major component in every native cellulosome thus far described, in contrast to the family 6 cellulases, which have been identified only in free-cellulase systems.
An important feature of the free-acting fungal and bacterial cellulases is that they contain a linker segment, often rich in prolines and threonines, that connects the catalytic module to the CBM (37). The role of such flexible linkers is thought to ensure independent action of the adjacent functional modules, thus allowing progressive and efficient hydrolysis of cellulose by the catalytic modules (6, 9, 10, 20, 25-27, 34, 36, 38, 40). The present communication focuses on the effect of linker length and dockerin position (relative to the catalytic module) on enzymatic activity within a designer cellulosome. For this purpose we have employed the highly active family 5 endoglucanase Cel5A from T. fusca (21, 22, 29), which was converted to the cellulosomal mode by replacement of its CBM with a dockerin module. Chimeric dockerin derivatives were prepared on either the N or C terminus of the Cel5A catalytic module, separated by linker segments of different lengths. In most cases, binary designer cellulosomes, comprising the respective Cel5A chimera together with a Cel48A chimera, were shown to be more efficient on crystalline cellulosic substrates than the combination of the wild-type free enzymes.
MATERIALS AND METHODS
Cloning of chimeric proteins.
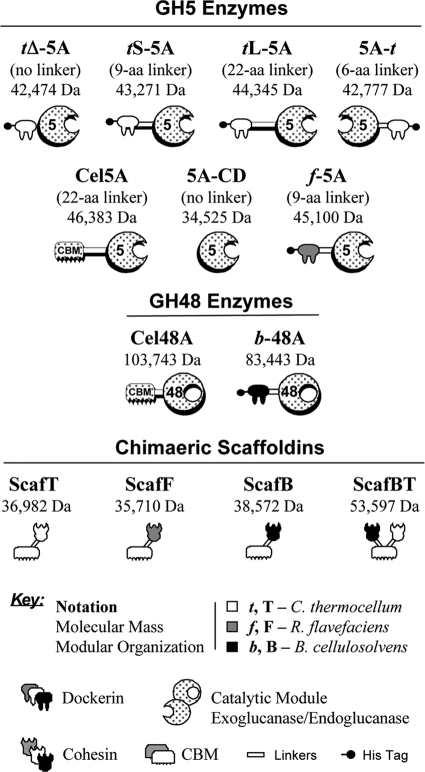
Fig. 1 provides a key to the abbreviations of all enzymes, scaffoldins, and chimeric derivatives used in this study.
FIG. 1.
Schematic representation of the recombinant proteins used in this study. The CBM2 of wild-type T. fusca enzymes (Cel5A or Cel48A) was replaced by a dockerin at the indicated position relative to the catalytic module. The shading of each symbol denotes the source of the module: stippled (T. fusca), white (C. thermocellum), dark gray (R. flavefaciens), black (B. cellulosolvens). The corresponding bacterial source—C. thermocellum, R. flavefaciens or B. cellulosolvens—is additionally indicated by the following notations: t, f, or b for the dockerins and T, F, or B for the cohesins, respectively. Chimeras f-5A, tL-5A, tS-5A, tΔ-5A, 5A-t, and b-48A included a His tag, attached distally to the respective dockerin module of the enzyme. The divergent species of cohesin-bearing scaffoldin (Scaf) included a C. thermocellum CBM3a, shown symbolically. The molecular mass is shown for each of the expressed proteins.
Cloning of the family 48 chimera, b-48A, with a dockerin from Bacteroides cellulosolvens (indicated by b in the designation) was described previously (11).
A standardized PCR amplification protocol was used for all modules used in this study. The reaction mixture (50 μl) contained ∼50 ng of genomic DNA, 1 μM of each primer, and 1× ReddyMix PCR Master Mix (catalog no. AB-0575; Abgene Ltd., Epsom, United Kingdom). The reaction was carried out for 25 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C for dockerins and cohesins (and 1 min for enzymes).
The DNA encoding the Ruminococcus flavefaciens scaffoldin B dockerin was amplified from R. flavefaciens genomic DNA using the primers 5′-AATTCCATGGCACACCATCACCATCACCATGCACCATCACCCGGCACAAAGC-3′ (NcoI site is in boldface, and the His tag underlined) and 5′-AATTGGTACCGCTTGAGGAAGTGTGATGAG-3′ (KpnI site is in boldface). The DNA encoding the Xyn10Z xylanase dockerin of Clostridium thermocellum was amplified from genomic DNA using the primers 5′-ATGGGGTACCTGAAAGCAGTTCCACAGG-3′ (KpnI site is in boldface) and 5′-TAATCTCGAGTCCGGGGAACTCTGTAATAATGC-3′ (XhoI site is in boldface).
Cloning of the Cel5A catalytic module for production of the catalytic module (lacking the CBM) 5A-CD was reported earlier (24).
The DNA encoding the Cel5A catalytic module for the f-5A construct (where f indicates the dockerin source as R. flavefaciens) was amplified from the Cel5A plasmid, using the primers 5′-ATATGGTACCCGGCACGCAGCCC-3′ (KpnI is site in boldface) and 5′-ATATCTCGAGtcaGGACTGGAGCTTGC-3′ (primer Cel5A*Rev; XhoI site is in boldface, and lowercase letters indicate an inserted stop codon).
For the Cel5A catalytic module with different linker lengths (the constructs with the C. thermocellulm dockerin), the following primers were used: for the native 22-residue linker (tL-5A, where L indicates the long linker and t indicates the dockerin source as C. thermocellulm), 5′-TTAAGGTACCGGGCGGCCCCGGCGG-3′ (KpnI site is in boldface) and Cel5A*Rev; for the short 9-residue linker (tS-5A, where S indicates the short linker), 5′-AATTGGTACCCGGCACGCAGCCCGGC-3′ (KpnI is site in boldface) and Cel5A*Rev; for the chimera lacking a linker (tΔ-5A), 5′-AATTGGTACCCGTCGAGCGGTACGGCAAAGTCC-3′ (KpnI site is in boldface) and Cel5A*Rev; and for the 5A-t chimera, 5′-AATTCCATGGTCGAGCGGTACGGCAAAGTCC-3′ (NcoI site is in boldface) and 5′-ATATGGTACCGACTGGAGCTTGCTCCGCACC-3′ (KpnI site is in boldface). The R. flavefaciens endoglucanase dockerin and the catalytic module of Cel5A were ligated into NcoI-XhoI-linearized pET28a to form pf-5A. The DNA segments encoding the dockerin of C. thermocellum xylanase Xyn10Z and the Cel5A catalytic module (with NcoI and KpnI restriction sites) were similarly ligated to form p5A-t. The three remaining Cel5A catalytic modules that were amplified (long linker, short linker, and no linker) were all ligated to pt-6B (12) and KpnI-XhoI linearized (replacing the Cel6B catalytic site) to form ptl-5A, pts-5A, and ptΔ-5A.
The DNA encoding the second cohesin of the C. thermocellum CipA scaffoldin was amplified from C. thermocellum genomic DNA using the primers 5′-AATTCCATGGCACACCATCACCATCACCATGTGGTAGTAGAAATTGGC-3′ (NcoI site is in boldface, and the His tag is underlined) and 5′-TATAGGTACCGCAACGTTAACACCACCG-3′ (KpnI site is in boldface).
The CohT construct was cloned as described previously (43), and the CohB and CohF constructs were constructed as reported recently (18). For the construction of the ScafBT plasmid (where ScafBT is a chimeric scaffoldin containing a central family 3a CBM [CBM3a] and B. cellulosolvens and C. thermocellum cohesins), the third B. cellulosolvens cohesin from the scaffoldin B subunit was amplified with 5′-GCAACCATGGCGGGGAAAAGTTCACCAG-3′ (NcoI site is in boldface) and 5′-GTAGGGTACCTTAGTTACAGTAATGCTTCC-3′ (KpnI site is in boldface) and ligated into NcoI-KpnI-linearized pET9d C-T scaffoldin (Scaf2 [16]), whereby the B. cellulosolvens cohesin replaced the Clostridium cellulolyticum cohesin.
Protein expression and purification.
Wild-type Cel48A and Cel5A, bearing a native CBM2 module, and 5A-CD (the Cel5A catalytic module alone, lacking the CBM2) were cloned, expressed, and purified as reported earlier (23). All His-tagged 5A protein chimeras were expressed and purified using Ni-nitrilotriacetic acid columns as previously described (12, 24). The CBM3a-containing proteins, CohF, CohB, CohT, and ScafBT, were expressed and purified on phosphoric acid-swollen cellulose (PASC), according to a previously described methodology (12). Purity of the proteins was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 12% acrylamide).
Affinity-based ELISA.
The affinity and specificity of binding of the dockerin bearing-chimeras to the cohesin-bearing chimeras were tested by an enzyme-linked immunosorbent assay (ELISA). Microtiter plates were coated with the target scaffoldin (3 nM in 0.1 M sodium carbonate, pH 9). Sample dockerin-containing enzymes (50 pM in Tris-buffered saline, pH 7.4, 10 mM CaCl2, and 0.05% Tween 20 in the presence of 2% bovine serum albumin) were then added, and the standard protocol was followed (1).
Nondenaturing PAGE.
Protein samples at 40 μM were mixed with sample buffer (lacking SDS) and subjected to PAGE (9% gels) as described earlier (11).
Enzyme activity.
In all cases, enzymatic chimeras were combined with their target CBM3a-Coh counterparts at 37°C for 1 h, as previously described (11, 12). Carboxymethyl cellulase and PASC assays were carried out as reported earlier (11, 22) in triplicate samples at enzyme concentrations of 0.02 μM for carboxymethyl cellulose (CMC) and 0.08 to 0.12 μM for PASC. Bacterial microcrystalline cellulose (BMCC) and Avicel hydrolysis activity were determined according to described procedures (11, 22). Reducing sugar concentrations were determined using dinitrosalicylic acid as described previously (30, 31), with glucose as a standard.
Cellulose binding assay.
Samples of 0.5 μM enzyme were mixed with increasing amounts of Avicel (1, 2, 5, 10, 20, and 30 mg/ml) and incubated at room temperature for 1 h. After the cellulose was spun down, the hydrolytic activity of the supernatant fluids was tested using CMC, as described above. A dockerin-bearing enzyme chimera, preattached to its matching CBM3a-Coh was used as a control for a CBM-restored enzyme.
RESULTS
Construction and expression of recombinant proteins.
A schematic representation of the different recombinant enzymes used in this study is shown in Fig. 1. The chimeric enzymes tL-5A, tS-5A, and tΔ-5A were designed to contain the catalytic module of T. fusca endoglucanase Cel5A with an N-terminal dockerin module, derived from C. thermocellum xylanase Xyn10Z, and the two modules were separated in the given chimeras by different lengths of the original T. fusca intermodular linker (the long 22-residue native linker or a shortened 9-residue truncated linker) or by the deletion of the entire T. fusca linker segment. In contrast, the chimeric enzyme 5A-t was designed to contain the same C. thermocellum dockerin attached to the C terminus of the Cel5A catalytic module by the short, native, 9-residue linker of C. thermocellum Xyn10Z. The chimeric enzyme, f-5A, was designed to be analogous to tS-5A with an R. flavefaciens dockerin instead of the C. thermocellum dockerin. This divergent dockerin-containing chimera was used as a negative control for interaction with the C. thermocellum cohesin or as a parallel positive control with its matching cohesin. The chimeras were purified by metal ion affinity chromatography via a His tag, located on the dockerin or cohesin end (i.e., distal to the catalytic module). Wild-type T. fusca Cel5A and its catalytic module (5A-CD) were cloned and purified as described previously, and the activities of these enzymes were compared to those of the different chimeras.
Three different single cohesin-containing constructs were used in this study, each located at the C terminus of C. thermocellum CBM3a: ScafF (with an R. flavefaciens cohesin), ScafT (with a C. thermocellum cohesin), and ScafB (with a B. cellulosolvens cohesin). All three were purified on PASC, via their CBMs, as described previously (12). In addition, a chimeric binary scaffoldin, ScafBT, containing the B. cellulosolvens cohesin followed by CBM3a and a cohesin from C. thermocellum, was constructed and purified on PASC in a similar manner.
All of the purified recombinant proteins showed a single major band in SDS-PAGE (data not shown), and in each case the mobility was consistent with the expected molecular mass.
Specificity of chimeric enzyme-borne dockerins.
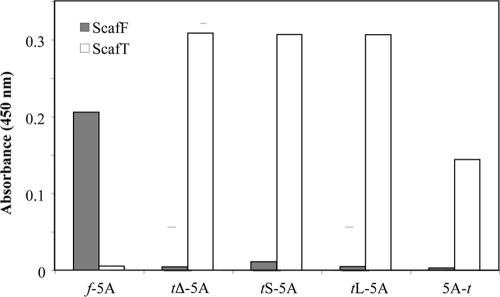
Species specificity of the five chimeric dockerin-bearing family 5A enzymes to the respective cohesins (ScafF and ScafT) was assayed by a semiquantitative ELISA procedure (Fig. 2). As expected, the f-5A chimera interacted exclusively with its matching cohesin (ScafF) and not with (ScafT). Likewise, chimeras tL-5A, tS-5A, tΔ-5A, and 5A-t were shown to interact only with ScafT and not with ScafF. Interestingly, the 5A-t chimera showed a somewhat lower affinity to ScafT, indicating that the dockerin placed on the nonnative end of the enzyme might be less able to interact with its matching cohesin.
FIG. 2.
Specificity of Cel5A chimeras for their cohesin targets. The interaction between 5A chimeras and their matching and nonmatching cohesins was examined by ELISA. Microtiter plates were coated by the cohesin-containing scaffoldins (ScafF, gray; ScafT, white) and reacted with 50 pM of the dockerin-linked enzymes. The resultant cohesin-dockerin interactions were detected by using primary antibody against the respective wild-type enzyme (T. fusca Cel5A) and HRP-labeled secondary antibody.
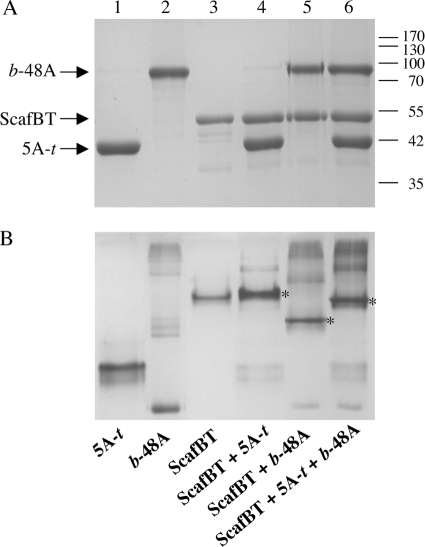
The efficiency and specificity of the interaction between the dockerin-bearing constructs to the cohesin-bearing chimeras were also demonstrated by combining stoichiometric amounts of the different proteins and subjecting the mixtures to nondenaturing PAGE analysis. The resultant mobility patterns (data not shown) showed clearly that each enzyme bound to the matching cohesin construct exclusively and completely but failed to interact with that of the other species. It was also shown that the chimeric scaffoldin ScafBT interacts selectively with each of the 5A-t and b-48a chimeras alone (Fig. 3, lanes 4 and 5) and as a binary complex with both chimeras (Fig. 3, lane 6). In each case, a major band, representing the respective multicomponent complex, was observed in the nondenaturing PAGE gel, which differed from the position of the ScafBT band alone (Fig. 3, lane 3). Residual banding patterns were also observed that indicated the presence of contaminating or noninteracting components (e.g., dockerins that failed to fold properly) in each enzyme preparation.
FIG. 3.
Species-specific interaction of recombinant dockerin-containing chimeras 5A-t and b-48A with the matching binary scaffoldin. Equimolar concentrations of the chimeric enzymes (5A chimeras and b-48A) with their matching cohesins were combined. Nonmatching cohesin-dockerin pairings were carried out as a negative control. (A) SDS-PAGE demonstrating the mixture of the binary chimeric scaffoldin ScafBT with the 5A-t and the b-48A enzyme chimeras. (B) Nondenaturing PAGE of the enzymatic mixtures, demonstrating the specific interactions of the dockerin-bearing chimeras with their target scaffoldins. Contents of lanes 1 to 6 for both gels are shown at the bottom of the figure. Major bands, indicating complexation of the chimeric scaffoldin with the specified enzyme components, are indicated by asterisks.
Cellulose binding.
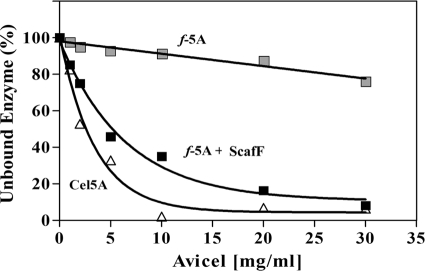
As expected, wild-type Cel5A (containing its CBM2) bound efficiently to cellulose (Fig. 4). Increasing amounts of cellulose (Avicel) resulted in decreased enzyme in the supernatant as a result of binding to the cellulose matrix. The chimeric f-5A, lacking CBM2, failed to bind to cellulose and remained in the supernatant fractions even at high concentrations of Avicel. However, the combination of the f-5A chimera with ScafF served to introduce cellulose binding at a level similar to that of Cel5A. This experiment indicates that the family 5 catalytic and dockerin modules do not bind to cellulose and that the cellulose-binding function of the cellulases is due to the CBM.
FIG. 4.
Cellulose binding assay: effect of the CBM. Cellulose affinity measurements for the native Cel5A enzyme (open triangles) and its f-5A chimera (gray squares) are shown. Each enzyme sample was mixed with increasing amounts of crystalline cellulose. After the cellulose was spun down, the hydrolytic activity of the supernatant fluids was tested using CMC. The f-5A chimera, attached to its matching scaffoldin (black squares), represented a CBM-restored enzyme complex.
Cellulose-degrading activity.
The family 5 enzyme derivatives were assayed on four different cellulosic substrates in order to compare their activities: CMC, a soluble substrate used for determining endoglucanase activity; PASC, an insoluble amorphous (noncrystalline) cellulose substrate; and two different crystalline forms of cellulose, BMCC and Avicel. Each chimera was tested in three different states: the free state, combined with its matching scaffoldin, and with a nonmatching scaffoldin as a negative control (Fig. 5).
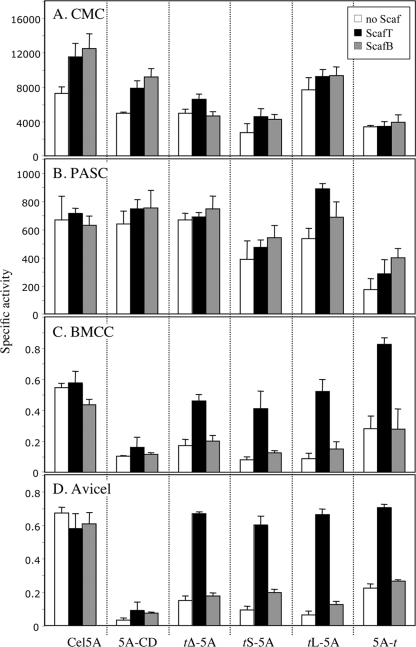
FIG. 5.
Activity of Cel5A and its various chimeric derivatives on different cellulosic substrates. Activity of wild-type Cel5A and its derivatives on CMC (A), PASC (B), BMCC (C), and Avicel (D) in the absence (white bars) and presence of matching (ScafT; black bars) or nonmatching (ScafB; gray bars) scaffoldin chimeras. The figure shows the mean of triplicate samples, examined in two independent experiments, with error bars indicating the standard deviations. Specific activity is defined as mol of reducing sugar per mol of enzyme per min.
On CMC, wild-type Cel5A showed relatively high endoglucanase activity (Fig. 5A) while decreased levels of carboxymethyl cellulase activity were observed for the different chimeras and the catalytic module alone (without the native CBM2). Among the dockerin-bearing chimeras, the long linker t-L5A demonstrated the highest level of endoglucanase activity.
Most of the enzymes tested on PASC (Fig. 5B) showed similar levels of activity. The presence of a CBM or a dockerin or matching or nonmatching scaffoldins had little or no effect on enzymatic activity, with the possible exception of the dockerin derivative containing the long linker (tL-5A), which displayed slightly elevated levels of activity in the presence of scaffoldin. The chimeric enzyme with the C-terminal dockerin (5A-t) exhibited significantly less activity on PASC, similar to its performance on CMC.
In the absence of matching scaffoldin, the activities of the Cel5A chimeras on the two crystalline cellulose substrates were significantly less than the activity of Cel5A (Fig. 5C and D). This clearly reflects the lack of the CBM and consequently a lack of targeting of the enzymes to the crystalline substrates. Indeed, when assembled on the matching scaffoldin, ScafT, all dockerin-bearing chimeras had a significant increase in crystalline cellulose degradation, approaching that of Cel5A. Linker length appeared to have no effect on the cellulose degradation by the parent enzyme. The results obtained for the divergent dockerin-containing chimera, f-5A, were essentially identical to its analogous chimera, tS-5A (data not shown).
Surprisingly, placement of the dockerin on the opposite (C-terminal) side of the catalytic module served to increase its activity on BMCC (Fig. 5C), despite its markedly lower levels of activity on CMC and PASC. In this case, the activity of 5A-t was significantly higher than that of the wild-type enzyme.
Activity of designer cellulosomes.
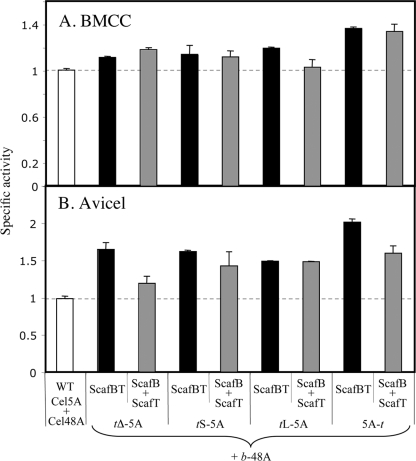
The different dockerin-bearing chimeras were combined with the chimeric T. fusca family 48 exoglucanase (b-48A) into simple designer cellulosome complexes using the binary chimeric scaffoldin, ScafBT. The cellulose-degrading activities of the resultant complexes were examined using BMCC (Fig. 6A) and Avicel (Fig. 6B), and their activity on these substrates was compared to that of a mixture of the wild-type enzymes. As observed for the individual dockerin-bearing chimeras alone, linker length had no effect on the activity. Interestingly, in all cases the designer cellulosome samples were more efficient in degrading the crystalline cellulose substrates than the mixture of wild-type enzymes. Designer cellulosomes containing the reversed chimera (5A-t) showed the highest activity on both crystalline substrates. The samples containing the combined individual chimeras assembled on their matching unary scaffoldins (containing a single matching cohesin and a CBM3a) were also more active than the wild-type enzyme mixture.
FIG. 6.
Enzymatic activity of the different Cel5A-derived chimeras in the designer cellulosomal mode, combined with a dockerin-containing chimeric exoglucanase. The four Cel5A chimeras, varying in their linker length and dockerin positions, were combined with the chimeric GH48 exoglucanase (b-48A) using ScafBT to assemble a binary designer cellulosome. The activities of the different assemblies were examined on the two crystalline substrates, BMCC (A) and Avicel (B). ScafBT (black bars) represents the two chimeras assembled on the binary scaffoldin. Samples designated ScafB+ScafT (gray bars) indicate the combined activity of the individual CBM-restored chimeras in the free state, i.e., b-48A and ScafB combined with the specified t-(5A) and ScafT. WT, combination of the wild-type enzymes, Cel5A and Cel48A (white bars). The figure shows the mean of triplicate samples, examined in two independent experiments, with error bars indicating the standard deviations. Specific activity is defined as mol of reducing sugar per mol of enzyme per min.
DISCUSSION
The ability to design and produce artificial cellulosomes of precise composition provides us with a way to address distinct aspects of cellulosome structure and function (5). Such designer cellulosomes may also provide precursors for improved cellulase systems for future industrial conversion of cellulosic biomass to biofuels (3-5, 8, 35). In previous work, we have produced designer cellulosomes of various compositions, in which different chimeric scaffoldins, bearing diverse permutations and combinations of CBMs and divergent cohesins, have been devised to assess the effect of targeting and enzyme position within the cellulosome complex (15). Using this approach, the comparative activities of various combinations of cellulases and a xylanase on pure and crude cellulosic substrates were assessed (17). This approach also enabled fabrication of novel cellulosome geometries, and their activities on crystalline cellulosic substrates were compared with those of more conventional designer cellulosomes (33). In one case, a fungus-derived cellulase was included into designer cellulosome modes together with standard cellulosomal enzymes (32).
One of our long-term objectives is to convert an entire free noncellulosomal cellulase system to a cellulosomal system by binding the cellulases to an artificial scaffoldin. Because T. fusca has a restricted number of highly active cellulases, its enzymes were selected for this effort. We have previously explored the properties of T. fusca endoglucanase Cel6A and exoglucanases Cel6B and Cel48A in preparation for their inclusion into designer cellulosome systems (11, 12). In all cases, the dockerin modules retained specific binding to their matching cohesins. Likewise, the catalytic modules retained their hydrolytic properties on various cellulosic substrates upon substitution of the dockerin for the CBM2 although removal of the CBM significantly affected the activity of the exoglucanases on crystalline cellulose.
In the present study, we have investigated the properties of a second T. fusca endoglucanase, Cel5A, and its conversion into the cellulosomal mode of action. We employed this enzyme to address the role of the intermodular linker segment on cellulase action within defined designer cellulosomes. In previous work (19), the linker connecting a dockerin module to the enzymatic subunit was studied by a small-angle X-ray scattering assay, and the conformational events likely to occur upon complexation between a dockerin and a cohesin were analyzed. The solution structure of the enzyme alone indicated that the linker is extended and flexible, but upon docking to the cohesin a pleating of the linker segment occurs, suggesting that the binding to the cognate cohesin includes a structural rearrangement of the enzyme, leading to optimal activity and cooperation with other catalytic subunits.
For this study, the native 22-residue linker that separates the N-terminal CBM2 from the catalytic module of Cel5A was either preserved (tL-5A), truncated to a 9-residue abridged version (tS-5A), or omitted altogether (tΔ-5A) during construction of the dockerin-appended derivatives. An alternative chimera (5A-t) was also prepared in which the dockerin was attached to the “nonnative” C terminus of the Cel5A catalytic module. These four different chimeric dockerin-containing, CBM-lacking forms of T. fusca Cel5A were examined for their activities on various crystalline and noncrystalline cellulosic substrates, either alone or in the cellulosomal mode together with a chimeric dockerin-containing form of exoglucanase, Cel48A.
The C. thermocellum dockerin from the bifunctional xylanase, Xyn10Z, was selected for this study. This dockerin is an “internal” dockerin that is positioned between two separate modules of this enzyme. The rationale was that we could attach this dockerin to either the N- or C-terminal portion of a given module since it is inherently attached within the native enzyme to modules on either end. Nevertheless, during the course of this study, we discovered that the dockerin from R. flavefaciens (which is originally positioned on the C terminus of its native parent protein) could be incorporated on the opposite (N-terminal) end of a chimeric protein (f-5A), with activity patterns equivalent to those of t-5A, thus indicating the versatility of the dockerin module in designer cellulosomes.
Like the two family 6 enzymes that were previously constructed and tested, all family 5 constructs bound efficiently and specifically to their target cohesin constructs, regardless of the dockerin type or linker length. Dockerin position relative to the catalytic module was shown to have an apparent influence on the binding properties of the enzyme but not on its binding specificity. The only chimera that exhibited somewhat lower levels of interaction with its matching cohesin was the 5A-t (the only chimera containing a C-terminal dockerin). Nevertheless, the latter dockerin-containing enzyme functioned in an equivalent manner to, and in some cases better than, the other dockerin-containing enzyme chimeras. The length of the linker that separates the dockerin from the catalytic module appeared to have little, if any, effect on the activity of the enzyme. On the other hand, the location of the dockerin at the N or C terminus appeared to have a significant effect. Despite the fact that the CBM2 is positioned on the N terminus of the native free Cel5A, the great majority of the known cellulosomal enzymes bear dockerins on the C-terminal sides of their catalytic modules. Taken together, our results suggest that the C terminus is the natural position for the dockerin in cellulosomes.
The results of this study demonstrate that free cellulase enzymes, unrelated to native cellulosome systems, can work in the cellulosomal mode and can act synergistically and more efficiently than the parallel wild-type enzyme mixture (Fig. 6). Although these findings are clearly not applicable to all enzymes, the studies show that dockerins can be freely attached to foreign enzymes with retention of component activity. In future studies, we will focus on the conversion of different enzyme systems to the cellulosomal mode of action in order to investigate the structural and functional properties of cellulosomes and their relationship to the parent bacterial cell and to various native and model cellulosic substrates.
Acknowledgments
We appreciate the technical assistance of Sarah Ouanounou and Yael Vazana.
This research was supported by a grant from the United States-Israel Binational Science Foundation, Jerusalem, Israel, and the Brazilian Friends of the Weizmann Institute of Science Alternative Energy Research Initiative and by the Israel Science Foundation (grants 966/09 and 159/07). E.A.B. is the incumbent of The Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry at the Weizmann Institute of Science.
Footnotes
Published ahead of print on 9 October 2009.
REFERENCES
- 1.Barak, Y., T. Handelsman, D. Nakar, A. Mechaly, R. Lamed, Y. Shoham, and E. A. Bayer. 2005. Matching fusion-protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18:491-501. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., J.-P. Belaich, Y. Shoham, and R. Lamed. 2004. The cellulosomes: multi-enzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58:521-554. [DOI] [PubMed] [Google Scholar]
- 3.Bayer, E. A., B. Henrissat, and R. Lamed. 2008. The cellulosome: a natural bacterial strategy to combat biomass recalcitrance, p. 407-426. In M. E. Himmel (ed.), Biomass recalcitrance. Blackwell, London, United Kingdom.
- 4.Bayer, E. A., R. Lamed, and M. E. Himmel. 2007. The potential of cellulases and cellulosomes for cellulosic waste management. Curr. Opin. Biotechnol. 18:237-245. [DOI] [PubMed] [Google Scholar]
- 5.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. [DOI] [PubMed] [Google Scholar]
- 6.Bayer, E. A., E. Morag, R. Lamed, S. Yaron, and Y. Shoham. 1998. Cellulosome structure: four-pronged attack using biochemistry, molecular biology, crystallography and bioinformatics, p. 39-65. In M. Claeyssens, W. Nerinckx, and K. Piens (ed.), Carbohydrases from Trichoderma reesei and other microorganisms. The Royal Society of Chemistry, London, United Kingdom.
- 7.Bayer, E. A., Y. Shoham, and R. Lamed. 2006. Cellulose-decomposing prokaryotes and their enzyme systems, p. 578-617. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 8.Bayer, E. A., Y. Shoham, and R. Lamed. 2008. Cellulosome-enhanced conversion of biomass: on the road to bioethanol, p. 75-96. In J. Wall, C. Harwood, and A. L. Demain (ed.), Bioenergy. ASM Press, Washington, DC.
- 9.Burton, J., S. G. Wood, M. Lynch, and A. G. Plaut. 1988. Substrate analogue inhibitors of the IgA1 proteinases from Neisseria gonorrhoeae. J. Med. Chem. 31:1647-1651. [DOI] [PubMed] [Google Scholar]
- 10.Bushuev, V. N., A. T. Gudkov, A. Liljas, and N. F. Sepetov. 1989. The flexible region of protein L12 from bacterial ribosomes studied by proton nuclear magnetic resonance. J. Biol. Chem. 264:4498-4505. [PubMed] [Google Scholar]
- 11.Caspi, J., D. Irwin, R. Lamed, H.-P. Fierobe, D. B. Wilson, and E. A. Bayer. 2008. Conversion of noncellulosomal Thermobifida fusca free exoglucanases into cellulosomal components: comparative impact on cellulose-degrading activity. J. Biotechnol. 135:351-357. [DOI] [PubMed] [Google Scholar]
- 12.Caspi, J., D. Irwin, R. Lamed, Y. Shoham, H.-P. Fierobe, D. B. Wilson, and E. A. Bayer. 2006. Thermobifida fusca family-6 cellulases as potential designer cellulosome components. Biocatal. Biotransformation 24:3-12. [Google Scholar]
- 13.Demain, A. L., M. Newcomb, and J. H. Wu. 2005. Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69:124-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doi, R. H., and A. Kosugi. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2:541-551. [DOI] [PubMed] [Google Scholar]
- 15.Fierobe, H.-P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Belaich, R. Lamed, Y. Shoham, and J.-P. Belaich. 2002. Degradation of cellulose substrates by cellulosome chimeras: substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 16.Fierobe, H.-P., A. Mechaly, C. Tardif, A. Belaich, R. Lamed, Y. Shoham, J.-P. Belaich, and E. A. Bayer. 2001. Design and production of active cellulosome chimeras: selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 17.Fierobe, H.-P., F. Mingardon, A. Mechaly, A. Belaich, M. T. Rincon, R. Lamed, C. Tardif, J.-P. Belaich, and E. A. Bayer. 2005. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined tri-functional scaffoldin. J. Biol. Chem. 280:16325-16334. [DOI] [PubMed] [Google Scholar]
- 18.Haimovitz, R., Y. Barak, E. Morag, M. Voronov-Goldman, R. Lamed, and E. A. Bayer. 2008. Cohesin-dockerin microarray: diverse specificities between two complementary families of interacting protein modules. Proteomics 8:968-979. [DOI] [PubMed] [Google Scholar]
- 19.Hammel, M., H. P. Fierobe, M. Czjzek, S. Finet, and V. Receveur-Brechot. 2004. Structural insights into the mechanism of formation of cellulosomes probed by small angle X-ray scattering. J. Biol. Chem. 279:55985-55994. [DOI] [PubMed] [Google Scholar]
- 20.Howard, M. B., N. A. Ekborg, L. E. Taylor, S. W. Hutcheson, and R. M. Weiner. 2004. Identification and analysis of polyserine linker domains in prokaryotic proteins with emphasis on the marine bacterium Microbulbifer degradans. Protein Sci. 13:1422-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, Y. J., and D. B. Wilson. 1988. Cloning of Thermomonospora fusca genes coding for beta 1-4 endoglucanases E1, E2 and E5. Gene 71:331-337. [DOI] [PubMed] [Google Scholar]
- 22.Irwin, D., L. Walker, M. Spezio, and D. Wilson. 1993. Activity studies of eight purified cellulases: specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42:1002-1013. [DOI] [PubMed] [Google Scholar]
- 23.Irwin, D. C., S. Zhang, and D. B. Wilson. 2000. Cloning, expression and characterization of a family 48 exocellulase, Cel48A, from Thermobifida fusca. Eur. J. Biochem. 267:4988-4997. [DOI] [PubMed] [Google Scholar]
- 24.Jung, H., D. B. Wilson, and L. P. Walker. 2003. Binding and reversibility of Thermobifida fusca Cel5A, Cel6B, and Cel48A and their respective catalytic domains to bacterial microcrystalline cellulose. Biotechnol. Bioeng. 84:151-159. [DOI] [PubMed] [Google Scholar]
- 25.Krieger, F., B. Fierz, and O. Bieri. 2003. Dynamics of unfolded polypeptide chains as model for the earliest steps in protein folding. J. Mol. Biol. 332:265-274. [DOI] [PubMed] [Google Scholar]
- 26.Ladurner, A. G., and A. R. Fersht. 1997. Glutamine, alanine or glycine repeats inserted into the loop of a protein have minimal effects on stability and folding rates. J. Mol. Biol. 273:330-337. [DOI] [PubMed] [Google Scholar]
- 27.Lamed, R., and E. A. Bayer. 1993. The cellulosome concept—a decade later, p. 1-12. In K. Shimada, S. Hoshino, K. Ohmiya, K. Sakka, Y. Kobayashi, and S. Karita (ed.), Genetics, biochemistry and ecology of lignocellulose degradation. Uni Publishers Co., Ltd., Tokyo, Japan.
- 28.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinnis, K., and D. B. Wilson. 1993. Disulfide arrangement and functional domains of β-1,4-endoglucanase E5 from Thermomonospora fusca. Biochemistry 32:8157-8161. [DOI] [PubMed] [Google Scholar]
- 30.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 31.Miller, G. L. R., W. E. Blum, and A. L. Burton. 1960. Measurements of carboxymethylcellulase activity. Anal. Biochem. 2:127-132. [Google Scholar]
- 32.Mingardon, F., A. Chanal, A. M. López-Contreras, C. Dray, E. A. Bayer, and H.-P. Fierobe. 2007. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl. Environ. Microbiol. 73:3822-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mingardon, F., A. Chanal, C. Tardif, E. A. Bayer, and H.-P. Fierobe. 2007. Exploration of new geometries in cellulosome-like chimeras. Appl. Environ. Microbiol. 73:7138-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nall, B. T. 1985. Proline isomerization and protein folding. Comments Mol. Cell. Biophys. 3:123-143. [Google Scholar]
- 35.Ohmiya, K., K. Sakka, T. Kimura, and K. Morimoto. 2003. Application of microbial genes to recalcitrant biomass utilization and environmental conservation. J. Biosci. Bioeng. 95:549-561. [PubMed] [Google Scholar]
- 36.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 37.Srisodsuk, M., T. Reinikainen, M. Penttilä, and T. T. Teeri. 1993. Role of the interdomain linker peptide of Trichoderma reesei cellobiohydrolase I in its interaction with crystalline cellulose. J. Biol. Chem. 268:20756-20761. [PubMed] [Google Scholar]
- 38.von Ossowski, I., J. T. Eaton, M. Czjzek, S. J. Perkins, T. P. Frandsen, M. Schülein, P. Panine, B. Henrissat, and V. Receveur-Bréchot. 2005. Protein disorder: conformational distribution of the flexible linker in a chimeric double cellulase. Biophys. J. 88:2823-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warren, R. A. J. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 40.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, D. B. 2004. Studies of Thermobifida fusca plant cell wall degrading enzymes. Chem. Rec. 4:72-82. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, D. B. 2008. Three microbial strategies for plant cell wall degradation. Ann. N. Y. Acad. Sci. 1125:289-297. [DOI] [PubMed] [Google Scholar]
- 43.Yaron, S., E. Morag, E. A. Bayer, R. Lamed, and Y. Shoham. 1995. Expression, purification and subunit-binding properties of cohesins 2 and 3 of the Clostridium thermocellum cellulosome. FEBS Lett. 360:121-124. [DOI] [PubMed] [Google Scholar]