Abstract
Essential membrane proteins are generally recognized as relevant potential drug targets due to their exposed localization in the cell envelope. Unfortunately, high-level production of membrane proteins for functional and structural analyses is often problematic. This is mainly due to their high overall hydrophobicity. To develop new concepts for membrane protein overproduction, we investigated whether the biogenesis of overproduced membrane proteins is affected by stress response-related proteolytic systems in the membrane. For this purpose, the well-established expression host Bacillus subtilis was used to overproduce eight essential membrane proteins from B. subtilis and Staphylococcus aureus. The results show that the σW regulon (responding to cell envelope perturbations) and the CssRS two-component regulatory system (responding to unfolded exported proteins) set critical limits to membrane protein production in large quantities. The identified sigW or cssRS mutant B. subtilis strains with significantly improved capacity for membrane protein production are interesting candidate expression hosts for fundamental research and biotechnological applications. Importantly, our results pinpoint the interdependent expression and function of membrane-associated proteases as key parameters in bacterial membrane protein production.
Membrane-embedded proteins are crucial for cellular homeostasis and life. Membrane proteins generally account for about 30% of the open reading frames in both prokaryotic and eukaryotic genomes (49), and they are involved in a wide range of different tasks. These include vital processes, such as energy transduction, phospholipid biosynthesis, protein translocation, cell wall biogenesis, cell division, and control of cell shape (52). Importantly, membrane proteins are partially exposed to the extracytoplasmic environment, which makes them readily accessible to drugs. For this reason, membrane proteins have become a major class of proteins in terms of current drug targets. Essential membrane proteins, which are indispensable for cell proliferation under specific conditions, are especially interesting from the pharmaceutical and biomedical perspectives because they represent prime targets for chemotherapy.
Unfortunately, progress in the area of membrane protein research has so far been slow. This has been attributed primarily to the high hydrophobicity of membrane proteins, which complicates high-level production, purification, and crystallization (25). Consequently, yields are often frustratingly low, as underscored by a series of elegant screens for membrane protein overproduction in Escherichia coli (10, 11, 15, 47). Moreover, the accumulation of overproduced proteins in biological membranes may affect bilayer integrity, which would be toxic for the producing cell (33). Additional limitations are potentially caused by saturation of the cellular machinery for insertion of proteins into the membrane or by saturation of the membrane itself, resulting in the cytoplasmic accumulation of overproduced membrane proteins as well as native membrane proteins (46). Such overproduced proteins are usually misfolded and/or inactive, and they have a high tendency to form insoluble (micro)aggregates. These practical problems focus attention on the fundamental question of which cellular mechanisms set the key limits to membrane protein production.
In the present studies, we show that important problems in membrane protein overproduction can be overcome by using different strains of the gram-positive bacterium Bacillus subtilis as the expression host, and we identify two key mechanisms that set limits to membrane protein production in this organism. B. subtilis is highly appreciated for biotechnological applications because it has a large capacity to secrete high-quality proteins into the culture medium and because it has the status of generally recognized as safe (18, 38, 50). Furthermore, B. subtilis is amenable to genetic engineering, and many expression systems are available (2, 16, 31, 40, 43, 44). This prompted us to investigate whether the secretion machinery of B. subtilis, which is also involved in membrane protein biogenesis (52), can be exploited for membrane protein overproduction. As model proteins for our studies, we selected essential membrane proteins that have a good potential to serve as targets for novel antimicrobial drugs. Accordingly, we not only overproduced B. subtilis membrane proteins but also their orthologues from the important human pathogen Staphylococcus aureus. Studies on these essential proteins are considered to be of major relevance, since S. aureus is rapidly gaining resistance against all available antibiotics and novel antibiotics against this pathogen are urgently needed (7, 17). The results of the present studies with homologous membrane proteins from B. subtilis and S. aureus show that, like in other expression hosts, bottlenecks in membrane protein production also do exist in B. subtilis. Importantly, however, at least some of the encountered bottlenecks can be overcome, because they relate to two dispensable membrane-associated stress-responsive systems: the σW regulon and the CssRS two-component regulatory system. Thus, the removal of at least one of these stress-responsive systems can result in drastically improved yields of particular membrane proteins.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
The plasmids and bacterial strains used in this study are listed in Table S1 of the supplemental material. B. subtilis and E. coli were grown with agitation in Luria broth (LB) medium (Difco Laboratories) at 37°C. Lactococccus lactis was grown at 30°C without agitation in M17 broth (Oxoid) supplemented with 0.5% (wt/vol) glucose and 0.5 M sucrose. S. aureus was grown at 37°C without agitation in beef heart infusion medium (Oxoid). Where appropriate, the growth medium was supplemented with antibiotics: ampicillin (100 μg/ml), erythromycin (2 μg/ml for B. subtilis and 5 μg/ml for L. lactis), chloramphenicol (5 μg/ml), kanamycin (20 μg/ml), phleomycin (5 μg/ml), spectinomycin (100 μg/ml), or tetracycline (10 μg/ml). For transformation of B. subtilis, Paris minimal medium was used as described by Kouwen et al. (28).
DNA techniques.
Chromosomal DNA was isolated from B. subtilis according to the methods of Bron and Venema (4), while chromosomal DNA from S. aureus was isolated using the GenElute genomic isolation kit (Sigma). B. subtilis was transformed as described by Kunst and Rapoport (30), E. coli was transformed using CaCl2-competent cells (37), and L. lactis was transformed as described by Leenhouts and Venema (32). Plasmids were isolated from E. coli and L. lactis using the High Pure plasmid isolation kit (Roche) or the Invisorb Spin Plasmid Mini Two kit (Invitek). For L. lactis, lysozyme was added at the first step, and the sample was incubated for 30 min at 50°C before continuing with the protocol supplied by the manufacturer. DNA purification, restriction, ligation, agarose gel electrophoresis, and PCR were performed as described by Sambrook et al. (37). Restriction enzymes were obtained from Roche Applied Science, New England Biolabs, or Sigma-Aldrich. Ligations were carried out using T4 DNA ligase (New England Biolabs), and PCR was performed using Pwo polymerase (Roche Applied Science). Constructed plasmids were checked by sequencing.
Sequence similarity searches and topology predictions.
Sequence similarity searches were performed using the protein-protein BLAST algorithm (BLASTP) of NCBI (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Topology predictions were performed using the Octopus (http://topcons.net/) and MEMSAT3 (http://bioinf.cs.ucl.ac.uk/psipred/) algorithms (24, 45).
Amplification of model genes and cloning into the subtilin-regulated expression (SURE) system.
The genes encoding selected model proteins were amplified using chromosomal DNA of B. subtilis 168 or S. aureus NCTC 8325 as a template and the primers listed in Table S2 of the supplemental material. A sequence encoding the Strep II tag was fused to the 3′ end of each gene. For B. subtilis rny, a silent point mutation was made to remove a restriction site for RcaI. To accomplish this, two separate PCRs were performed using primer sets B Rny F1/B Rny M1 or B Rny M2/B Rny R1. The PCR products were subsequently fused by PCR using primers B Rny F1 and B Rny R1. All PCR products were cloned in the pTOPO vector (Invitrogen) according to the protocol provided by the manufacturer. Cloned genes were subsequently transferred from pTOPO to pNZ8910 using restriction sites that were included in the start codon of each gene (see Table S2 in the supplemental material) and a restriction site originating from the pTOPO vector (B cdsA [PstI], S cdsA [XbaI], B pgsA [SpeI], S pgsA [SpeI], B rny [SpeI], S rny [SpeI], B plsY [XhoI], and S plsY [HindIII]). The pNZ8910 plasmids containing the different genes were subsequently checked by sequencing.
The amyE::spaRK construct was introduced into B. subtilis 1012 by transformation with genomic DNA of B. subtilis 168 NZ8900. Subsequently, pNZ8910-based plasmids containing genes for membrane proteins were introduced into B. subtilis 168 amyE::spaRK and B. subtilis 1012 amyE::spaRK.
Construction of a cssRS deletion mutant.
To delete cssRS, the flanking regions of the genes were amplified by PCR with primer sets CssRS d1+d2 and CssRS d3+d4. Next, these regions were fused by PCR to a spectinomycin cassette that was amplified from pDG1726 (primers are listed in Table S2 of the supplemental material). B. subtilis was subsequently transformed with the final PCR product. The correct removal of the cssRS genes was verified by Western blotting with antibodies against CssS.
Induction of membrane protein overproduction.
Subtilin was prepared as described by Bongers et al. (2). Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.15 in LB medium and incubated until mid-exponential growth (OD600 of 0.9 to 1.1). Subsequently, 1% (vol/vol) subtilin was added to part of the culture, and both the induced and noninduced cultures were further incubated for 2 h. The OD600 was measured at the end of the induction, and cells were subsequently lysed in NuPAGE LDS sample preparation buffer (Invitrogen) by using a minibeadbeater 16 (Biospec Inc.). To test whether the SURE system was functional in B. subtilis rasP, induction of expression of green fluorescent protein (GFP) from plasmid pNZ8907 was performed. GFP was detected using fluorescence microscopy.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Protein samples were heated for 5 min at 95°C and loaded on precast NuPAGE 10% bis-Tris gels (Invitrogen Life Technologies), with a correction for differences in OD600s of the cell cultures (an OD600 of 2 was taken as 10 μl). Semidry Western blotting was performed with Protran nitrocellulose membranes (Schleicher & Schuell). Strep II-tagged proteins were detected using StrepMAB Classic (IBA GmbH), diluted 1:1,000 in blocking buffer (LiCor Biosciences). Rabbit antibodies were used to detect Pbp4*, HtrA, HtrB, and CssS. These primary antibodies were visualized with fluorescent IRDye goat anti-rabbit or goat anti-mouse secondary antibodies (LiCor Biosciences) at 1:5,000 dilutions. The Odyssey infrared imaging system (LiCor Biosciences) was used to record the fluorescence at 700 or 800 nm, depending on the IRDye fluorescent label used. To verify gel loading, control blots were stained with ink (0.1% Pelikan ink; Königsblau number 4001; in phosphate-buffered saline with 0.05% Tween and 1% acetic acid).
Quantification of Rny production.
Purified Rny was a kind gift from Jörg Stülke (8). Dilution series of purified Rny and of cell lysates from noninduced and induced B. subtilis 168 amyE::spaRK pNZ::Bacillus Rny were loaded on precast NUPAGE 10% bis-Tris gels (Invitrogen Life Technologies). For the cell lysates, the amounts loaded on gels were corrected for OD600 differences (10 μl lysate when the OD600 was 2). After SDS-PAGE, Western blotting, and immunodetection with Rny-specific polyclonal antibodies raised in rabbits (Eurogentec), the signal in each lane of the Western blot was quantified using the ImageJ gel analyzer. For the purified Rny protein, a calibration plot was made. Similarly, for the dilution series of the induced and noninduced samples the signal was plotted against the dilutions. Subsequently, the calibration plot was used to determine the amount of protein in each of the dilution series.
Subcellular localization of the model proteins.
Fractionation experiments were performed to localize the YolF protein and the control proteins LipA, TrxA, SipS, and BdbD in B. subtilis. Cells were grown overnight in LB medium, collected by centrifugation, and resuspended in protoplast buffer (100 mM Tris-HCl, pH 8.2, 20 mM MgCl2, 20% sucrose, 1 mg/ml lysozyme, 0.01% DNase, and Complete protease inhibitors TM263). After a 30-min incubation at 37°C, proteins released from the cells by protoplasting (i.e., the cell wall fraction) were separated from the protoplasts by centrifugation (10 min, 4,000 × g, 4°C). The protoplasts were resuspended in disruption buffer (50 mM Tris-HCl, pH 8.2, 2.5 mM EDTA) and disrupted with glass beads by using a bead beater. Cellular debris and unbroken protoplasts were removed by centrifugation (10 min, 4,000 × g, 4°C), and the supernatant was ultracentrifuged (30 min, 200,000 × g, 4°C). Next, the supernatant fraction with the cytosolic proteins was collected. The pellet was resuspended in solubilization buffer (20 mM Tris, pH 8.0, 10% glycerol, 50 mM NaCl, 0.03% DDM [n-dodecyl-β-maltoside]) and incubated overnight at 4°C. Nonsolubilized membranes and solubilized membrane proteins were subsequently separated by centrifugation (15 min, 100,000 × g, 4°C), and the supernatant fraction with the solubilized membrane proteins was collected. The subcellular fractions thus obtained were analyzed by SDS-PAGE, Western blotting, and immunodetection with specific antibodies.
Sedimentation centrifugation.
An aliquot of each of the isolated membrane protein fractions was treated with 8 M urea. Forty-microliter aliquots of the original and the urea-treated samples were loaded on top of 100 μl of protoplast buffer with 20% sucrose and, for the urea-treated samples, with 8 M urea. These samples were centrifuged for 30 min at 100,000 × g in a swing-out rotor. From the resulting sample an upper fraction (40 μl), middle fraction (50 μl), and a lower fraction (approximately 70 μl) were taken, and 4× LDS sample buffer was added. Subsequently, the pellet was resuspended in 20 μl of 1× LDS loading buffer. The samples were heated at 95°C for 10 min before starting SDS-PAGE with 20 μl of each fraction. Via Western blotting, the amounts of the Strep-tagged model protein, as well as the marker proteins YolF (12.1 kDa), BdbD (24.8 kDa), and Rny (58.7 kDa), were determined in each fraction. In the calculations of the amounts of protein in each sample, a correction was made for the different volumes of each fraction.
RESULTS
Overproduction of essential model proteins.
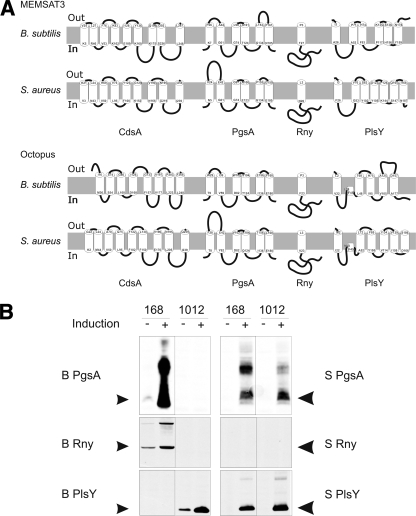
Previous studies have shown that 271 genes of B. subtilis are essential for growth under favorable laboratory conditions (26). These include 29 genes for membrane proteins. From these 29 we selected four proteins with differing predicted membrane topologies as model proteins for the present studies, namely, CdsA, PgsA, Rny, and PlsY (Fig. 1A). These proteins raised our interest because they are highly conserved in other bacteria but not in humans and because they are known to be essential for viability in other bacterial species (14, 23, 42). Additionally, their biochemical properties are poorly characterized, and structural information is completely lacking.
FIG. 1.
Predicted topologies of membrane proteins and their overproduction in B. subtilis. (Panel A) Topologies of the selected membrane proteins from B. subtilis and S. aureus were assessed with the MEMSAT3 and Octopus algorithms. (Panel B) Induced overproduction of membrane proteins from B. subtilis (B) and S. aureus (S) in B. subtilis strains 168 and 1012. Black arrowheads indicate the predicted electrophoretic mobilities according to the masses of the proteins.
For controlled expression of the selected membrane proteins, we applied the SURE system (2) in the two commonly used B. subtilis strains, 168 and 1012. Although these strains are generally considered to be highly similar, substantial differences were observed with regard to production of the model B. subtilis membrane proteins and their orthologues from S. aureus (Fig. 1B). To detect overproduced proteins by Western blotting, all genes were provided with the coding sequences for the Strep II tag. Only two proteins, namely, PgsA and PlsY from S. aureus, were overproduced in both strains. Three other proteins were readily produced in one strain but not in the other (i.e., PgsA, Rny, and PlsY from B. subtilis), and three proteins could not be detected in either of the strains (CdsA from B. subtilis and S. aureus and Rny from S. aureus) (Fig. 1B and data not shown). At present, we do not know whether this is due to lack of overproduction of these proteins or degradation of the Strep II tag. However, Strep II tag degradation seems unlikely in the case of S. aureus Rny, since the detection of this tag correlated consistently with the detection of Rny with specific polyclonal antibodies.
As demonstrated by quantitative immunoblotting, using purified B. subtilis Rny for calibration, B. subtilis 168 amyE::spaRK pNZ8910::B Rny overproduced Rny ∼45-fold to a level of about 4 mg per liter upon subtilin induction (see Fig. S1 in the supplemental material). Judged by the signal intensity, we believe that the production levels of other Strep II-tagged proteins were at least in the same range as shown for Rny or higher (Fig. 1B). Without induction, Rny was overproduced ∼7-fold, to about 0.63 mg per liter. These findings show that some, but not all, membrane proteins can be readily overproduced in B. subtilis and that there are strain-specific differences in productivity. Because of these differences, all further expression experiments were conducted using both the 168 and 1012 strains.
To pinpoint possible bottlenecks in membrane protein overproduction, we first tested a mutant strain that overproduced cytoplasmic chaperones. This was accomplished through deletion of the hrcA gene, resulting in upregulation of GrpE, DnaK, DnaJ, GroEL, and GroES. Unfortunately, this mutation only had negative effects on membrane protein overproduction (data not shown). Additionally, the effects of a lowered protease production by deletion of the genes for eight extracellular proteases were compared to the effects of growing cells in medium supplemented with Complete protease inhibitors (Roche) to inhibit protease activity. Interestingly, only growth in medium supplemented with protease inhibitors had a mildly positive effect on the amounts of membrane proteins produced, whereas knockout of the eight major extracellular proteases resulted in reduced productivity (data not shown). These findings directed our attention to stress-responsive systems in the B. subtilis cell envelope that are known to involve protease activity: the CssRS two-component regulatory system and the σW regulon.
The CssRS system sets limits to membrane protein overproduction.
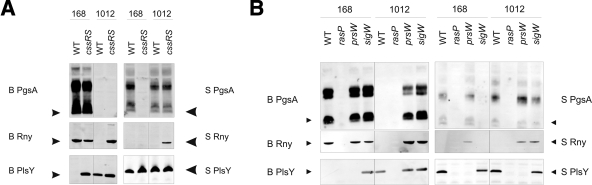
The CssS sensor and CssR regulator proteins form a two-component regulatory system that responds to the accumulation of malfolded secretory proteins at the membrane-cell wall interface (22). Stimulation of CssRS results in upregulation of two membrane-associated proteases, HtrA and HtrB, which have catalytic sites located at the extracytoplasmic side of the membrane (29, 41).To investigate possible interference of the CssRS system with membrane protein overproduction, cssRS deletion mutants of B. subtilis 168 and 1012 were used. Indeed, deletion of cssRS resulted in major improvements in membrane protein production (Fig. 2A). Most strikingly, knockout of cssRS now permitted the production of S. aureus Rny, albeit only in the 1012 strain. Furthermore, deletion of cssRS resulted in the production of large amounts of B. subtilis Rny in the 1012 strain and B. subtilis PlsY in the 168 strain. On the other hand, the cssRS deletion resulted in production of CdsA from neither B. subtilis nor S. aureus (data not shown). This deletion even interfered with the production of S. aureus PgsA in the 168 strain (Fig. 2A). Taken together, these findings show that the CssRS system impacts significantly on membrane protein overproduction and that, depending on the membrane protein studied, a cssRS deletion can have either highly beneficial effects, no effects, or even adverse effects on this process.
FIG. 2.
Membrane protein overproduction in cssRS, rasP, prsW, and sigW mutant strains. Overproduction of membrane proteins was assessed in the parental B. subtilis strains 168 and 1012 (WT) and in strains with specific mutations. (A) Strains containing a cssRS::spec deletion mutation; (B) strains carrying rasP::tet, prsW::bleo, or sigW::bleo deletion mutations. Black arrowheads indicate the predicted electrophoretic mobilities according to the masses of the proteins.
To further specify the mechanism by which CssRS impacts membrane protein overproduction, the complete htrA or htrB gene was deleted. However, membrane protein overproduction in these single knockouts was severely impaired (data not shown), which was probably due to the fact that htrA and htrB are cross-regulated. A double knockout by combination of both single knockouts could not be obtained, which is consistent with previously reported findings of Noone and Devine (34).
Roles of PrsW, RasP, and the σW regulon in membrane protein overproduction.
Regulatory intramembrane proteolysis is a process in which a membrane-bound regulatory protein is released to the cytoplasm by degradation of the transmembrane segment (20, 36). Two proteases of B. subtilis that are important for RIP are PrsW and RasP. PrsW represents a novel site 1 protease of the membrane-embedded metalloprotease superfamily (13, 35), whereas RasP is an intramembrane cleaving protease that belongs to the site 2 protease family of zinc metalloproteases (5). Both proteins are involved in degradation of RsiW, an anti-sigma factor that modulates the activity of σW (20, 39). Since both PrsW and RasP are capable of cleaving membrane proteins, we investigated whether overproduction of our eight model proteins was influenced in rasP or prsW deletion strains. Remarkably, deletion of prsW mostly enhanced membrane protein overproduction in both B. subtilis 168 and 1012, whereas deletion of rasP compromised membrane protein overproduction (Fig. 2B). Positive effects of the prsW deletion were observed for PgsA of B. subtilis and Rny of B. subtilis and S. aureus. Importantly, deletion of rasP did not per se preclude induction of the SURE system, as was verified by effective induction of a GFP control (data not shown).
The effects of PrsW on membrane protein overproduction could relate either directly to its proteolytic activity or indirectly to various roles of the σW regulon in membrane protein degradation. Therefore, the effects of prsW and sigW mutations were compared (Fig. 2B). Interestingly, these two mutations had largely overlapping, but not identical, effects on membrane protein overproduction. For example, a sigW deletion allowed the overproduction of B. subtilis PlsY in the 168 strain but interfered with S. aureus Rny production in this strain, whereas these effects were not observed upon deletion of prsW. Despite the highly beneficial effect of the sigW mutation on membrane protein overproduction, combining knockouts of sigW and rasP resulted in a loss of model protein production (data not shown), indicating that the adverse effect of rasP on membrane protein production was dominant over the positive effect of sigW.
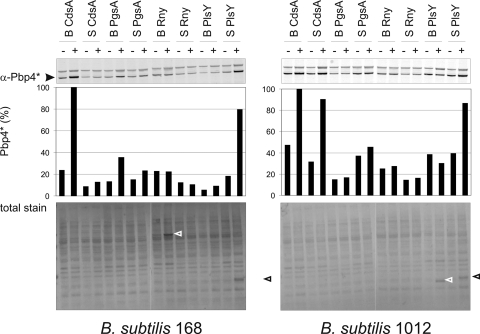
The results obtained with sigW and prsW mutants suggest a pivotal role for the σW regulon in the control of membrane protein overproduction. Therefore, activation of the σW regulon upon induced membrane protein production was monitored using antibodies against Pbp4*, a convenient standard marker for σW activation (Fig. 3) (39). The results thus obtained were subsequently confirmed with a transcriptional fusion between the sigW promoter and GFP (data not shown). Notably, induction of cdsA resulted in the strongest σW responses, even though CdsA was not detectably overproduced. In contrast, S. aureus plsY induction caused a strong σW response in the 1012 strain, while the PlsY protein was readily overproduced (Fig. 2B and 3), and this was in some experiments also observed for B. subtilis plsY induction (data not shown). Importantly, in those cases where sigW deletion resulted in strong overproduction of particular membrane proteins, their induction did not trigger a significant σW response. These findings show that there is no direct correlation between improved membrane protein production due to sigW deletion and induction of the σW regulon by overproduction of membrane proteins. This is consistent with our observation that the σW regulon is not the only limiting determinant in membrane protein overproduction and that deletions of prsW, rasP, and sigW can have very different effects on this process. The addition of subtilin itself did not lead to a detectable activation of the σW regulon at any time point during subsequent cultivation, indicating that the observed activation of the σW regulon is induced by the membrane protein expression rather than the exposure of cells to the subtilin. To assess whether a simultaneous deletion of sigW and cssRS would further enhance the overproduction of certain membrane proteins, we constructed a sigW cssRS double mutant strain. Unfortunately, none of our model proteins was overproduced in this double mutant.
FIG. 3.
Activation of the σW regulon. Activation of the σW regulon was monitored by assessment of the cellular Pbp4* levels in the presence (+) or absence (-) of subtilin-induced overexpression of genes for membrane proteins in B. subtilis strains 168 and 1012. As a control for protein loading, all blots were stained with ink (total stain). Black and white arrowheads mark bands of highly overproduced membrane proteins.
Increased levels of HtrA and HtrB in rasP mutant strains.
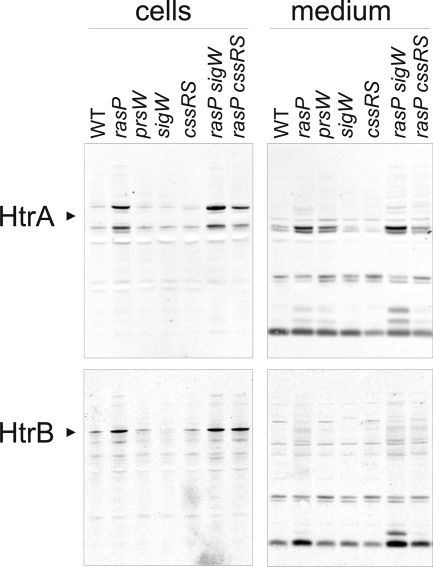
Since in several cases deletion of cssRS, prsW, or sigW resulted in improved membrane protein overproduction, the possibility of cross-interactions between the CssRS and σW stress-responsive systems was examined. First, we verified that no induction of σW occurred in prsW mutant strains, which was the case irrespective of membrane protein overproduction. Likewise, deletion of cssRS had no detectable effect on induction of the σW regulon as reflected by the cellular Pbp4* levels, irrespective of membrane protein overproduction (data not shown). Subsequently, the activity of the CssRS two-component system was monitored in B. subtilis prsW, rasP, or sigW mutant strains by monitoring the levels of cell-associated and extracellular HtrA or HtrB. Interestingly, the levels of HtrA and HtrB detected in B. subtilis rasP mutant cells were significantly increased compared to the parental strains, whereas levels of HtrA and HtrB in B. subtilis prsW or sigW mutants seemed generally slightly decreased compared to the parental strains (Fig. 4; only the results of B. subtilis 168 are shown). Notably, we detected for the first time an extracellular degradation product for HtrB, the amounts of which correlated well with the intracellular amounts of HtrB, as was the case for extracellular HtrA (Fig. 4). Importantly, the effect of rasP deletion on HtrA/B levels was dominant over the sigW mutation, and even over a cssRS mutation.
FIG. 4.
Induced overproduction of HtrA and HtrB. Cellular (“cells”) and extracellular (“medium”) levels of HtrA and HtrB were assessed by Western blotting for B. subtilis strain 168 (WT) and 168-derived strains with rasP::tet, prsW::bleo, sigW::bleo, and/or cssRS::spec single or double mutations. Cells were cultivated overnight. Black arrowheads indicate the predicted mobilities of HtrA and HtrB. All detectable major protein bands correspond to HtrA or HtrB, as was verified by Western blotting experiments with htrA or htrB mutant strains (data not shown).
Subcellular localization of overproduced membrane proteins.
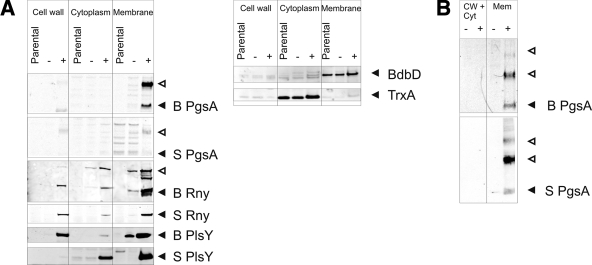
Conceivably, the removal of the CssRS and σW stress-responsive systems might lead to an accumulation of malfolded proteins in the cytoplasm, instead of the intended insertion of correctly folded proteins into the membrane. We therefore assessed whether the overproduced membrane proteins were indeed inserted into the membrane. For this purpose, the best-producing strains were selected for each model protein (in the cases of CdsA, PgsA, and Rny from B. subtilis the wild-type strains were used). As shown in Fig. 5, significant amounts of each overproduced protein fractionated with the membrane. In the case of Rny and PlsY, some overproduced protein was also detected in the cell wall and cytoplasmic fractions, suggesting that some overproduced protein was mislocalized or released from the membranes during cell disruption. To investigate whether the proteins detected in the membrane fractions were soluble, sedimentation centrifugation was used to separate soluble proteins from aggregates. Significant amounts of urea-soluble aggregates could only be detected for overproduced B. subtilis Rny (see Fig. S2 in the supplemental material). These observations show that most overproduced membrane proteins are correctly targeted to the membrane.
FIG. 5.
Subcellular localization of overproduced membrane proteins. (A) The subcellular localization of overproduced membrane proteins was determined by fractionation of cells lacking the SURE plasmid (parental) or cells containing the SURE plasmid with (+) or without (-) subtilin-induced expression of membrane proteins. For each model protein the best-producing strain was used. As a control for correct fractionation, the presence of BdbD (membrane protein) and TrxA (cytosolic protein) was determined in the samples of the S PlsY-producing strain. (B) For low-abundance proteins, the fractionation was repeated without lysozyme treatment of the cells, to avoid the (low) background signal caused by cross-reactivity of the secondary antibody to lysozyme. Filled and open arrowheads mark bands of overproduced membrane proteins.
DISCUSSION
In the present studies, we identified several strains of B. subtilis that have significantly improved capacities for membrane protein overproduction, and we pinpointed two stress-responsive systems as key bottlenecks in membrane protein production. To accomplish this, we used a subtilin-regulated (SURE) system to induce overproduction of Strep II-tagged membrane proteins. Induced expression of eight different proteins in two different strains of B. subtilis initially led to detectable overproduction of five of these proteins. These observations are in accordance with findings in E. coli that some membrane proteins are readily produced, whereas others cannot be detected at all (10, 11, 47). Strikingly, overproduction of membrane proteins under identical conditions in the laboratory strains B. subtilis 168 and 1012, which are generally considered to be highly similar, resulted in entirely different expression patterns. From recent studies, it has become clear that there is considerable genomic diversity among different laboratory strains of B. subtilis (12, 51), which probably provides a molecular basis for the observed differences in membrane protein production. In this respect, it is relevant that the 1012 strain seems to be a hybrid of the 168 and 23 strains (T. Wiegert, unpublished data).
Several of the tested modifications in stress-responsive and proteolytic systems of B. subtilis, such as deletion of multiple extracellular proteases or rasP, had a negative impact on membrane protein overproduction. These findings show that membrane protein overproduction is a delicate process which is easily compromised. This may relate to particular stresses provoked by such mutations rather than direct effects on membrane protein biogenesis. If so, the addition of protease inhibitors during induction of membrane protein overproduction appears to be a milder approach for the cells than deleting eight extracellular proteases. Clearly, deletion of rasP resulted in elevated levels of the HtrA and HtrB quality control proteases, which is possibly contraproductive for membrane proteins. Notably, even though removal of certain genes (e.g., rasP) has negative effects on membrane protein overproduction, it is still possible that the corresponding protein sets a limit to this process. This idea is supported by the fact that RasP was shown to degrade the RsiW and FtsL membrane proteins, thereby setting limits to their cellular levels (3, 39), and that RasP also impacts on secretion of the α-amylase AmyQ (19). We therefore conclude that the absence of RasP has pleiotropic effects on the stability of overproduced and native membrane proteins.
Three mutations were identified that had a positive effect on membrane protein production. These involved inactivation of the CssRS two-component system or removal of the prsW or sigW genes. Most likely, CssRS impacts on membrane protein overproduction through the HtrA and HtrB proteases, as the corresponding genes are main targets of CssRS regulation (9). This idea is consistent with the observation that impediment of membrane protein overproduction upon deletion of rasP is accompanied by elevated production of HtrA and HtrB. At present, it is not clear whether the impact of PrsW on membrane protein overproduction is due to a direct effect via its protease activity or an indirect effect via activation of the σW regulon, or both. For example, deletion of sigW had similar, but occasionally even stronger, effects on membrane protein yields than deletion of prsW. Furthermore, there appears to be no clear-cut correlation between the activity of the σW regulon and membrane protein overproduction; deletion of rasP, which precludes activation of the σW regulon like the prsW and sigW mutations, had a dominant negative effect on membrane protein overproduction, while the prsW and sigW mutations had generally positive effects on membrane protein overproduction. Within the σW regulon there are at least three putative proteases that might be involved in membrane protein degradation, namely, SppA and YqeZ, which are similar to E. coli signal peptide peptidase (SppA) (1), and YjoB, which belongs to the AAA family and might be involved in modulating the activity of one or more proteases (27). Which of these proteases are actually involved in membrane protein degradation will be a subject for further studies.
Remarkably, for each individual membrane protein tested, the effects of cssRS, prsW, or sigW deletion on the final yield varied significantly. This indicates that the yields of particular membrane proteins are determined to different extents by CssRS- and σW-dependent processes. More noteworthy, in some cases the interference with either CssRS or σW resulted in significantly improved yields, showing that the interference with two seemingly independent stress-responsive processes can have identical effects on membrane protein overproduction. These observations imply that there is some degree of interdependence between the CssRS two-component system and the σW regulon. Consistent with this view, we observed that deletion of rasP, and possibly also prsW and sigW, differentially influenced the CssRS two-component system, as evidenced by the cellular HtrA and HtrB levels. Interestingly, such effects have not yet been reported in transcriptional studies on the σW regulon (6, 21), indicating that the effect of RasP on HtrA and HtrB levels is probably not mediated through the σW regulon. Instead, it is very well possible that RasP is directly involved in the proteolysis of HtrA and HtrB, which might explain why the levels of these proteins are elevated in a rasP cssRS double mutant.
In conclusion, the present studies show that membrane-associated stress-responsive systems set major limits to membrane protein overproduction in B. subtilis. The removal of such bottlenecks is possible and significantly improved the yields of six out of eight tested membrane proteins. We are confident that our findings form an excellent starting point for further improvement of B. subtilis as a cell factory for membrane proteins and that they set the stage for further mechanistic studies on the interdependent expression and function of membrane-associated proteases in bacterial membrane protein production. In this respect, it will be of interest to investigate whether lower levels of induction will have beneficial effects on the membrane protein production levels, the quality of the overproduced membrane proteins, or the health of the producer cells, as was recently shown in E. coli (48). Such studies could then be extended to other B. subtilis strains, like strain 23, to explore their exploitation potentials for membrane protein production.
Supplementary Material
Acknowledgments
We thank Kevin Devine and David Noone for providing antibodies against CssS, HtrA, and HtrB.
J.M.V.D. and J.C.Z. were supported through CEU projects LSHG-CT-2004-503468, LSHG-CT-2004-005257, LSHM-CT-2006-019064, LSHG-CT-2006-037469, and PITN-GA-2008-215524, the transnational SysMO Initiative through Project BACELL SysMO, the European Science Foundation under the EUROCORES Programme EuroSCOPE, and grant 04-EScope 01-011 from the Research Council for Earth and Life Sciences of The Netherlands Organization for Scientific Research. T.W. was supported by the Deutsche Forschungsgemeinschaft (Schu 414/21-1).
Footnotes
Published ahead of print on 9 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bolhuis, A., A. Matzen, H. L. Hyyrylainen, V. P. Kontinen, R. Meima, J. Chapuis, G. Venema, S. Bron, R. Freudl, and J. M. van Dijl. 1999. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis required for efficient translocation and processing of secretory proteins. J. Biol. Chem. 274:24585-24592. [DOI] [PubMed] [Google Scholar]
- 2.Bongers, R. S., J. W. Veening, M. Van Wieringen, O. P. Kuipers, and M. Kleerebezem. 2005. Development and characterization of a subtilin-regulated expression system in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl. Environ. Microbiol. 71:8818-8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bramkamp, M., L. Weston, R. A. Daniel, and J. Errington. 2006. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol. Microbiol. 62:580-591. [DOI] [PubMed] [Google Scholar]
- 4.Bron, S., and G. Venema. 1972. Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat. Res. 15:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Brown, M. S., J. Ye, R. B. Rawson, and J. L. Goldstein. 2000. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100:391-398. [DOI] [PubMed] [Google Scholar]
- 6.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, H. F., and S. S. Hegde. 2007. Combating the growing problem of methicillin-resistant Staphylococcus aureus: do the newer antibiotics represent a better alternative to vancomycin? Expert Rev. Anti Infect. Ther. 5:333-335. [DOI] [PubMed] [Google Scholar]
- 8.Commichau, F. M., F. M. Rothe, C. Herzberg, E. Wagner, D. Hellwig, M. Lehnik-Habrink, E. Hammer, U. Volker, and J. Stulke. 2009. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics 8:1350-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drew, D., M. Lerch, E. Kunji, D. J. Slotboom, and J. W. de Gier. 2006. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 3:303-313. [DOI] [PubMed] [Google Scholar]
- 11.Drew, D., D. J. Slotboom, G. Friso, T. Reda, P. Genevaux, M. Rapp, N. M. Meindl-Beinker, W. Lambert, M. Lerch, D. O. Daley, K. J. van Wijk, J. Hirst, E. Kunji, and J. W. de Gier. 2005. A scalable, GFP-based pipeline for membrane protein overexpression screening and purification. Protein Sci. 14:2011-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earl, A. M., R. Losick, and R. Kolter. 2008. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 16:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellermeier, C. D., and R. Losick. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 20:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, C. Kg, P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 15.Geertsma, E. R., M. Groeneveld, D. J. Slotboom, and B. Poolman. 2008. Quality control of overexpressed membrane proteins. Proc. Natl. Acad. Sci. USA 105:5722-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissendorfer, M., and W. Hillen. 1990. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 33:657-663. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein, F. W. 2007. Combating resistance in a challenging, changing environment. Clin. Microbiol. Infect. 13(Suppl. 2):2-6. [DOI] [PubMed] [Google Scholar]
- 18.Harwood, C. R., and R. Cranenburgh. 2008. Bacillus protein secretion: an unfolding story. Trends Microbiol. 16:73-79. [DOI] [PubMed] [Google Scholar]
- 19.Heinrich, J., T. Lunden, V. P. Kontinen, and T. Wiegert. 2008. The Bacillus subtilis ABC transporter EcsAB influences intramembrane proteolysis through RasP. Microbiology 154:1989-1997. [DOI] [PubMed] [Google Scholar]
- 20.Heinrich, J., and T. Wiegert. 2006. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol. Microbiol. 62:566-579. [DOI] [PubMed] [Google Scholar]
- 21.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 22.Hyyrylainen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Pragai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 23.Ji, Y., B. Zhang, S. F. Van Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 24.Jones, D. T. 2007. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23:538-544. [DOI] [PubMed] [Google Scholar]
- 25.Keyes, M. H., D. N. Gray, K. E. Kreh, and C. R. Sanders. 2008. Solubilizing detergents for membrane proteins, p. 15-38. In S. Iwata (ed.), Methods and results in crystallization of membrane proteins. International University Line, La Jolla, CA.
- 26.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotschwar, M., S. Diermeier, and W. Schumann. 2004. The yjoB gene of Bacillus subtilis encodes a protein that is a novel member of the AAA family. FEMS Microbiol. Lett. 230:241-249. [DOI] [PubMed] [Google Scholar]
- 28.Kouwen, T. R., A. van der Groot, R. Dorenbos, T. Winter, H. Antelmann, M. C. Plaisier, W. J. Quax, J. M. van Dijl, and J. Y. Dubois. 2007. Thiol-disulphide oxidoreductase modules in the low-GC gram-positive bacteria. Mol. Microbiol. 64:984-999. [DOI] [PubMed] [Google Scholar]
- 29.Krojer, T., M. Garrido-Franco, R. Huber, M. Ehrmann, and T. Clausen. 2002. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 416:455-459. [DOI] [PubMed] [Google Scholar]
- 30.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 31.Lam, K. H., K. C. Chow, and W. K. Wong. 1998. Construction of an efficient Bacillus subtilis system for extracellular production of heterologous proteins. J. Biotechnol. 63:167-177. [DOI] [PubMed] [Google Scholar]
- 32.Leenhouts, K. J., and G. Venema. 1993. Lactococcal plasmid vectors, p. 65-94. In K. G. Hardy (ed.), Plasmids, a practical approach. Oxford University Press, Oxford, United Kingdom.
- 33.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 34.Noone, D., A. Howell, R. Collery, and K. M. Devine. 2001. YkdA and YvtA, HtrA-like serine proteases in Bacillus subtilis, engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J. Bacteriol. 183:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pei, J., and N. V. Grishin. 2001. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 26:275-277. [DOI] [PubMed] [Google Scholar]
- 36.Rudner, D. Z., P. Fawcett, and R. Losick. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA 96:14765-14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Schallmey, M., A. Singh, and O. P. Ward. 2004. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50:1-17. [DOI] [PubMed] [Google Scholar]
- 39.Schobel, S., S. Zellmeier, W. Schumann, and T. Wiegert. 2004. The Bacillus subtilis σW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 52:1091-1105. [DOI] [PubMed] [Google Scholar]
- 40.Schumann, W. 2007. Production of recombinant proteins in Bacillus subtilis. Adv. Appl. Microbiol. 62:137-189. [DOI] [PubMed] [Google Scholar]
- 41.Skorko-Glonek, J., K. Krzewski, B. Lipinska, E. Bertoli, and F. Tanfani. 1995. Comparison of the structure of wild-type HtrA heat shock protease and mutant HtrA proteins. A Fourier transform infrared spectroscopic study. J. Biol. Chem. 270:11140-11146. [DOI] [PubMed] [Google Scholar]
- 42.Thanassi, J. A., S. L. Hartman-Neumann, T. J. Dougherty, B. A. Dougherty, and M. J. Pucci. 2002. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 30:3152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thuy Le, A. T., and W. Schumann. 2007. A novel cold-inducible expression system for Bacillus subtilis. Protein Expr. Purif. 53:264-269. [DOI] [PubMed] [Google Scholar]
- 44.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 45.Viklund, H., and A. Elofsson. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662-1668. [DOI] [PubMed] [Google Scholar]
- 46.Wagner, S., L. Baars, A. J. Ytterberg, A. Klussmeier, C. S. Wagner, O. Nord, P. A. Nygren, K. J. van Wijk, and J. W. de Gier. 2007. Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteomics 6:1527-1550. [DOI] [PubMed] [Google Scholar]
- 47.Wagner, S., M. L. Bader, D. Drew, and J. W. de Gier. 2006. Rationalizing membrane protein overexpression. Trends Biotechnol. 24:364-371. [DOI] [PubMed] [Google Scholar]
- 48.Wagner, S., M. M. Klepsch, S. Schlegel, A. Appel, R. Draheim, M. Tarry, M. Hogbom, K. J. van Wijk, D. J. Slotboom, J. O. Persson, and J. W. de Gier. 2008. Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. USA 105:14371-14376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallin, E., and G. von Heijne. 1998. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7:1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Westers, L., H. Westers, and W. J. Quax. 2004. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Biochim. Biophys. Acta 1694:299-310. [DOI] [PubMed] [Google Scholar]
- 51.Zeigler, D. R., Z. Pragai, S. Rodriguez, B. Chevreux, A. Muffler, T. Albert, R. Bai, M. Wyss, and J. B. Perkins. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190:6983-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zweers, J. C., I. Barak, D. Becher, A. J. Driessen, M. Hecker, V. P. Kontinen, M. J. Saller, L. Vavrova, and J. M. van Dijl. 2008. Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb. Cell Fact. 7:10.:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.