Abstract
Rickettsia parkeri, a recently recognized pathogen of human, is one of several Rickettsia spp. in the United States that causes a spotted fever rickettsiosis. To gain insights into its biology and pathogenesis, we applied the proteomics approach to establish a two-dimensional gel proteome reference map and combined this technique with cell surface biotinylation to identify surface-exposed proteins of a low-passage isolate of R. parkeri obtained from a patient. We identified 91 proteins by matrix-assisted laser desorption ionization-tandem time of flight mass spectrometry. Of these, 28 were characterized as surface proteins, including virulence-related proteins (e.g., outer membrane protein A [OmpA], OmpB, β-peptide, and RickA). Two-dimensional immunoblotting with serum from the R. parkeri-infected index patient was utilized to identify the immunoreactive proteins as potential targets for diagnosis and vaccine development. In addition to the known rickettsial antigens, OmpA and OmpB, we identified translation initiation factor 2, cell division protein FtsZ, and cysteinyl-tRNA synthetase as immunoreactive proteins. The proteome map with corresponding cell surface protein analysis and antigen detection will facilitate a better understanding of the mechanisms of rickettsial pathogenesis.
Rickettsia parkeri, a member of the spotted fever group Rickettsia (SFGR), was first isolated from the Gulf Coast tick, Amblyomma maculatum, in 1937 (29). In 2004, the first confirmed human infection with R. parkeri was reported in a 40-year-old man from the Tidewater area of coastal Virginia. The agent was isolated in cell culture from an eschar biopsy specimen and designated the Portsmouth strain (28). Recently, the first recognized case of tick bite-associated human infection was described (43); however, the epidemiology of R. parkeri is not well defined. In the United States, R. parkeri has been detected in A. maculatum and A. americanum; the geographical overlap between R. parkeri and these ticks with that of the vectors of R. rickettsii (the etiological agent of Rocky Mountain spotted fever [RMSF]) suggests that many cases of R. parkeri infection have been misidentified as RMSF (27, 35). For example, Western blot analysis of serum specimens from 15 U.S. patients previously diagnosed with RMSF identified four serum specimens reactive with a 120-kDa protein of R. parkeri, suggesting infection with R. parkeri rather than R. rickettsii (30). However, a serologic test specific for this pathogen is not available (43), and little is known about its biology.
Due to their obligate intracellular nature, genetic manipulation of Rickettsia has proven difficult. Alternatively, protein expression profiles (proteomes) are utilized to identify the mechanisms of pathogenesis and differentiate rickettsial species recognizing host immune response specificity to cell surface molecules, referred to as outer membrane proteins (Omps). The presence or absence of some Omps allows for differentiation between the typhus group and the SFGR, and the response to some species within the SFGR is specific (2). Proteomes have been developed for R. prowazekii (7), R. conorii (31), and R. felis (26) by using two-dimensional liquid chromatography-tandem mass spectrometry (2D LC-MS/MS), two-dimensional polyacrylamide gel electrophoresis (2D PAGE) and MS, or sodium dodecyl sulfate (SDS)-PAGE and nanoLC-MS/MS, respectively. More recently, an emphasis has been placed on better understanding surface protein expression profiles for obligate intracellular bacteria in the family Anaplasmataceae since it is well recognized that Omps for these bacteria are critical for host cell invasion (12, 13). Likewise, the rickettsial Omps are critical for bacterial attachment and invasion of host cells (21, 23, 40).
To better understand the molecular basis of virulence of R. parkeri, we utilized 2D PAGE with a pH 3-10 immobilized pH gradient (IPG) coupled with matrix-assisted laser desorption ionization-tandem time of flight (MALDI-TOF/TOF) MS in order to establish the protein expression profile of a low-passage strain of R. parkeri isolated from an infected patient. This reference map will be useful for comparative analyses of protein profiling of R. parkeri as it is maintained under differing microenvironments (e.g., in the arthropod vector and vertebrate host). Biotinylation of cell surface proteins and 2D immunoblotting analysis were also used to identify the surface-exposed proteins and immunoreactive proteins as potential targets for diagnosis and vaccine development.
MATERIALS AND METHODS
Rickettsial culture and purification.
R. parkeri strain Portsmouth, isolated from a skin biopsy specimen obtained from the index case of R. parkeri rickettsiosis (28), were grown in African green monkey kidney cell line (Vero E6) in Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (HyClone, Logan, UT) in a humidified 5% CO2 incubator at 34°C. All rickettsiae used in subsequent purifications were obtained from 3 to 11 passes of the initial isolate in Vero E6 cells. When more than 90% of the cells were infected, as determined by Diff-Quik (Dade Behring, Deerfield, IL) staining according to the manufacturer's protocol, rickettsiae were purified from cell cultures as previously described by Weiss et al. (42), with modifications. Cells were collected and centrifuged at 500 × g for 15 min at 4°C. The resulting pellet was resuspended in K36 buffer (0.1 M potassium chloride, 0.015 M sodium chloride, 0.05 M potassium phosphate buffer [pH 7.0]), and the cells were lysed by passing the suspension several times through a 25- and a 27-gauge needle attached to a 10-ml syringe. Large cell debris was removed by centrifugation at 100 × g for 5 min at 4°C, and the supernatant was filtered through a 5-μm-pore size syringe filter (Millex-SV; Millipore, Billerica, MA). The rickettsiae in the filtrate were harvested by centrifugation at 9,500 × g for 30 min at 4°C. The obtained pellet was washed and resuspended in K36 buffer. The rickettsial suspension was layered over the discontinuous Renografin (Merry X-Ray Corp., Lake Charles, LA) gradient (15 to 37.5%) and centrifuged in an OptimaXL-100K ultracentrifuge at 90,000 × g using an SW41-Ti rotor (Beckman Coulter, Fullerton, CA) for 1.5 h at 4°C. The rickettsial band was drawn into a syringe through a 27-gauge needle and washed twice with K36 buffer by centrifugation at 13,000 × g for 10 min at 4°C. The homogeneity of Rickettsia was examined by Diff-Quik staining, and the Rickettsia pellet was either immediately used for biotinylation of rickettsial surface proteins or stored in protease inhibitor cocktail (Roche, Indianapolis, IN) at −80°C until being subjected to protein preparation.
Biotinylation of surface proteins.
Freshly purified R. parkeri were enumerated by using a BacLight viability stain kit (Molecular Probes, Carlsbad, CA) as described previously (37). Viable rickettsiae corresponding to 5 × 1010 cells were washed three times with phosphate-buffered saline (PBS; pH 8.0) by centrifugation at 13,000 × g for 10 min at 4°C. The bacteria were surface labeled by incubation with 417 μM sulfosuccinimidyl-6-[biotin-amido]hexanoate (Sulfo-NHS-LC-Biotin; Pierce, Rockford, IL) in PBS (pH 8.0) or in PBS alone (negative control) at 4°C for 5 min. Excess Sulfo-NHS-LC-Biotin was quenched and removed by three washes in 100 mM glycine-PBS with incubation for 15 min at 4°C for the first wash. The bacterial pellet was washed with PBS (pH 8.0) and stored in protease inhibitor cocktail at −80°C until used for protein extraction.
Protein preparation.
The rickettsial pellet was resuspended in lysis buffer (8 M urea, 2 M thiourea, 60 mM dithiothreitol [DTT], 0.2% Triton X-100, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS]) (5) and incubated for 15 min at room temperature. The suspension was subjected to cell lysis by continuous sonication for 5 min at 4 to 5°C in an ice-bath sonicator (Tru-Sweep model 275TA; Crest Ultrasonics, Trenton, NJ), followed by centrifugation at 13,000 × g for 10 min at 4°C. The procedure was repeated at least two more times to ensure complete cell lysis. Proteins in the supernatant were precipitated overnight with an equal volume of methanol and 4 volumes of acetone at −20°C. The protein pellet was collected by centrifugation at 13,000 × g for 15 min at 4°C, air dried, and dissolved in lysis buffer. The protein concentration was determined by a Bio-Rad protein assay (Bio-Rad, Hercules, CA) using bovine serum albumin as the standard. Samples were divided into aliquots and stored at −80°C.
2D PAGE.
An IPG strip (7 cm, linear pH 3 to 10; Bio-Rad) was passively rehydrated for 12 h with either 40 μg (for construction of proteome and cell surface proteome map), 20 μg (for detection of surface proteins), or 30 μg (for detection of immunoreactive proteins) of total protein in lysis buffer with an addition of 0.25% (vol/vol) Bio-Lyte 3/10 ampholyte (Bio-Rad) and 0.001% bromophenol blue. The first-dimension separation by isoelectric focusing (IEF) was then carried out in a Protean IEF cell apparatus (Bio-Rad) at 20°C with the following parameters: for 40 μg of total protein, 100 V for 1 h, 250 V for 1 h, 1,000 V for 30 min, 6,000 V for 2.5 h, and 6,000 V for 50,000 V·h; for 20 and 30 μg of total protein, 100 V for 30 min, 250 V for 30 min, 1,000 V for 30 min, 6,000 V for 6,000 V·h; and a final focusing step (20 μg of protein, 6,000 V for 30,000 V·h; 30 μg of protein, 6,000 V for 40,000 V·h). The focused IPG strip was incubated in 1× NuPAGE LDS sample buffer (Invitrogen) containing 1% (wt/vol) DTT for 15 min, followed by incubation for 15 min in 1× NuPAGE LDS sample buffer containing 2.5% (wt/vol) iodoacetamide. The second-dimension SDS-PAGE was conducted on the XCell SureLock Mini-Cell System (Invitrogen) using NuPAGE Novex 4 to 12% Bis-Tris Zoom gels (Invitrogen) at 100 V until the tracking dye font reached the bottom of the gel. After the electrophoretic run, gels were fixed in 10% methanol-7% acetic acid for 1 h and overnight stained with SYPRO Ruby protein gel stain (Bio-Rad). Gels were digitized by using Molecular Imager Gel Doc XR System (Bio-Rad) and kept at 4°C for spot excision and protein identification.
Western blot analysis.
Unlabeled or biotin-labeled R. parkeri proteins separated by 2D PAGE were electroblotted onto Immun-Blot polyvinylidene difluoride (PVDF) membranes (Bio-Rad) by using an XCell II blot module (Invitrogen) according to the manufacturer's protocol. After protein transfer, membranes were blocked for 2 h with 3% skim milk in Tris-buffered saline-0.1% Tween 20 (TBST; 20 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% Tween 20). For identification of immunoreactive proteins, the membranes were probed with a convalescent-phase serum sample obtained from the index patient with R. parkeri rickettsiosis (28) at a dilution of 1:200 for 2 h at room temperature, followed by a secondary antibody (horseradish peroxidase [HRP]-conjugated rabbit anti-human immunoglobulin G [IgG]; Sigma, St. Louis, MO) at a dilution of 1:80,000 for 1 h at room temperature. The 2D blot probed with secondary antibody alone served as the negative control. Rickettsial surface proteins were detected by incubation of the membrane with streptavidin-HRP conjugate (Invitrogen) at a dilution of 1:6,000 for 1 h at room temperature. Before and after addition of the secondary antibody, the membranes were rinsed and washed twice for 10 min each time with TBST. The membranes were developed by using a SuperSignal West Pico chemiluminescent substrate kit (Pierce). The developed membranes were stained with an MemCode reversible protein stain kit (Pierce), according to the manufacturer's protocol, to match the location of proteins on the membrane with the Western blot signals. Protein spots on a SYPRO Ruby-stained gel were aligned with the positive signals on a 2D immunoblot by using ImageMaster 2D Platinum software (version 5.0; Amersham Biosciences, Piscataway, NJ). The immunoreactive spots were excised by using the EXQuest spot cutter (Bio-Rad) and identified by MALDI-TOF/TOF MS.
Protein digestion, MALDI-TOF/TOF MS, and data analysis.
Protein digestion and MS were performed by the Nevada Proteomics Center, University of Nevada (Reno, NV) as follows: excised protein spots were digested on an Investigator Proprep (Genomic Solutions, Ann Arbor, MI) using a previously described protocol (33) with some modifications. Samples were washed twice with 25 mM ammonium bicarbonate and 100% acetonitrile, reduced and alkylated using 10 mM DTT and 100 mM iodoacetamide, and incubated with 75 ng of trypsin in 25 mM ammonium bicarbonate for 6 h at 37°C. Samples were prepared and spotted onto a MALDI target with ZipTipu-C18 (Millipore). Samples were aspirated, dispensed three times, eluted with 70% acetonitrile and 0.2% formic acid, and then overlaid with 0.5 μl of a 5-mg/ml MALDI matrix (α-cyano-4-hydroxycinnamic acid) and 10 mM ammonium phosphate. All MS data were collected by using an ABI 4700 MALDI TOF/TOF apparatus (Applied Biosystems, Foster City, CA). The data were acquired in reflector mode from a mass range of 700 to 4,000 Da, and 1,250 laser shots were averaged for each mass spectrum. Each sample was internally calibrated on trypsin's autolysis peaks. The eight most intense ions from the MS analysis, which were not on the exclusion list, were subjected to MS/MS. For MS/MS analysis, the mass range was 70 to precursor ion with a precursor window of −1 to 3 Da, with an average 5,000 laser shots for each spectrum. The data were stored in an Oracle database. The data were extracted from the Oracle database and a peak list was created by using GPS Explorer software (Applied Biosystems) from the raw data generated from the ABI 4700. This peak list was based on signal-to-noise filtering and an exclusion list and included deisotoping. The resulting file was then searched by Mascot (Matrix Science, Boston, MA) with database search parameters including a mass tolerance of 20 or 50 ppm, one missed cleavage, oxidation of methionines, and carbamidomethylation of cysteines. Only matched proteins with significant scores (P < 0.05) were reported.
In silico analyses.
The identified proteins were grouped according to the clusters of orthologous groups (COGs) functional classification (http://www.ncbi.nlm.nih.gov/COG/) (38). The signal peptide at the N terminus of the protein was predicted by the programs SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) (3) and LipoP 1.0 (http://www.cbs.dtu.dk/services/LipoP/) (20).
RESULTS
Proteome reference map of R. parkeri strain Portsmouth.
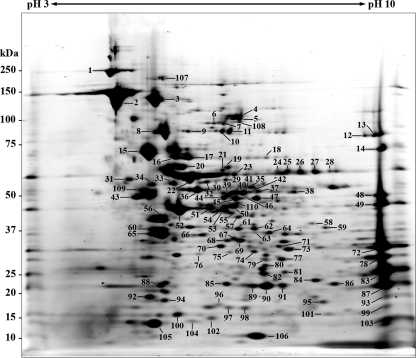
To establish the proteome map of R. parkeri, a 2D PAGE of rickettsial protein extract was performed by using a 7-cm pH 3 to 10 IPG strip, followed by a 4 to 12% Bis-Tris gel. Approximately 270 protein spots with isoelectric points (pIs) ranging from 4.5 to 9.5 and molecular masses ranging from 10 to 240 kDa could be visualized by SYPRO Ruby protein gel stain. The majority of proteins were located in the pI range of 5 to 8 and molecular-mass range of 20 to 100 kDa (Fig. 1). The intense protein spots of various pIs and molecular masses were excised and identified by MALDI-TOF/TOF MS. The 110 identified spots represent 91 unique proteins (Table 1). Of these, 12 proteins with predicted molecular masses of >70 kDa, 54 proteins with predicted molecular masses of 30 to 70 kDa, and 25 proteins with predicted molecular masses of <30 kDa were identified. Fourteen of the gene products, including OmpB (spots 2 and 107), preprotein translocase subunit SecA (spots 4 and 5), polyribonucleotide nucleotidyltransferase (spots 10 and 11), Omp1 (spots 12 and 13), chaperonin GroEL (spots 21 and 22), methyltransferase (spots 24 to 28), trigger factor (spots 40 to 42), heat shock protease (spots 46 and 47), hypothetical protein (spots 48 and 49), CapD protein (spots 62 and 63), 30S ribosomal protein S2 (spots 68 and 69), 3-deoxy-manno-octulosonate cytidylyltransferase (spots 79 and 80), thioredoxin peroxidase 1 (spots 89 and 90), and single-stranded DNA-binding protein (spots 97 and 98) were found in multiple spots indicating isoforms (Table 1). These isoforms are apparent as either a vertical or horizontal pattern of spots on a 2D gel. The sequence coverage of the identified proteins ranged from 4% (spot 1, cell surface antigen rOmpA) to 98% (spot 106, 10-kDa chaperonin). OmpB, protein PS 120, elongation factor G, DnaK protein, and elongation factor Tu were the highly abundant proteins on the R. parkeri proteome map. In most cases, observed molecular mass values were in good agreement with that of predicted, except for spot 72, which was identified as OmpB. This protein spot had a molecular mass of 32 kDa, much lower than its predicted molecular mass (164 kDa), suggesting protein cleavage. It has been demonstrated that the whole molecule of OmpB is processed into the mature 120-kDa protein and the 32-kDa fragment, known as β-peptide (16). Because the Mascot search result of this spot showed that all six matched peptides were located in the carboxy-terminal region of the full-length OmpB, a finding consistent with the β-peptide, we identified spot 72 as the OmpB β-peptide. Among the identified protein spots, 12 spots were identified as 11 different unknown or hypothetical proteins in which seven of them had pIs ranging from 5 to 8, and the remaining exhibited basic pIs (pH > 9).
FIG. 1.
2D gel proteome reference map of R. parkeri strain Portsmouth. IEF was performed with total protein extract of R. parkeri using a 7-cm pH 3 to 10 immobilized gradient strip, followed by SDS-PAGE on a 4 to 12% Bis-Tris gel and stained with SYPRO Ruby protein gel stain. The numbers refer to the protein identities shown in Table 1. The molecular masses of the Precision Plus Protein Kaleidoscope standards (Bio-Rad) are indicated on the left.
TABLE 1.
R. parkeri (Portsmouth) proteins identified by MALDI-TOF/TOF MS
| Category and spot no.a | GenInfo identifier no. | Protein descriptionb | Score | No. of peptides matched | Predicted molecular mass (kDa) | Predicted pI | Sequence coverage (%) |
|---|---|---|---|---|---|---|---|
| Translation, ribosomal structure, and biogenesis (J) | |||||||
| 8 | 15892097 | Elongation factor G | 1160 | 60 | 78.00 | 5.22 | 76 |
| 10 | 15619751 | Polyribonucleotide nucleotidyltransferase* | 120 | 28 | 82.07 | 6.15 | 35 |
| 11 | 34580431 | Polyribonucleotide nucleotidyltransferase* | 1040 | 50 | 82.17 | 6.31 | 47 |
| 16 | 20139895 | Aspartyl-tRNA synthetase | 274 | 40 | 67.97 | 5.35 | 51 |
| 20 | 15619842 | 30S ribosomal protein S1 | 226 | 28 | 63.59 | 5.49 | 28 |
| 23 | 34580946 | Arginyl-tRNA synthetase | 91 | 14 | 65.22 | 6.24 | 28 |
| 30 | 34581409 | Glutamyl-tRNA synthetase | 471 | 25 | 58.43 | 5.81 | 32 |
| 33 | 157828067 | Aspartyl/glutamyl-tRNA amidotransferase subunit B | 555 | 28 | 54.47 | 5.29 | 33 |
| 51 | 15893139 | Seryl-tRNA synthetase | 884 | 33 | 48.66 | 5.62 | 54 |
| 52 | 22087329 | Elongation factor Tu | 306 | 8 | 42.97 | 5.50 | 22 |
| 59 | 15892504 | Phenylalanyl-tRNA synthetase subunit alpha | 582 | 31 | 40.48 | 7.77 | 56 |
| 65 | 15619155 | Elongation factor EF-Ts | 810 | 35 | 33.72 | 5.21 | 81 |
| 67 | 157828584 | Tryptophanyl-tRNA synthetase | 519 | 22 | 37.54 | 6.42 | 32 |
| 68 | 34580930 | 30S ribosomal protein S2* | 472 | 21 | 32.94 | 6.34 | 58 |
| 69 | 15619153 | 30S ribosomal protein S2* | 231 | 20 | 32.84 | 6.34 | 46 |
| 84 | 34580453 | Probable sigma(54) modulation protein | 101 | 12 | 21.88 | 7.71 | 37 |
| 86 | 157828071 | Ribosome recycling factor | 481 | 16 | 20.91 | 7.88 | 50 |
| 93 | 15892102 | 50S ribosomal protein L10 | 263 | 15 | 18.16 | 9.55 | 51 |
| 99 | 167471261 | 50S ribosomal protein L19 | 251 | 16 | 15.86 | 10.15 | 58 |
| 104 | 161723851 | 30S ribosomal protein S6 | 364 | 17 | 13.92 | 5.82 | 84 |
| 105 | 42453334 | Ribosomal protein L7/L12 | 257 | 15 | 15.01 | 6.41 | 78 |
| 108 | 15892739 | Translation initiation factor IF-2† | 438 | 35 | 91.07 | 6.52 | 40 |
| 110 | 15892034 | Cysteinyl-tRNA synthetase† | 179 | 18 | 53.25 | 6.23 | 24 |
| Transcription (K) | |||||||
| 34 | 34581537 | N utilization substance protein A | 900 | 41 | 56.57 | 4.99 | 48 |
| 60 | 34581393 | DNA-directed RNA polymerase alpha chain | 387 | 29 | 38.23 | 5.10 | 60 |
| 92 | 34581077 | Transcription elongation factor EF | 319 | 23 | 18.13 | 4.99 | 67 |
| 103 | 15619757 | Unknown (RC0668) | 126 | 7 | 14.39 | 9.48 | 29 |
| Replication, recombination, and repair (L) | |||||||
| 18 | 34581050 | DNA mismatch repair protein MutL | 382 | 25 | 69.19 | 7.04 | 24 |
| 56 | 15619665 | DNA polymerase III beta chain | 214 | 31 | 42.21 | 5.27 | 65 |
| 61 | 15893105 | Recombinase A | 802 | 35 | 39.65 | 6.93 | 63 |
| 97 | 157829131 | Single-strand DNA-binding protein* | 236 | 11 | 17.44 | 6.07 | 57 |
| 98 | 157829131 | Single-strand DNA-binding protein* | 157 | 11 | 17.44 | 6.07 | 48 |
| Signal transduction mechanisms (T) | |||||||
| 82 | 15619141 | Transcriptional activator protein czcR | 88 | 12 | 26.59 | 6.85 | 39 |
| 101 | 13235447 | Hypothetical protein | 82 | 10 | 17.17 | 7.83 | 47 |
| Cell wall/membrane biogenesis (M) | |||||||
| 12 | 34580844 | Outer membrane protein Omp1* | 195 | 41 | 86.90 | 8.61 | 41 |
| 13 | 34580844 | Outer membrane protein Omp1* | 144 | 47 | 86.90 | 8.61 | 49 |
| 37 | 67004914 | Carboxyl-terminal protease | 91 | 21 | 50.46 | 6.18 | 38 |
| 57 | 15619530 | Putative UDP-N-acetylglucosamine 2-epimerase | 92 | 22 | 43.33 | 6.25 | 62 |
| 62 | 15619529 | CapD protein* | 218 | 27 | 38.39 | 6.68 | 52 |
| 63 | 15619529 | CapD protein* | 66 | 10 | 38.39 | 6.68 | 18 |
| 77 | 157828329 | Putative dTDP-4-dehydrorhamnose reductase | 760 | 30 | 32.27 | 7.64 | 52 |
| 79 | 34580558 | 3-Deoxy-manno-octulosonate cytidylyltransferase* | 173 | 18 | 27.45 | 6.45 | 58 |
| 80 | 34580558 | 3-Deoxy-manno-octulosonate cytidylyltransferase* | 280 | 23 | 27.45 | 6.45 | 50 |
| 83 | 34581125 | Hypothetical protein | 464 | 24 | 26.36 | 9.54 | 53 |
| 87 | 34581124 | Hypothetical protein | 471 | 20 | 24.02 | 9.38 | 60 |
| Intracellular trafficking, secretion, and vesicular transport (U) | |||||||
| 4 | 167472009 | Preprotein translocase subunit SecA* | 110 | 21 | 103.51 | 6.28 | 28 |
| 5 | 53732210 | Preprotein translocase subunit SecA* | 87 | 31 | 103.60 | 6.17 | 31 |
| Posttranslational modification, protein turnover, chaperones (O) | |||||||
| 6 | 34580983 | ClpB protein | 117 | 24 | 95.98 | 5.93 | 32 |
| 15 | 34580814 | DnaK protein | 493 | 52 | 67.94 | 5.00 | 62 |
| 17 | 34581106 | Heat shock protein HtpG | 289 | 35 | 70.76 | 5.61 | 44 |
| 21 | 53732249 | Chaperonin GroEL* | 163 | 20 | 58.59 | 5.62 | 32 |
| 22 | 53732249 | Chaperonin GroEL* | 294 | 36 | 58.59 | 5.62 | 60 |
| 31 | 34580880 | Periplasmic serine protease | 688 | 25 | 55.58 | 4.71 | 34 |
| 40 | 34581103 | Trigger factor* | 505 | 32 | 50.98 | 6.06 | 48 |
| 41 | 34581103 | Trigger factor* | 178 | 24 | 50.98 | 6.06 | 33 |
| 42 | 34581103 | Trigger factor* | 135 | 29 | 50.98 | 6.06 | 58 |
| 44 | 42453569 | ATP-dependent protease HslVU (ClpYQ), ATPase subunit | 115 | 27 | 49.62 | 6.05 | 49 |
| 46 | 15892157 | Heat shock protease* | 87 | 16 | 55.54 | 7.68 | 33 |
| 47 | 34580813 | Heat shock protease* | 175 | 34 | 55.53 | 7.68 | 42 |
| 64 | 15892560 | Thioredoxin reductase | 517 | 23 | 37.52 | 7.15 | 40 |
| 78 | 34581487 | Protein export protein prsA precursor | 94 | 19 | 31.40 | 9.04 | 50 |
| 88 | 15620090 | GrpE protein | 159 | 13 | 20.16 | 5.18 | 47 |
| 89 | 157828322 | Thioredoxin peroxidase 1* | 444 | 21 | 22.49 | 6.62 | 62 |
| 90 | 15892374 | Thioredoxin peroxidase 1* | 493 | 24 | 22.74 | 6.62 | 67 |
| 91 | 157828616 | ATP-dependent Clp protease proteolytic subunit | 393 | 19 | 22.86 | 6.60 | 42 |
| 95 | 34581294 | Bacterioferritin comigratory protein | 336 | 15 | 17.90 | 7.74 | 51 |
| 100 | 34580701 | Heat shock protein | 211 | 18 | 18.95 | 6.34 | 76 |
| 106 | 34581406 | 10-kDa chaperonin | 429 | 21 | 10.53 | 6.74 | 98 |
| Cell division and chromosome partitioning (D) | |||||||
| 109 | 157828869 | Cell division protein FtsZ† | 525 | 26 | 48.45 | 4.89 | 56 |
| Energy production and conversion (C) | |||||||
| 7 | 157829065 | Aconitate hydratase | 78 | 16 | 97.35 | 5.88 | 15 |
| 9 | 15892430 | Malic enzyme | 88 | 20 | 84.42 | 5.61 | 21 |
| 19 | 157964195 | Succinate dehydrogenase flavoprotein subunit | 237 | 17 | 65.95 | 6.23 | 23 |
| 32 | 161723840 | F0F1 ATP synthase subunit alpha | 1150 | 45 | 56.13 | 5.85 | 51 |
| 43 | 53732375 | F0F1-type ATP synthase, beta subunit | 296 | 31 | 51.07 | 5.00 | 62 |
| 45 | 167471897 | Dihydrolipoamide dehydrogenase | 761 | 40 | 49.32 | 6.17 | 57 |
| 53 | 15619278 | Dihydrolipoamide acetyltransferase component | 301 | 25 | 42.77 | 6.89 | 60 |
| 55 | 157829141 | Type II citrate synthase | 413 | 28 | 49.40 | 6.20 | 39 |
| 58 | 53732427 | NAD/NADP transhydrogenase alpha subunit | 311 | 26 | 40.60 | 7.66 | 54 |
| 66 | 34580715 | Pyruvate dehydrogenase e1 component alpha subunit precursor | 100 | 23 | 36.67 | 5.76 | 49 |
| 70 | 34580561 | Malate dehydrogenase | 623 | 25 | 33.64 | 6.01 | 62 |
| 73 | 34580487 | Succinyl-CoA synthetase alpha chain | 143 | 18 | 30.24 | 6.66 | 48 |
| Carbohydrate transport and metabolism (G) | |||||||
| 76 | 34581482 | Hypothetical protein | 77 | 14 | 34.45 | 5.44 | 40 |
| Amino acid transport and metabolism (E) | |||||||
| 36 | 15619233 | Aminopeptidase A | 342 | 28 | 53.99 | 5.69 | 39 |
| 38 | 167471346 | Thermostable carboxypeptidase | 462 | 27 | 57.24 | 6.73 | 33 |
| 39 | 34580709 | Isocitrate dehydrogenase | 115 | 22 | 53.89 | 6.10 | 30 |
| 71 | 157828466 | Dihydrodipicolinate synthase | 221 | 12 | 32.81 | 7.08 | 30 |
| 81 | 157828064 | Dihydrodipicolinate reductase | 275 | 15 | 26.58 | 7.05 | 44 |
| Nucleotide transport and metabolism (F) | |||||||
| 75 | 15892328 | FAD-dependent thymidylate synthase | 483 | 23 | 34.58 | 6.23 | 48 |
| 102 | 34580958 | Nucleoside diphosphate kinase | 134 | 12 | 14.71 | 5.77 | 59 |
| Coenzyme transport and metabolism (H) | |||||||
| 50 | 34581632 | Hypothetical protein | 523 | 26 | 51.09 | 6.24 | 44 |
| Lipid transport and metabolism (I) | |||||||
| 35 | 34581415 | Propionyl-CoA carboxylase beta chain precursor | 136 | 17 | 57.05 | 6.58 | 26 |
| 54 | 167472306 | 3-Oxoacyl-(acyl carrier protein) synthase II | 365 | 21 | 45.58 | 5.81 | 38 |
| Inorganic ion transport and metabolism (P) | |||||||
| 85 | 34580412 | Superoxide dismutase | 310 | 17 | 24.83 | 6.25 | 52 |
| 96 | 157828723 | Hypothetical protein A1G_04785 | 207 | 15 | 19.51 | 5.50 | 41 |
| General function prediction only (R) | |||||||
| 24 | 165933779 | Methyltransferase* | 332 | 23 | 63.38 | 7.68 | 22 |
| 25 | 165933779 | Methyltransferase* | 748 | 34 | 63.38 | 7.68 | 37 |
| 26 | 165933779 | Methyltransferase* | 95 | 16 | 63.38 | 7.68 | 21 |
| 27 | 165933779 | Methyltransferase* | 764 | 40 | 63.38 | 7.68 | 44 |
| 28 | 165933779 | Methyltransferase* | 641 | 38 | 63.38 | 7.68 | 42 |
| 29 | 34581008 | Hypothetical protein | 261 | 26 | 61.54 | 6.18 | 36 |
| 74 | 34580794 | Hypothetical protein | 623 | 29 | 35.61 | 6.37 | 56 |
| Function unknown (S) | |||||||
| 1 | 62861417 | Cell surface antigen rOmpA† | 94 | 9 | 210.38 | 5.27 | 4 |
| 2 | 6969950 | OmpB*† | 287 | 14 | 164.16 | 5.20 | 12 |
| 72 | 6969950 | OmpB β-peptide | 322 | 6 | 164.16 | 5.20 | 4 |
| 107 | 6969950 | OmpB*† | 289 | 26 | 164.16 | 5.20 | 12 |
| Not in COGs | |||||||
| 3 | 13568657 | Protein PS 120 | 460 | 67 | 110.63 | 5.18 | 60 |
| 14 | 34581459 | Hypothetical WASP N-WASP MENA proteins | 286 | 33 | 59.45 | 9.27 | 44 |
| 48 | 15892021 | Hypothetical protein RC0098* | 173 | 6 | 48.05 | 9.24 | 12 |
| 49 | 34580943 | Hypothetical protein* | 551 | 39 | 48.07 | 9.24 | 60 |
| 94 | 15619114 | Unknown (RC0076) | 225 | 20 | 21.14 | 5.56 | 78 |
The COG category abbreviations are given in parentheses.
*, Protein isoforms; †, immunoreactive proteins recognized by serum from the R. parkeri-infected index patient. CoA, coenzyme A.
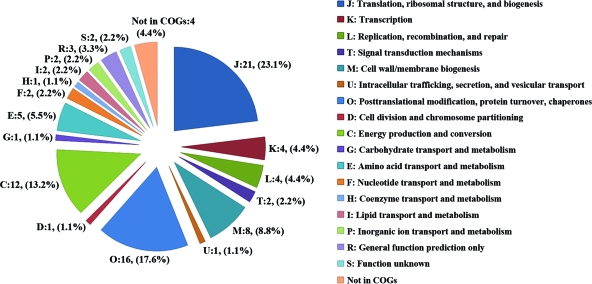
The identified proteins were classified into different functional categories according to COGs and found to be distributed in 17 different orthologous groups (Fig. 2). The majority of these proteins are involved in translation, ribosomal structure, and biogenesis (COG:J, 23.1%); posttranslational modification, protein turnover, and chaperones (COG:O, 17.6%); energy production and conversion (COG:C, 13.2%); and cell wall/membrane biogenesis (COG:M, 8.8%). There were 4.4% of proteins identified that were not in COGs, and 2.2% belong to the unknown function orthologous group (COG:S).
FIG. 2.
Functional distribution of identified R. parkeri proteins. The pie chart displays the proportion of identified proteins assigned to different functional categories according to the COGs functional classification (http://www.ncbi.nlm.nih.gov/COG). The number and percentage of identified proteins associated with each COG functional category are shown.
Identification of R. parkeri surface-exposed proteins by cell surface biotinylation.
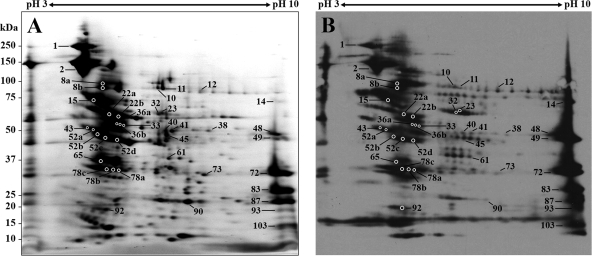
We performed biotinylation of viable R. parkeri using water-soluble and membrane-impermeable reagent, Sulfo-NHS-LC-Biotin, to identify surface-exposed proteins. The protein products resolved by 2D PAGE were visualized by SYPRO Ruby protein gel stain or transferred to a PVDF membrane. The biotinylated proteins were detected by using HRP-linked streptavidin and enhanced chemiluminescence. The 2D blot of unlabeled rickettsial proteins incubated with streptavidin-HRP, which served as the negative control to check for endogenous biotin-containing polypeptides, showed only one positive spot with an apparent molecular mass of ∼68 kDa and a pI of 5.6 (data not shown). This spot was not included in the analysis of rickettsial surface proteins. A total of 59 intense protein spots in a SYPRO Ruby-stained gel corresponding to Western blot signals were subjected to MALDI-TOF/TOF analysis. Among the 59 selected spots, 40 of which represent 28 proteins including known immunodominant surface-exposed proteins, Omps A and B, were successfully identified in the present study (Table 2). Some of the labeled proteins appeared as chains of spots with slightly different pIs (Fig. 3) that were not observed in 2D gel of unlabeled sample (Fig. 1). SignalP and LipoP analyses revealed a putative N-terminal signal peptide sequence with the cleavage site for signal peptidase I (SpI) in seven identified proteins including OmpA, OmpB, Omp1, protein export protein prsA precursor, and three hypothetical proteins (spots 48, 83, and 87).
TABLE 2.
Surface-exposed proteins of R. parkeri (Portsmouth) identified by MALDI-TOF/TOF MS
| Spot no. | GenInfo identifier no. | Protein description | Score | No. of peptides matched | Predicted molecular mass (kDa) | Predicted pI | Sequence coverage (%) |
|---|---|---|---|---|---|---|---|
| 1 | 1778893 | rOmpA | 50 | 1 | 110.11 | 5.30 | 1 |
| 2 | 6969950 | OmpB | 298 | 14 | 164.21 | 5.20 | 10 |
| 8a-b | 15892097 | Elongation factor G | 163 | 30 | 78.00 | 5.22 | 35 |
| 10 | 34580431 | Polyribonucleotide nucleotidyltransferase | 254 | 28 | 82.17 | 6.31 | 27 |
| 11 | 34580431 | Polyribonucleotide nucleotidyltransferase | 293 | 32 | 82.17 | 6.31 | 36 |
| 12 | 34580844 | Outer membrane protein Omp1 | 161 | 22 | 86.90 | 8.61 | 15 |
| 14 | 34581459 | Hypothetical WASP N-WASP MENA proteins | 191 | 16 | 59.45 | 9.27 | 19 |
| 15 | 34580814 | DnaK protein | 618 | 46 | 68.05 | 5.00 | 48 |
| 22a-b | 15892891 | Chaperonin GroEL | 475 | 39 | 58.68 | 5.62 | 60 |
| 23 | 15892018 | Arginyl-tRNA synthetase | 48 | 2 | 65.25 | 6.24 | 5 |
| 32 | 34581170 | ATP synthase alpha chain | 253 | 22 | 56.11 | 5.74 | 30 |
| 33 | 34580853 | Glutamyl-tRNA amidotransferase subunit B | 221 | 24 | 54.37 | 5.35 | 23 |
| 36a-b | 34580862 | Aminopeptidase A | 95 | 14 | 54.25 | 5.62 | 19 |
| 38 | 15892151 | Thermostable carboxypeptidase | 129 | 15 | 57.23 | 6.70 | 17 |
| 40 | 34581103 | Trigger factor | 192 | 27 | 50.98 | 6.06 | 48 |
| 41 | 34581103 | Trigger factor | 634 | 42 | 50.98 | 6.06 | 66 |
| 43 | 34581172 | ATP synthase beta chain | 442 | 28 | 51.21 | 4.87 | 46 |
| 45 | 34581573 | Dihydrolipoamide dehydrogenase precursor | 220 | 17 | 49.31 | 6.38 | 29 |
| 48 | 15892021 | hypothetical protein RC0098 | 252 | 25 | 48.05 | 9.24 | 45 |
| 49 | 15892021 | hypothetical protein RC0098 | 186 | 21 | 48.05 | 9.24 | 41 |
| 52a-d | 61223562 | Elongation factor Tu (EF-Tu) | 429 | 28 | 43.02 | 5.50 | 53 |
| 61 | 15893105 | Recombinase A | 212 | 22 | 39.65 | 6.93 | 45 |
| 65 | 15892036 | Elongation factor Ts | 226 | 21 | 33.78 | 5.21 | 60 |
| 72 | 6969950 | OmpB β-peptide | 293 | 23 | 164.21 | 5.20 | 9 |
| 73 | 34580487 | Succinyl-CoA synthetase alpha chain | 255 | 14 | 30.58 | 6.66 | 35 |
| 78a-c | 34581487 | Protein export protein prsA precursor | 148 | 11 | 31.40 | 9.04 | 28 |
| 83 | 15893204 | Hypothetical protein RC1281 | 486 | 23 | 26.92 | 9.58 | 51 |
| 87 | 34581124 | Hypothetical protein | 549 | 21 | 24.02 | 9.38 | 51 |
| 90 | 15892374 | Thioredoxin peroxidase 1 | 478 | 23 | 22.74 | 6.62 | 64 |
| 92 | 34581077 | Transcription elongation factor EF | 96 | 12 | 18.13 | 4.99 | 46 |
| 93 | 15892102 | 50S ribosomal protein L10 | 61 | 1 | 18.16 | 9.55 | 4 |
| 103 | 15892591 | Hypothetical protein RC0668 | 54 | 1 | 14.50 | 9.48 | 8 |
FIG. 3.
2D gel and blot of biotin-labeled R. parkeri surface proteins. The biotinylated proteins resolved by 2D PAGE were stained with SYPRO Ruby protein gel stain (A) or transferred to a PVDF membrane and detected using streptavidin-HRP conjugate (B). The numbers refer to the protein identities shown in Table 2. The molecular masses of the Precision Plus Protein Kaleidoscope standards (Bio-Rad) are indicated on the left.
R. parkeri immunoreactive proteins recognized by patient serum.
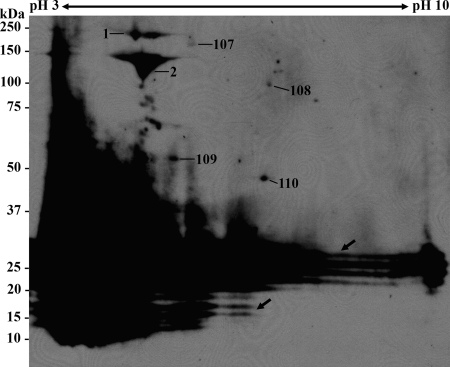
To identify immunoreactive antigens of R. parkeri, proteins separated by 2D PAGE were electroblotted onto a PVDF membrane and probed with a serum specimen obtained from the patient from whom R. parkeri strain Portsmouth was also isolated. The 2D immunoblots were then incubated with anti-IgG antibody, which was tested to have no reactivity with R. parkeri proteins (data not shown). Protein spots on a SYPRO Ruby-stained gel, which aligned to antigenic spots on a 2D immunoblot, were excised and analyzed by MALDI-TOF/TOF MS. Serum from the index patient reacted with seven protein spots with the observed molecular masses ranging from 50 to 240 kDa (Fig. 4). Six immunoreactive spots corresponding to five proteins—OmpA (spot 1), OmpB (spots 2 and 107), translation initiation factor IF-2 (spot 108), cell division protein FtsZ (spot 109), and cysteinyl-tRNA synthetase (spot 110)—were successfully identified (see Table 1). Moreover, the typical ladder pattern of lipopolysaccharide was recognized by this serum.
FIG. 4.
2D immunoblot of R. parkeri protein extract. The proteins separated by 2D PAGE were transferred to a PVDF membrane and probed with R. parkeri index patient serum. The numbers refer to the protein identities shown in Table 1. Arrows indicate the typical ladder pattern of lipopolysaccharide. The molecular masses of the Precision Plus Protein Kaleidoscope standards (Bio-Rad) are indicated on the left.
DISCUSSION
Although R. parkeri was first identified more than 70 years ago, there are relatively few data that describe its biology, and none that identify molecular constituents involved in its pathogenic behavior in human hosts. In the present study, a 2D gel proteome reference map of R. parkeri strain Portsmouth was constructed. A total of 110 spots representing 91 proteins were identified by MALDI-TOF/TOF. A variety of analytical methods have been used for rickettsial proteome analyses. The number of identified proteins in the present study was less than that of R. prowazekii analyzed by a 2D LC-MS/MS technique (7) and R. felis characterized by using two proteomic approaches: 2D PAGE coupled with MALDI-TOF and SDS-PAGE with nanoLC-MS/MS (26). However, our identification rate was comparable to that of R. felis analyzed only by 2D PAGE and MS. Among the 91 R. parkeri proteins that we identified, 60 were orthologs not reported in the R. felis 2D proteome map by this technique (26). The theoretical and experimentally observed molecular mass and pI values of the identified proteins were in general agreement, except for spot 72, which was identified as OmpB β-peptide. A good correlation between the predicted and observed molecular mass and pI of R. parkeri β-peptide was found when the molecular mass and pI values were calculated based on the amino acid sequence reported in the GenBank database (accession number FJ644549) using the pI/molecular-mass tool in the Expasy proteomic server (http://www.expasy.org/tools/pi_tool.html). The presence of several protein isoforms as either a vertical or horizontal pattern of spots on the 2D map of R. parkeri was likely due to posttranslational modifications (PTMs). Similar observations were made in other rickettsial proteomic studies (26, 31). In bacteria, PTMs play important roles in protein stability, signaling process, and host-pathogen interaction and in determining antigenicity (44). Identifying proteins that undergo PTMs, as reported here, facilitates future studies designed to decipher the biological significance of PTMs in Rickettsia spp.
Several of the R. parkeri proteins that we identified have been implicated in the virulence of other Rickettsia spp. The WASP N-WASP MENA proteins or RickA are involved in actin-based motility through activation of the Arp2/3 complex utilized by SFGR to exit from the host cell (15, 18, 19). The ability of R. parkeri to form actin tails was reported by Heinzen et al. (17). The role of methyltransferase in the pathogenesis of R. prowazekii has been suggested. This protein encoded by open reading frames RP028 and RP027 was expressed in the virulent Breinl strain but not in the avirulent Madrid E strain (8). The finding was supported by a frameshift mutation of the gene only in the avirulent Madrid E strain (46). BLASTP analysis showed that a hypothetical protein of R. parkeri (spot 29) shared 86 and 87% amino acid sequence identity to R. prowazekii proteins encoded by RP027 and RP028, respectively, suggesting the expression of methyltransferase in R. parkeri. We also identified three surface cell antigen proteins, including OmpA, OmpB, and protein PS 120. OmpA and OmpB are involved in rickettsial adhesion to and invasion of host cells (21, 23, 40). In contrast to other rickettsial proteome analyses (7, 26), only one protein involved in the secretion system, preprotein translocase subunit SecA, was successfully identified in the current study. In addition, we were unable to detect any type IV secretion system (T4SS) proteins. T4SS genes have been identified in all Rickettsia genomes analyzed to date, including the earliest diverging species, R. bellii (25), and the closely related R. africae (10). In this context, it is likely that one or more functional T4SS genes also exist in R. parkeri. Although we identified >90 R. parkeri-associated proteins, it is possible that some others elaborated by this pathogen were not detected because of the amount of bacterial protein used in the analyses or low-level expression during the particular growth conditions. However, when the complete genome sequence of R. parkeri becomes available, these data will allow for prediction of these and other genes and better assessment of the complete guild of proteins associated with this SFGR.
Two additional hypothetical proteins detected in our proteome analysis, as well as the β-peptide of R. parkeri, are orthologs of putative rickettsial adhesins. The β-peptide and R. conorii protein encoded by RC1281, which has sequence similarities to R. parkeri proteins of unknown function (spots 83 and 87), act as adhesin molecules that bind to surface proteins of Vero cells (32). The confirmed expression of these virulence determinants is consistent with other pathogenic rickettsial species and the ability of R. parkeri to cause disease in humans.
All identified proteins were analyzed for their COGs functional classifications. We observed a similar expression profile to previously reported rickettsial proteomes in which a large portion of identified R. parkeri proteins belongs to the functional category of translation, ribosomal structure and biogenesis (7, 26). Moreover, the most common genes identified in the genomes of Rickettsia, Orientia, and Wolbachia are involved in translation (11, 14). Further analysis of the unique requirements for protein synthesis associated with arthropod versus vertebrate host should illuminate novel mechanisms of pathogenesis.
We further applied cell surface biotinylation and a proteomics approach to identify 28 distinct surface proteins of R. parkeri. Of these, seven proteins, including OmpA, OmpB, Omp1, protein export protein prsA precursor, and three hypothetical proteins, were predicted to have the signal peptide sequences with the cleavage site for SpI. The findings of the present study corroborate the results reported by Ammerman et al. (1) in which an Escherichia coli-based alkaline phosphatase assay identified OmpB, Omp1, protein export protein prsA, and proteins of unknown function encoded by open reading frames RT0064, RT0815, and RT0816, which are the orthologs of three R. parkeri hypothetical proteins identified in the present study, to be Sec-dependent extracytoplasmic proteins. The surface expression of two R. parkeri hypothetical proteins (spots 83 and 87) and β-peptide is supported by previous work showing that orthologs of these proteins function as putative adhesins in R. conorii (32).
The rest of the surface proteins identified in the present study lack a putative N-terminal signal sequence and are generally considered cytosolic proteins. These proteins could be secreted by an unknown mechanism or released from bacteria with damaged cell membranes, an artifact of the purification step, and subsequently bound to the surface of intact cells. However, homologs of these proteins, including elongation factor G, polyribonucleotide nucleotidyltransferase, DnaK protein, chaperonin GroEL, two tRNA synthetases, aminopeptidase A, trigger factor, ATP synthase beta chain, dihydrolipoamide dehydrogenase, elongation factor Tu, recombinase A, and elongation factor Ts, were detected in the membrane fraction of R. conorii and other gram-negative bacteria (4, 12, 13, 22, 31, 34). The localization of the WASP N-WASP MENA proteins or RickA on the cell surface of R. conorii has been demonstrated. This protein activates Arp2/3 and stimulates actin polymerization (15). Because surface proteins are known to play crucial roles in host cell adhesion and invasion, further studies should be conducted to examine the functions of identified surface proteins in the virulence of R. parkeri.
In addition to the proteome map and surface-associated protein identification, the immunoreactive proteins of R. parkeri were identified by 2D immunoblotting analysis. Five proteins reacted with a convalescent-phase serum sample from the index patient. As expected, OmpA and OmpB were identified as major antigens and as surface-exposed proteins in the present study. It has been shown that both proteins are able to stimulate protective immunity against rickettsiosis in laboratory animals (6, 9, 36, 41). To the best of our knowledge, the immunogenicity of the remaining three antigenic proteins—translation initiation factor IF-2, cell division protein FtsZ, and cysteinyl-tRNA synthetase—has not been described for other Rickettsia spp.; it is unknown whether these represent immunologically reactive proteins unique to R. parkeri or whether antigenic homologs exist among other SFGR. Further studies will require screening with serum specimens from additional patients. However, the antigenicity of translation initiation factor IF-2 and cell division protein FtsZ homologs has been reported in previous studies of several bacteria (4, 24, 45). Moreover, the FtsZ-like protein has been suggested to be involved in pathogenesis of Bacillus anthracis (39).
In summary, we established a 2D reference map of proteins expressed in R. parkeri and identified 91 distinct proteins by MALDI-TOF/TOF. Of these, 28 were characterized as surface-exposed proteins by using cell surface biotinylation technique, including virulence-related proteins. Our data provide a basis for understanding the pathogenesis of R. parkeri. The proteome reference map will facilitate comparative analyses of differential protein expression under various environmental conditions or during the infection process. Finally, we identified novel immunoreactive proteins recognized by serum from the index patient which may serve as potential targets for diagnosis and disease prevention.
Acknowledgments
We thank J. A. Macaluso for helpful comments.
Protein identification at the Nevada Proteomics Center, University of Nevada, Reno, was supported by National Institutes of Health grant P20 RR-016464 from the INBRE Program of the National Center for Research Resources. This research was supported by the National Institute of Allergy and Infectious Diseases (AI070705).
The findings and conclusions presented in the present study are those of the authors and do not necessarily represent the views of the U.S. Department of Health and Human Services.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 21 September 2009.
REFERENCES
- 1.Ammerman, N. C., M. S. Rahman, and A. F. Azad. 2008. Characterization of Sec-translocon-dependent extracytoplasmic proteins of Rickettsia typhi. J. Bacteriol. 190:6234-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anacker, R. L., R. E. Mann, and C. Gonzales. 1987. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J. Clin. Microbiol. 25:167-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen, J. D., H. Nielsen, H. G. von, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 4.Boonjakuakul, J. K., H. L. Gerns, Y. T. Chen, L. D. Hicks, M. F. Minnick, S. E. Dixon, S. C. Hall, and J. E. Koehler. 2007. Proteomic and immunoblot analyses of Bartonella quintana total membrane proteins identify antigens recognized by sera from infected patients. Infect. Immun. 75:2548-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourchookarn, A., P. O. Havanapan, V. Thongboonkerd, and C. Krittanai. 2008. Proteomic analysis of altered proteins in lymphoid organ of yellow head virus-infected Penaeus monodon. Biochim. Biophys. Acta 1784:504-511. [DOI] [PubMed] [Google Scholar]
- 6.Bourgeois, A. L., and G. A. Dasch. 1981. The species-specific surface protein antigen of Rickettsia typhi: immunogenicity and protective efficacy in guinea pigs, p. 71-80. In W. Burgdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial diseases. Academic Press, Inc., New York, NY.
- 7.Chao, C. C., D. Chelius, T. Zhang, L. Daggle, and W. M. Ching. 2004. Proteome analysis of Madrid E strain of Rickettsia prowazekii. Proteomics 4:1280-1292. [DOI] [PubMed] [Google Scholar]
- 8.Chao, C. C., D. Chelius, T. Zhang, E. Mutumanje, and W. M. Ching. 2007. Insight into the virulence of Rickettsia prowazekii by proteomic analysis and comparison with an avirulent strain. Biochim. Biophys. Acta 1774:373-381. [DOI] [PubMed] [Google Scholar]
- 9.Crocquet-Valdes, P. A., C. M. az-Montero, H. M. Feng, H. Li, A. D. Barrett, and D. H. Walker. 2001. Immunization with a portion of rickettsial outer membrane protein A stimulates protective immunity against spotted fever rickettsiosis. Vaccine 20:979-988. [DOI] [PubMed] [Google Scholar]
- 10.Fournier, P. E., K. K. El, Q. Leroy, C. Robert, B. Giumelli, P. Renesto, C. Socolovschi, P. Parola, S. Audic, and D. Raoult. 2009. Analysis of the Rickettsia africae genome reveals that virulence acquisition in Rickettsia species may be explained by genome reduction. BMC Genomics 10:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuxelius, H. H., A. Darby, C. K. Min, N. H. Cho, and S. G. Andersson. 2007. The genomic and metabolic diversity of Rickettsia. Res. Microbiol. 158:745-753. [DOI] [PubMed] [Google Scholar]
- 12.Ge, Y., and Y. Rikihisa. 2007. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J. Bacteriol. 189:7819-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, Y., and Y. Rikihisa. 2007. Surface-exposed proteins of Ehrlichia chaffeensis. Infect. Immun. 75:3833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie, J. J., K. Williams, M. Shukla, E. E. Snyder, E. K. Nordberg, S. M. Ceraul, C. Dharmanolla, D. Rainey, J. Soneja, J. M. Shallom, N. D. Vishnubhat, R. Wattam, A. Purkayastha, M. Czar, O. Crasta, J. C. Setubal, A. F. Azad, and B. S. Sobral. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS ONE 3:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gouin, E., C. Egile, P. Dehoux, V. Villiers, J. Adams, F. Gertler, R. Li, and P. Cossart. 2004. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature 427:457-461. [DOI] [PubMed] [Google Scholar]
- 16.Hackstadt, T., R. Messer, W. Cieplak, and M. G. Peacock. 1992. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun. 60:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinzen, R. A., S. F. Hayes, M. G. Peacock, and T. Hackstadt. 1993. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect. Immun. 61:1926-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hybiske, K., and R. S. Stephens. 2008. Exit strategies of intracellular pathogens. Nat. Rev. Microbiol. 6:99-110. [DOI] [PubMed] [Google Scholar]
- 19.Jeng, R. L., E. D. Goley, J. A. D'Alessio, O. Y. Chaga, T. M. Svitkina, G. G. Borisy, R. A. Heinzen, and M. D. Welch. 2004. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell. Microbiol. 6:761-769. [DOI] [PubMed] [Google Scholar]
- 20.Juncker, A. S., H. Willenbrock, H. G. von, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H., and D. H. Walker. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 24:289-298. [DOI] [PubMed] [Google Scholar]
- 22.Lopez, J. E., W. F. Siems, G. H. Palmer, K. A. Brayton, T. C. McGuire, J. Norimine, and W. C. Brown. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73:8109-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez, J. J., S. Seveau, E. Veiga, S. Matsuyama, and P. Cossart. 2005. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell 123:1013-1023. [DOI] [PubMed] [Google Scholar]
- 24.Nowalk, A. J., R. D. Gilmore, Jr., and J. A. Carroll. 2006. Serologic proteome analysis of Borrelia burgdorferi membrane-associated proteins. Infect. Immun. 74:3864-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogata, H., S. B. La, S. Audic, P. Renesto, G. Blanc, C. Robert, P. E. Fournier, J. M. Claverie, and D. Raoult. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogawa, M., P. Renesto, S. Azza, D. Moinier, P. Fourquet, J. P. Gorvel, and D. Raoult. 2007. Proteome analysis of Rickettsia felis highlights the expression profile of intracellular bacteria. Proteomics 7:1232-1248. [DOI] [PubMed] [Google Scholar]
- 27.Paddock, C. D., R. W. Finley, C. S. Wright, H. N. Robinson, B. J. Schrodt, C. C. Lane, O. Ekenna, M. A. Blass, C. L. Tamminga, C. A. Ohl, S. L. McLellan, J. Goddard, R. C. Holman, J. J. Openshaw, J. W. Sumner, S. R. Zaki, and M. E. Eremeeva. 2008. Rickettsia parkeri rickettsiosis and its clinical distinction from Rocky Mountain spotted fever. Clin. Infect. Dis. 47:1188-1196. [DOI] [PubMed] [Google Scholar]
- 28.Paddock, C. D., J. W. Sumner, J. A. Comer, S. R. Zaki, C. S. Goldsmith, J. Goddard, S. L. McLellan, C. L. Tamminga, and C. A. Ohl. 2004. Rickettsia parkeri: a newly recognized cause of spotted fever rickettsiosis in the United States. Clin. Infect. Dis. 38:805-811. [DOI] [PubMed] [Google Scholar]
- 29.Parker, R. R., G. M. Kohls, G. W. Cox, and G. E. Davis. 1939. Observations on an infectious agent from Amblyomma maculatum. Public Health Rep. 54:1482-1484. [Google Scholar]
- 30.Raoult, D., and C. D. Paddock. 2005. Rickettsia parkeri infection and other spotted fevers in the United States. N. Engl. J. Med. 353:626-627. [DOI] [PubMed] [Google Scholar]
- 31.Renesto, P., S. Azza, A. Dolla, P. Fourquet, G. Vestris, J. P. Gorvel, and D. Raoult. 2005. Proteome analysis of Rickettsia conorii by two-dimensional gel electrophoresis coupled with mass spectrometry. FEMS Microbiol. Lett. 245:231-238. [DOI] [PubMed] [Google Scholar]
- 32.Renesto, P., L. Samson, H. Ogata, S. Azza, P. Fourquet, J. P. Gorvel, R. A. Heinzen, and D. Raoult. 2006. Identification of two putative rickettsial adhesins by proteomic analysis. Res. Microbiol. 157:605-612. [DOI] [PubMed] [Google Scholar]
- 33.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 34.Seo, G. M., C. Cheng, J. Tomich, and R. R. Ganta. 2008. Total, membrane, and immunogenic proteomes of macrophage- and tick cell-derived Ehrlichia chaffeensis evaluated by liquid chromatography-tandem mass spectrometry and MALDI-TOF methods. Infect. Immun. 76:4823-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stothard, D. R., and P. A. Fuerst. 1995. Evolutionary analysis of the spotted fever and typhus groups of Rickettsia using 16S rRNA gene sequences. Syst. Appl. Microbiol. 18:52-61. [Google Scholar]
- 36.Sumner, J. W., K. G. Sims, D. C. Jones, and B. E. Anderson. 1995. Protection of guinea-pigs from experimental Rocky Mountain spotted fever by immunization with baculovirus-expressed Rickettsia rickettsii rOmpA protein. Vaccine 13:29-35. [DOI] [PubMed] [Google Scholar]
- 37.Sunyakumthorn, P., A. Bourchookarn, W. Pornwiroon, C. David, S. A. Barker, and K. R. Macaluso. 2008. Characterization and growth of polymorphic Rickettsia felis in a tick cell line. Appl. Environ. Microbiol. 74:3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tinsley, E., and S. A. Khan. 2006. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 188:2829-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchiyama, T. 2003. Adherence to and invasion of Vero cells by recombinant Escherichia coli expressing the outer membrane protein rOmpB of Rickettsia japonica. Ann. N. Y. Acad. Sci. 990:585-590. [DOI] [PubMed] [Google Scholar]
- 41.Vishwanath, S., G. A. McDonald, and N. G. Watkins. 1990. A recombinant Rickettsia conorii vaccine protects guinea pigs from experimental boutonneuse fever and Rocky Mountain spotted fever. Infect. Immun. 58:646-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss, E., J. C. Coolbaugh, and J. C. Williams. 1975. Separation of viable Rickettsia typhi from yolk sac and L cell host components by renografin density gradient centrifugation. Appl. Microbiol. 30:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitman, T. J., A. L. Richards, C. D. Paddock, C. L. Tamminga, P. J. Sniezek, J. Jiang, D. K. Byers, and J. W. Sanders. 2007. Rickettsia parkeri infection after tick bite, Virginia. Emerg. Infect. Dis. 13:334-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, H. J., A. H. Wang, and M. P. Jennings. 2008. Discovery of virulence factors of pathogenic bacteria. Curr. Opin. Chem. Biol. 12:93-101. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, A., C. Xie, H. Chen, and M. Jin. 2008. Identification of immunogenic cell wall-associated proteins of Streptococcus suis serotype 2. Proteomics 8:3506-3515. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, J. Z., J. F. Hao, D. H. Walker, and X. J. Yu. 2006. A mutation inactivating the methyltransferase gene in avirulent Madrid E strain of Rickettsia prowazekii reverted to wild type in the virulent revertant strain Evir. Vaccine 24:2317-2323. [DOI] [PubMed] [Google Scholar]