Abstract
The ferric enterobactin receptor CfrA not only is responsible for high-affinity iron acquisition in Campylobacter jejuni but also is essential for C. jejuni colonization in animal intestines. In this study, we determined the feasibility of targeting the iron-regulated outer membrane protein CfrA for immune protection against Campylobacter colonization. Alignment of complete CfrA sequences from 15 Campylobacter isolates showed that the levels of amino acid identity for CfrA range from 89% to 98%. Immunoblotting analysis using CfrA-specific antibodies demonstrated that CfrA was dramatically induced under iron-restricted conditions and was widespread and produced in 32 Campylobacter primary strains from various sources and from geographically diverse areas. The immunoblotting survey results were highly correlated with the results of an enterobactin growth promotion assay and a PCR analysis using cfrA-specific primers. Inactivation of the cfrA gene also impaired norepinephrine-mediated growth promotion, suggesting that CfrA is required for C. jejuni to sense intestinal stress hormones during colonization. Complementation of the cfrA mutant with a wild-type cfrA allele in trans fully restored the production and function of CfrA. A growth assay using purified anti-CfrA immunoglobulin G demonstrated that specific CfrA antibodies could block the function of CfrA, which diminished ferric enterobactin-mediated growth promotion under iron-restricted conditions. The inhibitory effect of CfrA antibodies was dose dependent. Immunoblotting analysis also indicated that CfrA was expressed and immunogenic in chickens experimentally infected with C. jejuni. Amino acid substitution mutagenesis demonstrated that R327, a basic amino acid that is highly conserved in CfrA, plays a critical role in ferric enterobactin acquisition in C. jejuni. Together, these findings strongly suggest that CfrA is a promising vaccine candidate for preventing and controlling Campylobacter infection in humans and animal reservoirs.
Campylobacter species, including Campylobacter jejuni and Campylobacter coli, are the most common bacterial causes of human enteritis in many industrialized countries (24). Human Campylobacter illnesses are caused primarily by C. jejuni (∼90%) and secondarily by C. coli (∼10%). Campylobacter is also associated with Guillain-Barré syndrome, an acute flaccid paralysis that may lead to respiratory muscle compromise and death (37). Parallel to its increased prevalence, Campylobacter has become increasingly resistant to clinical antibiotics (e.g., fluoroquinolones and macrolides), which greatly compromises the effectiveness of antibiotic treatment (15). Despite the growing need for new antibiotics due to the increasing drug resistance of Campylobacter and other bacteria, many pharmaceutical companies have been getting out of the antibiotic discovery field recently (41, 48). Therefore, development of alternative intervention strategies, such as vaccination, to prevent and control Campylobacter infections in humans and animal reservoirs is urgently needed. However, no commercial vaccine against C. jejuni is available. Information concerning protective antigens as candidates for a vaccine against C. jejuni is limited, primarily due to a lack of understanding of the pathogenesis mechanisms and due to the antigenic complexity of this organism.
Bacterial outer membrane proteins (OMPs) are considered the major mediators of host-pathogen interactions and are promising candidates for the design of protective vaccines (30). Iron-regulated OMPs are important virulence factors in bacteria and play a critical role in bacterial adaptation to host niches, primarily by mediating iron uptake (30). All gram-negative bacteria have an absolute requirement for iron. However, the levels of free iron in vivo are well below the levels required for growth of gram-negative bacteria (46). For example, in the intestine, the principal site of colonization by C. jejuni, there are two potential sources of iron, the mucosa and food (46). However, iron from these sources (e.g., lactoferrin in mucosal secretions and heme in food) is normally not available to invading gram-negative bacteria in the intestine (46). Iron binding proteins in the intestine sequester free ferric iron at concentrations as low as 10−24 M and make it unavailable to most bacteria, which require at least 10−7 M iron for normal growth (7). Therefore, to obtain sufficient iron for survival and multiplication, enteric gram-negative bacteria have evolved sophisticated genetic systems for iron uptake. The most efficient strategy is to utilize high-affinity iron uptake systems, in which iron-regulated OMPs (e.g., the ferric enterobactin receptor FepA in Escherichia coli) can bind to an iron-siderophore complex and promote iron-siderophore transport across the membrane into cells (35). Thus, iron-regulated OMPs directly interact with the iron-restricted environment encountered by pathogenic bacteria and function as “gatekeepers” for iron assimilation in bacteria. Enterobactin is a siderophore that is particularly interesting because it has the highest affinity for ferric iron of the natural siderophore compounds that have been tested and can effectively capture iron from other iron complexes in neutral and alkaline conditions (46). On the other hand, enterobactin is produced by a wide variety of commensal bacteria in the intestine, and significant amounts of it are likely to be produced by the resident microflora in the intestine (46). Thus, ferric enterobactin may be a significant source of iron for enteric pathogens, including C. jejuni, during intestinal colonization, and the surface-exposed ferric enterobactin receptor is a potential target for vaccine development.
Despite its inability to synthesize enterobactin, C. jejuni can express the ferric enterobactin receptor CfrA and other essential components of the ferric enterobactin uptake system (e.g., TonB and the ceu ABC transport system) for utilization of ferric enterobactin as a sole iron source (46). The C. jejuni CfrA sequence exhibits a high level of similarity with the sequence of BfeA, a ferric enterobactin receptor produced by Bordetella bronchiseptica, which also does not have genes involved in the synthesis of enterobactin (3, 46). CfrA was induced under iron-restricted conditions and was responsible for high-affinity enterobactin-mediated iron acquisition in Campylobacter (18, 21, 39, 47). Strikingly, inactivation of the cfrA gene alone not only impaired enterobactin-mediated iron assimilation in C. jejuni but also completely eliminated colonization of chickens by C. jejuni, although the parent strain colonized all chickens at a level of >107 CFU/g cecal contents (39). This finding indicates that another iron uptake system(s) in C. jejuni cannot replace the function of CfrA and that CfrA plays an essential role in colonization of chickens by C. jejuni. It was suggested that CfrA may also have another novel function(s) important for intestinal colonization of chickens by C. jejuni (39). Together, these previous studies strongly suggest that CfrA is a potential candidate for a vaccine against C. jejuni. However, there is little information concerning the sequence homology, immunogenicity, prevalence, and novel functions of CfrA in C. jejuni. In addition, it is also not known if CfrA is expressed and antigenic in vivo and if CfrA-specific antibodies can block the function of CfrA. Addressing these issues is crucial for understanding the role of CfrA in C. jejuni pathogenesis and the feasibility of targeting CfrA for immune protection against Campylobacter colonization.
In this study, we examined the sequence homology, immunogenicity, prevalence, and novel functions of CfrA in C. jejuni. Our studies indicated that CfrA is highly conserved and produced in almost all Campylobacter primary isolates tested, which were obtained from various sources and from geographically diverse areas. Interestingly, norepinephrine (NE), a stress hormone present in the intestine, can promote the growth of C. jejuni in a CfrA-dependent manner. We also observed that CfrA antibodies can inhibit the enterobactin-mediated growth promotion of C. jejuni and that CfrA is expressed and immunogenic in vivo. These findings support our hypothesis that CfrA is a promising candidate for a subunit vaccine against Campylobacter infection.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The major bacterial strains and plasmids used in this study and their sources are listed in Table 1. Seven C. jejuni isolates (JL10, 12, 36, 78, 81, 83, and 118 [Table 1]) were used for amplification and sequencing of the full-length cfrA gene. Of the 32 strains used for the CfrA prevalence assay, 30 are C. jejuni isolates and 2 (JL96 and JL170) are C. coli isolates. These primary Campylobacter strains were isolated from human (15 isolates), bovine (5 isolates), chicken (5 isolates), turkey (1 isolate), pig (1 isolate), and ovine (1 isolate) sources, as well as from farm environments, including a lagoon (1 isolate), a bird dropping (2 isolates), and a mouse trap (1 isolate). These Campylobacter strains were obtained from geographically diverse areas, including Ohio (14 isolates), Tennessee (5 isolates), Georgia (3 isolates), Michigan (2 isolates), Alabama (1 isolate), Iowa (1 isolate), Maryland (1 isolate), Minnesota (1 isolate), Colorado (1 isolate), France (2 isolates), and the United Kingdom (1 isolate). The C. jejuni strains were routinely grown in Mueller-Hinton (MH) broth (Difco) or on MH agar at 42°C under microaerophilic conditions, which were generated using a CampyGen Plus (Oxoid) gas pack in an enclosed jar. Forty micromolar ferric sulfate was added when iron-rich conditions were required. Twenty micromolar deferoxamine mesylate (DFO) was added to the medium when iron-limited conditions were required. E. coli was grown in Luria-Bertani broth with shaking (250 rpm) or on Luria-Bertani agar at 37°C overnight. When necessary, culture media were supplemented with kanamycin (30 μg/ml) or chloramphenicol (4 or 20 μg/ml).
TABLE 1.
Key bacterial plasmids and strains used in this study
| Plasmid or strain | Description | Source or reference |
|---|---|---|
| Plasmids | ||
| pGEM-T Easy | PCR cloning vector, Ampr | Promega |
| pCA | pGEM-T Easy containing 1.6-kb N-terminal cfrA fragment, Ampr | This study |
| pmCA | pCA with chloramphenicol resistance cassette inserted in cfrA gene, Ampr Cmr | This study |
| pRY107 | E. coli-C. jejuni shuttle vector, Kanr | 51 |
| pQE-30 | N-His6 fusion recombinant protein vector | Qiagen |
| pCFRA-NHIS | pQE-30 ligated with cfrA segment encoding mature CfrA | This study |
| pCfrA | pRY107 derivative containing a 2.5-kb cfrA open reading frame plus its promoter region | This study |
| pCfrA (K297A) | pCfrA derivative with K297A mutation in CfrA | This study |
| pCfrA (R327A) | pCfrA derivative with R327A mutation in CfrA | This study |
| pCfrA (Q271A) | pCfrA derivative with Q271A mutation in CfrA | This study |
| pCfrA (F337A) | pCfrA derivative with F337A mutation in CfrA | This study |
| C. jejuni strains | ||
| JL10 | ATCC 33291, human isolate | ATCC |
| JL12 | 15046764, bovine isolate | 52 |
| JL36 | S3B, chicken isolate | 43 |
| JL78 | W42606, human isolate | 22 |
| JL81 | F34078, human isolate | 22 |
| JL83 | M76297, human isolate | 22 |
| JL118 | CVM20088, chicken isolate | Chickena |
| JL241 | NCTC 11168, human isolate | 40 |
| JL242 | 81-176, human isolate | 6 |
| JL364 | 81-176 CJJ81176_0471 isogenic mutant, Kanr | This study |
| JL324 | JL241 derivative, cfrA::cm | This study |
| JL335 | JL324 containing pCfrA | This study |
| JL442 | JL324 containing pCfrA (K297A) | This study |
| JL446 | JL324 containing pCfrA (R327A) | This study |
| JL494 | JL324 containing pCfrA (Q271A) | This study |
| JL495 | JL324 containing pCfrA (F337A) | This study |
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 lacIqZΔM15] | Promega |
| JL48 | Conjugation helper strain, DH5α containing plasmid RK2013 | 1 |
| JL122 | E. coli AN102, enterobactin transport mutant for enterobactin purification | 2 |
| JL275 | JM109 containing pCFRA-NHIS | This study |
| JL334 | DH5α containing pCfrA | This study |
Isolated from chicken breast and kindly provided by Shaohua Zhao (Center for Veterinary Medicine, Food and Drug Administration).
PCR.
The major PCR primers used in this study and the expected sizes of the products are listed in Table 2. Each PCR was performed with a 50-μl mixture containing each deoxynucleoside triphosphate at a concentration of 200 μM, each primer at a concentration of 200 nM, 2.5 mM MgSO4, 50 ng of template DNA, and 5 U of Taq DNA polymerase (Promega) or PfuUltra high-fidelity DNA polymerase (Stratagene). The cycling conditions varied based on the estimated annealing temperatures of the primers and the expected sizes of the products (available upon request). PCR products were purified with a QIAquick PCR purification kit (Qiagen) when necessary for cloning or sequencing analysis. Notably, to examine the prevalence of the cfrA gene in various Campylobacter isolates, primers CfrAF and CfrAR (Table 2) were designed based on the highly conserved regions of the cfrA gene.
TABLE 2.
Major primers used in this study
| Primer | DNA sequence (5′-3′)a | Product size (bp) | Target gene or function |
|---|---|---|---|
| PF1 | AAAGGATCCCAAAATGTAGAACTAGATAGC (BamHI) | 2,043 | cfrA (no signal peptide region) |
| PR1 | AAACCCGGGAAAGTTACCATTGATAGAAAT (SmaI) | ||
| CHFL1 | TTTGCTAGCTGCTCGGCGGTGTTCCTTT (NheI) | 802 | cat |
| CHFR1 | TTTGCTAGCGCGCCCTTTAGTTCCTAAAG (NheI) | ||
| CfrAF | GAGATGTTGCAGAGGCTATCG | 527 | cfrA |
| CfrAR | TGCCTTTGTAGGACTTTGAGC | ||
| CfrAF1 | TCAATATTTAACAAAAGGAGAAAAATG | 2,151 | cfrA (complete open reading frame) |
| CfrAR1 | AAGCCTTTGAAAGCTCTTTGG | ||
| CfrAF2 | TTTCATTGGGTTGTATGTGTAAAAA | 1,631 | Part of cfrA plus 445-bp upstream region |
| CfrAR2 | TCTGCAAAAATTGCCAATAAA | ||
| CfrAR3 | TCTGCAAAAATTGCCAATAAA | NAb | cfrA sequencing primer |
| Q271A_F | GATAATAAACAAGGTGCATTAGGAACCATCACAAGTCCAGGTAGAACACC | 9,300 | Create Q271A mutation in CfrA |
| Q271A_R | GGTTCCTAATGCACCTTGTTTATTATCATAATGATTTC | ||
| K297A_F | GAAGTTGATGCATTTGTGACTTATTTAAGTCATG | 9,300 | Create K297A mutation in CfrA |
| K297A_R | GTCACAAATGCATCAACTTCCATAATATCTGC | ||
| R327A_F | GATGGCGCAGAAGTCGTAGGGCAATCTACACAGCCG | 9,300 | Create R327A mutation in CfrA |
| R327A_R | GACTTCTGCGCCATCATTGCTCACTCTATTATATTG | ||
| F337A_F | CACAGCCGGCTTTGGGAGAAAATAGAGATATAGTC | 9,300 | Create F337A mutation in CfrA |
| F337A_R | CTCCCAAAGCCGGCTGTGTAGATTGCCCTACGACTTCGCGGCC |
Restriction sites in the primer sequences are underlined, and the restriction enzymes are indicated in parentheses. Bold type indicates nucleotides designed for amino acid substitution mutagenesis.
NA, not applicable.
Sequence analysis of CfrA.
The complete open reading frame of cfrA from C. jejuni was PCR amplified using primers CfrAF1 and CfrAR1 (Table 2). The purified PCR products were subjected to sequencing and alignment. Multiple-sequence alignment was performed by using the ClustalW program in MEGA 4.0 (25). A CfrA-based dendrogram was constructed by using neighbor-joining methods in MEGA 4.0 (25). To identify the conserved amino acids in CfrA that may play a critical role in ferric enterobactin assimilation, multiple-sequence alignment of the FepA sequences from E. coli (33), Salmonella enterica (4), Pseudomonas aeruginosa (13), and Bordetella pertussis (5) and the CfrA sequences from different Campylobacter strains was also carried out by using ClustalW in MEGA 4.0 (25).
Production of rCfrA and generation of polyclonal antisera.
Full-length histidine-tagged recombinant CfrA (rCfrA) was produced in E. coli by using the pQE-30 vector of the QIAexpress expression system (Qiagen). Briefly, primers PF1 and PR1 (Table 2) were used to amplify a 2,043-bp fragment encoding mature 676-amino-acid CfrA (amino acids 21 to 696) from C. jejuni NCTC 11168. Restriction sites (BamHI and SmaI) were attached to the 5′ ends of the primers to facilitate the directional cloning of the amplified PCR product into the pQE-30 vector. The amplified PCR product was digested with BamHI and SmaI and was ligated into the pQE-30 vector, which previously had been digested with the same enzymes. Cloning, expression, and purification of rCfrA were performed by using procedures described previously (1, 26, 31, 52). Plasmid pCFRA-NHIS in E. coli JM109 clone JL275 producing rCfrA was sequenced, and no frameshift or other mutations in the coding sequence of cfrA were detected.
Approximately 2 mg of highly purified rCfrA obtained from JL275 was used for production of rabbit polyclonal antisera against CfrA. The rabbit CfrA polyclonal antisera were prepared by Pacific Immunology Corp. (Ramona, CA). Pre- and postimmune serum samples were analyzed by immunoblotting, using both the rCfrA and membrane proteins of NCTC 11168 grown under iron-restricted conditions. The postimmune sera reacted specifically with CfrA, while the preimmune sera were negative with CfrA.
SDS-PAGE and immunoblotting.
Various C. jejuni isolates were grown in iron-rich medium (MH medium with 40 μM ferric sulfate) or iron-limited medium (MH medium with 20 μM DFO) to late log phase at 42°C under microaerophilic conditions. To prepare whole-cell lysates, Campylobacter cells grown in the iron-rich or iron-limited conditions described above were harvested and solubilized by boiling them for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. SDS-PAGE and immunoblotting were performed as described previously (31).
Insertional mutation of cfrA and complementation in trans.
An isogenic cfrA mutant of C. jejuni NCTC 11168 was constructed by insertional mutagenesis using a protocol described previously (1, 26, 32). Briefly, primers CfrAF2 and CfrAR2 (Table 2) were used to amplify a 1.6-kb cfrA gene fragment. The PCR product with a unique NheI restriction site was cloned into pGEM-T Easy (Promega), resulting in the construct pCA. The chloramphenicol resistance (Cmr) cassette was PCR amplified from plasmid pUOA18 (49) using primers CHLF1 and CHLR1(Table 2), which have an NheI restriction site at the 5′ end. The 0.8-kb PCR product containing the Cmr cassette was digested by NheI and ligated to NheI-digested pCA to obtain the construct pmCA (Table 1). Sequence analysis of the construct indicated that the Cmr cassette was inserted into cfrA with the same transcriptional direction. Plasmid pmCA, which served as a suicide vector, was transferred into C. jejuni NCTC 11168 by natural transformation. One Cmr mutant (JL324) was selected on MH agar containing 4 μg/ml of chloramphenicol. Inactivation of the cfrA gene in JL324 was confirmed by PCR (data not shown) and immunoblotting using CfrA-specific antibodies.
To complement the cfrA mutation in JL324, a 2.5-kb fragment spanning from the promoter region of cfrA to 36 bp downstream of the cfrA stop codon was PCR amplified from NCTC 11168 using primers CfrAF2 and CfrAR1 (Table 2). The PCR was performed using Pfu DNA polymerase (Stratagene), and the blunt-ended PCR product was purified and ligated to the shuttle vector pRY107 (51), which was digested with SmaI prior to ligation. The ligation mixture was introduced into DH5α by transformation. A transformant with a plasmid bearing the intact cfrA gene (designated pCfrA) was identified (JL334). The pCfrA plasmid from JL334 was then transferred to JL324, a cfrA isogenic mutant, by triparental conjugation using DH5α/pRK2013 as a helper strain (1). The complementation strain was designated JL335 and was confirmed by immunoblotting using CfrA-specific antibodies.
Construction of JL364.
An open reading frame (CJJ81176_0471) of C. jejuni 81-176 was also annotated as a TonB-dependent receptor responsible for ferric enterobactin assimilation (http://www.ncbi.nlm.nih.gov/protein/121612217). The genomic DNA extracted from an isogenic CJJ81176_0471 mutant of C. jejuni 81-176 was kindly provided by Julian M. Ketley and Richard Haigh (University of Leicester, United Kingdom). This genomic DNA was used to transform JL242 (Table 1) by natural transformation as described by Wang and Taylor (50). The transformation procedure introduced the insertional mutation of CJJ81176_0471 into JL242, creating isogenic mutant JL364 (Table 1). The CJJ81176_0471 mutation in JL364 was confirmed by PCR (data not shown).
Purification of enterobactin.
An enterobactin transport mutant of E. coli AN102 (JL122) (Table 1) was kindly provided by Sandra K. Armstrong (University of Minnesota) and used for enterobactin purification. Enterobactin was purified from E. coli AN102 as described previously (2). The purified enterobactin was dissolved in a methanolic solution and stored at 4°C until it was used.
Growth promotion assay.
Briefly, C. jejuni log-phase cells grown in MH broth (∼2 × 107 cells) were mixed with melted MH agar containing the chelator DFO at a final concentration of 20 μM (for the enterobactin growth promotion test) or 5 μM (for the NE growth promotion test). Each mixture was immediately poured into petri dishes for solidification. A sterile disk containing 10 μl of enterobactin (5 mM), NE bitartrate (100 mM), or H2O was placed on the surface of the agar in each dish. The growth zones around the disks were observed and measured after 24 h of incubation at 42°C under microaerophilic conditions.
Inhibition of enterobactin uptake by anti-CfrA IgG.
The ability of specific anti-CfrA immunoglobulin G (IgG) to inhibit CfrA-mediated ferric enterobactin assimilation was assessed by using a modified growth promotion assay (36). The IgG fraction of heat-inactivated rabbit serum samples (preimmune control serum and postimmune anti-CfrA serum) was purified by the ammonium sulfate precipitation method (27). The purified IgG was dialyzed against phosphate-buffered saline (PBS), sterilized by membrane filtration, aliquoted, and stored at −20°C prior to use. The IgG concentration was measured by using the bicinchoninic acid protein assay reagent (Pierce). To evaluate the inhibitory effect of anti-CfrA IgG on CfrA function, 1 ml of a log-phase culture of C. jejuni strain JL241 grown in MH broth was centrifuged, washed once with PBS, and resuspended in 0.5 ml PBS. The bacterial suspension was mixed with 0.5 ml of purified anti-CfrA IgG (16 mg/ml), purified preimmune IgG (16 mg/ml), or PBS and then added to 24 ml MH agar containing 20 μM DFO and poured into petri dishes for solidification. A sterile disk containing 10 μl of enterobactin (5 mM) or H2O was placed on the surface of the agar in each dish. The growth zone around the disk was observed and measured after 24 h of incubation at 42°C under microaerophilic conditions. To measure dose-dependent inhibition of CfrA by CfrA antibodies, anti-CfrA IgG was added to agar plates at final concentrations of 0, 12.8, 32, 64, and 320 μg/ml. All assays were performed in three independent experiments with triplicate measurements in each independent experiment. The significance of differences in growth inhibition was determined using the Student t test.
Immunoblot analysis of CfrA expression in vivo.
rCfrA was separated by SDS-PAGE and then electrophoretically transferred to a nitrocellulose membrane as described previously (32). The blots were incubated with different chicken serum samples (after 1:100 dilution in blocking buffer), including two negative control serum samples from 3-week-old Campylobacter-free broiler chickens, one serum sample from a 2-day-old Campylobacter-free broiler chicken that displayed a high titer of maternally derived antibodies to C. jejuni (44), and one serum sample from 6-week-old broiler chickens experimentally infected with C. jejuni S3B (43). The immunoblotting procedure used has been described previously (32). All chicken sera were kindly provided by Qijing Zhang (Iowa State University).
Site-directed amino acid substitution mutagenesis.
The predicted basic or aromatic amino acids in CfrA that may be important for ferric enterobactin assimilation were replaced by alanine using partial overlapping PCR (53). Briefly, the partial overlapping primer pairs containing the desired mutations (Table 2) were used for PCR amplification of the pCfrA plasmid. PfuUltra high-fidelity DNA polymerase (Stratagene) was used in PCR, and the cycling conditions were as follows: 95°C for 2 min, followed by 18 cycles of 94°C for 1 min, 56°C for 1 min, and 68°C for 9.5 min and then 68°C for 30 min. The 9.5-kb PCR product was purified, concentrated, and treated with DpnI (New England Biolabs) for 2 h to digest the methylated, nonmutated parental DNA template. The digested product was transformed into DH5α. A plasmid with a specific amino acid substitution in CfrA was selected and confirmed by sequencing using primer CfrAR3 (Table 2). Four plasmids with different amino acid substitutions (Table 1) were generated and then transferred to an isogenic cfrA mutant of C. jejuni NCTC 11168 (JL324), creating the constructs JL442, JL446, JL494, and JL495 (Table 1). These constructs and control strain JL335 were used for the enterobactin growth promotion assay to determine the roles of specific amino acids in ferric enterobactin assimilation in C. jejuni.
Nucleotide sequence accession numbers.
The full coding sequences of CfrA of seven C. jejuni strains have been deposited in the GenBank database under accession numbers FJ771033 (JL10), FJ771034 (JL12), FJ771035 (JL36), FJ771036 (JL78), FJ771037 (JL81), FJ771038 (JL83), and FJ771039 (JL118).
RESULTS
Sequence homology of CfrA.
The full-length cfrA gene from seven C. jejuni isolates was sequenced in this study. The new sequences were translated into protein sequences, which aligned with the CfrA sequences from eight Campylobacter strains (six C. jejuni strains and two C. coli strains) that were deposited in the NCBI public database (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). ClustalW analysis in MEGA 4.0 showed that the levels of amino acid identity for CfrA range from 89% to 98%. According to secondary structure prediction, CfrA consists of 22 β-strands with long loops on the external side of the membrane and short turns facing the periplasmic space. The highly conserved regions were predicted to form the β-strands, while the variable regions were located in the putative loop structure (data not shown). Interestingly, the CfrA sequences of two C. coli strains (C. coli VC167 and RM2228) exhibit a high level of amino acid identity (98%) to the CfrA sequence of C. jejuni NCTC 11168. Together, the results of the CfrA sequence analysis showed that CfrA is highly conserved among Campylobacter isolates which possess the cfrA gene.
CfrA is widely expressed and immunogenically conserved among various Campylobacter strains.
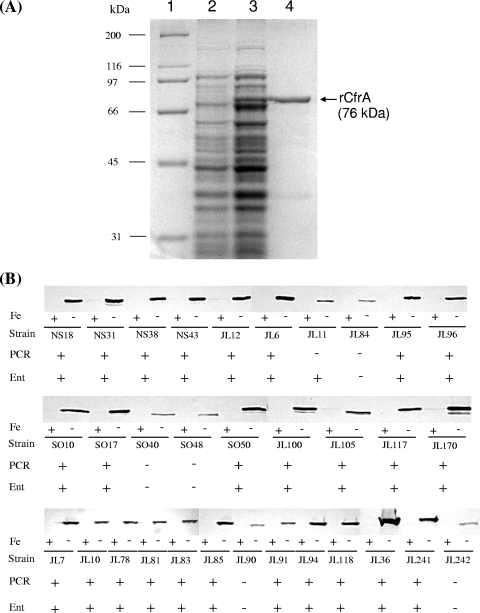
To determine the prevalence and antigenicity of CfrA, full-length histidine-tagged rCfrA was produced in E. coli and purified for preparation of CfrA-specific antiserum. rCfrA had a molecular mass of approximately 76 kDa as determined by SDS-PAGE (Fig. 1A), consistent with the calculated molecular mass based on the deduced amino acid sequence of CfrA. The expression and prevalence of CfrA in 32 Campylobacter primary strains were first examined by immunoblotting using CfrA-specific antibodies. As shown in Fig. 1B, CfrA was dramatically induced under iron-limited conditions, while iron-replete conditions suppressed the expression of CfrA. Each strain produced a significant band that was reactive with specific anti-CfrA antibodies, suggesting that CfrA is highly conserved in Campylobacter strains. However, C. jejuni 81-176 (JL242), which does not contain a cfrA gene (20), also produced a band that reacted with CfrA antibodies, but this band migrated slightly faster than typical CfrA of NCTC 11168 (JL241) (Fig. 1B). This finding suggests that the CfrA antibodies may cross-react with other iron-regulated OMPs in Campylobacter, which could confound interpretation of data if only the CfrA antibodies are used. Thus, we conducted several experiments to overcome this limitation. First, we designed highly cfrA gene-specific PCR primers based on the sequence alignment described above. PCR analysis demonstrated that 26 of the 32 Campylobacter strains tested contain the cfrA gene. Second, we determined the abilities of these strains to assimilate ferric enterobactin. As shown in Fig. 1B, all 26 PCR-positive strains displayed significant enterobactin-mediated growth promotion, while 5 of the 6 PCR-negative strains (JL84, SO40, SO48, JL90, and JL242) did not utilize enterobactin. These findings indicated that the presence and expression of the cfrA gene are highly correlated with the enterobactin utilization phenotype in C. jejuni. Interestingly, JL11, which lacks the typical cfrA gene, still can effectively utilize enterobacin (Fig. 1B) and even produced significantly larger growth zones than other Campylobacter strains tested (data not shown). Finally, we determined the identity of the iron-regulated OMP that was reactive with CfrA antibodies in C. jejuni 81-176 (JL242) (Fig. 1B). BLAST searches revealed a gene (CJJ81176_0471) in the genome of 81-176 that encodes a putative iron-regulated OMP sharing 34% amino acid identity and 54% amino acid similarity with CfrA. The homolog of CJJ81176_0471 in 81-176 was annotated as a pseudogene (Cj0444) in NCTC 11168. We hypothesized that the iron-regulated protein detected in 81-176 (JL242) (Fig. 1B) was CJJ81176_0471, which can cross-react with CfrA antibodies. To test this hypothesis, we constructed JL364, an isogenic CJJ81176_0471 mutant of C. jejuni 81-176, for immunoblotting using CfrA-specific antibodies. We observed that inactivation of CJJ81176_0471 in JL364 completely abolished production of the specific band observed with wild-type strain JL242 (data not shown). Complementation of JL364 with the complete CJJ81176_0471 open reading frame fully restored production of a band that reacts with CfrA antibodies, as observed for wild-type strain 81-176 (data not shown), further indicating that the CJJ81176_0471 protein is an iron-regulated OMP that cross-reacts with CfrA-specific antibodies in C. jejuni 81-176.
FIG. 1.
Prevalence and expression of CfrA in different Campylobacter strains. (A) SDS-PAGE analysis of rCfrA production in E. coli. Lane 1, molecular weight marker (Bio-Rad); lane 2, whole-cell lysate of noninduced E. coli; lane 3, whole-cell lysate of E. coli induced for 1 h with 1 mM isopropyl-β-d-thiogalactopyranoside; lane 4, rCfrA purified by Ni-nitrilotriacetic acid affinity chromatography. (B) Immunoblotting, PCR, and functional survey of ferric enterobactin uptake systems in 32 Campylobacter strains from various sources and from various geographically diverse areas. For immunoblot analysis, each strain was grown in iron-replete (+) or iron-limited (−) MH broth, and whole-cell lysates were loaded onto an SDS-PAGE gel for blotting. The cfrA-specific primers were used in PCR to amplify the cfrA-specific sequence from each strain; successful PCR amplification of the cfrA sequence from a specific strain is indicated by a plus sign, while a minus sign indicates the absence of a specific cfrA sequence. For the enterobactin growth promotion assay (Ent), a strain showing significant growth promotion in response to enterobactin treatment is indicated by a plus sign.
CfrA is also responsible for NE-mediated growth promotion.
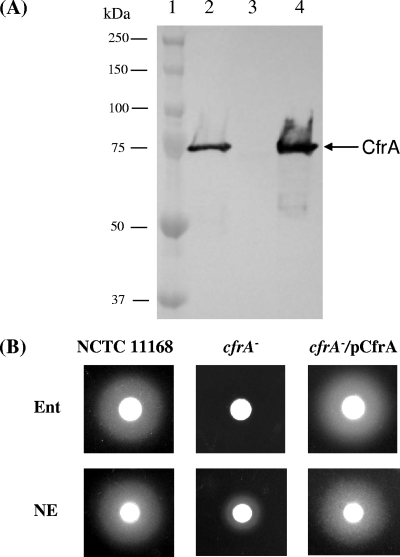
To confirm the role of CfrA in ferric enterobactin assimilation observed in a previous study (39) and to further examine the novel functions of CfrA, a cfrA isogenic mutant (JL324) was constructed in this study. In contrast to a previous study (39), we also constructed a complemented strain (JL335) to preclude any potential polar effect due to the inactivation of cfrA in JL324. As shown in Fig. 2A, insertional inactivation of cfrA abolished production of CfrA in NCTC 11168 under iron-restricted conditions. Complementation of the cfrA mutation in trans completely restored production of normal CfrA in JL335 (Fig. 2A). As expected, the standard growth promotion assay showed that the cfrA mutant did not utilize enterobactin for growth, while complementation of the cfrA mutation resulted in a significant growth zone whose size was similar to the size of the growth zone for wild-type strain NCTC 11168 (Fig. 2B, top panel), consistent with the immunoblot analysis (Fig. 2A).
FIG. 2.
Functional analysis of CfrA. (A) Immunoblot analysis of CfrA production in C. jejuni wild-type strain NCTC 11168 (lane 2), isogenic cfrA mutant JL324 (lane 3), and complemented strain JL335 (lane 4). All of the C. jejuni strains were grown in iron-restricted medium, and similar amounts of bacterial cells were loaded in the lanes for immunoblotting using specific antibodies against CfrA. Prestained molecular mass markers (Bio-Rad) (lane 1) were coelectrophoresed and blotted to estimate of the size of the protein. (B) Role of CfrA in enterobactin (Ent)- and NE-mediated growth promotion. The procedure used for growth promotion assays is described in Materials and Methods.
Recent evidence has demonstrated that catecholamine hormones, such as NE, may fill a nutritional need and/or serve as host environmental cues to promote growth and enhance the virulence of a panel of enteric bacterial pathogens, including C. jejuni (12, 23, 34). Since an early study indicated that the growth response of E. coli to NE requires a functional enterobactin uptake system (9), we determined the role of CfrA in the growth response of C. jejuni to NE using a modified growth promotion assay. Compared to the wild-type strain, which produced a growth zone with a diameter of 2.50 ± 0.08 cm, the cfrA mutant exhibited the much weaker response to NE and produced a significantly smaller growth zone (1.43 ± 0.10 cm) (Fig. 2B, bottom panel). Complementation of the cfrA mutation restored NE-mediated growth promotion (growth zone diameter, 2.65 ± 0.13 cm) (Fig. 2B) to a level comparable that observed for wild-type strain NCTC 11168.
CfrA-specific IgG inhibited enterobactin-mediated growth promotion.
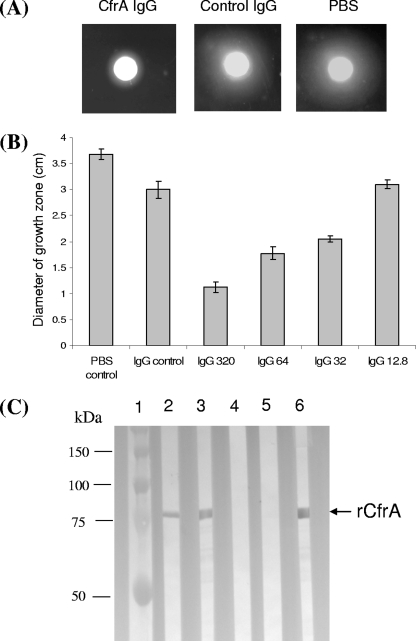
To test whether CfrA antibodies can inhibit enterobactin-mediated growth promotion, IgG was purified from heat-inactivated rabbit preimmune serum (as a control IgG) or from CfrA-specific antiserum by ammonium sulfate precipitation. The purified IgG was dialyzed against PBS, the final concentration was adjusted to 16 mg/ml, and the IgG was sterilized by membrane filtration for growth assays. As shown in Fig. 3A, the diameter of the growth halo in the presence of anti-CfrA IgG (1.13 ± 0.08 cm) is significantly less (P < 0.01) than the diameter in the presence of the same concentration of control IgG (2.87 ± 0.12 cm) or PBS alone (3.50 ± 0.18 cm), strongly suggesting that CfrA-specific IgG blocks the ligand binding site of CfrA and consequently inhibits ferric enterobactin assimilation for growth promotion. The inhibition of CfrA by the specific anti-CfrA IgG was also dose dependent; a concentration of anti-CfrA IgG as low as 32 μg/ml had a significant inhibitory effect (P < 0.01) when the growth was compared to the growth in the presence of a high concentration of control IgG (320 μg/ml) (Fig. 3B).
FIG. 3.
Functional and immunological characteristics of specific CfrA antibodies. (A) Inhibitory effect of anti-CfrA IgG on enterobactin-mediated growth promotion in C. jejuni. Purified CfrA IgG (320 μg/ml), control IgG (320 μg/ml, from preimmune serum), or PBS was mixed with C. jejuni NCTC 11168 cells for a modified growth promotion assay as described in Materials and Methods. (B) Dose-dependent inhibitory effect of anti-CfrA IgG on enterobactin-mediated growth promotion in C. jejuni. Approximately 1 × 108 C. jejuni cells were inoculated onto iron-restricted MH agar supplemented with PBS (PBS control), 320 μg/ml of IgG from preimmune serum (IgG control), or anti-CfrA IgG at various concentrations, including 320 μg/ml (IgG 320), 64 μg/ml (IgG 64), 32 μg/ml (IgG 32), and 12.8 μg/ml (IgG 12.8). The bars and error bars indicate the means ± standard deviations of the growth zone diameter for four independent measurements. (C) Immunoblot analysis of in vivo antibody responses to CfrA. rCfrA was blotted with individual chicken serum samples (lanes 2 to 5) or with the rabbit anti-CfrA antibody (positive control) (lane 6). Lane 2, serum sample from a 7-week-old broiler chicken infected with C. jejuni S3B; lane 3, serum sample from a 2-day-old broiler chicken with a high level of maternal antibodies against C. jejuni; lanes 4 and 5, serum samples from 3-week-old broiler chickens which were free of Campylobacter. Prestained molecule mass markers (Bio-Rad) (lane 1) were coelectrophoresed and blotted to estimate the sizes of the proteins.
In vivo expression and immunogenicity of CfrA.
To determine if CfrA is expressed and immunogenic in vivo, multiple chicken sera were tested for reactivity with rCfrA (Fig. 3C). One serum from a C. jejuni S3B (JL36)-infected chicken showed a vivid antibody reaction to rCfrA (Fig. 3C, lane 2). Serum from a 2-day-old broiler that was Campylobacter free but showed a high titer of maternally derived antibodies (43, 44) to C. jejuni also strongly reacted with rCfrA (Fig. 3C, lane 3). Two sera from 21-day-old Campylobacter-free chickens were negative with rCfrA (Fig. 3C, lanes 4 and 5). This result indicates that CfrA is expressed during in vivo Campylobacter infection and elicits a specific antibody response in chickens. In addition, the anti-CfrA maternal antibodies could be transferred from yolks to hatchlings.
Region and amino acids in CfrA critical for ferric enterobactin assimilation.
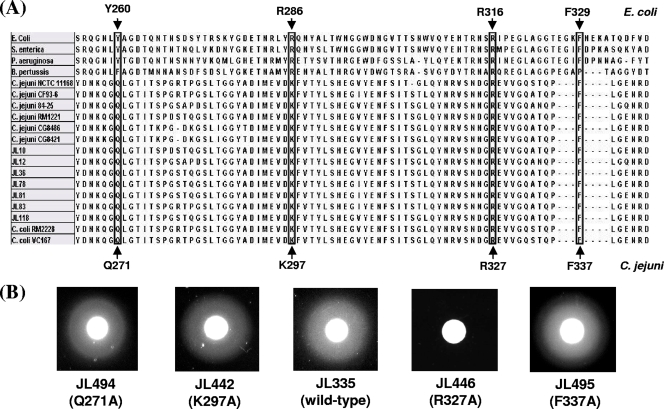
Identification of a functionally important region and/or amino acids is critical for vaccine optimization. To predict which region and amino acids in CfrA are likely important for ferric enterobactin binding, we performed a cross-species sequence alignment of the FepA sequences from four other bacteria and the CfrA sequences of 15 Campylobacter strains. It has been reported that the FepA region between residues 255 and 336 (Fig. 4A) contributes to the high-affinity binding of FepA to its substrate, ferric enterobactin, in E. coli (36). As shown in Fig. 4A, the CfrA region between residues 265 and 343, which covers the third and fourth extracellular loops, is highly conserved in CfrA and thus may be actively involved in the interaction between CfrA and ferric enterobactin. Since K297 and R327, two positively charged amino acids in CfrA, aligned with R286 and R316, two critical amino acids in E. coli FepA (38), K297 and R327 may contribute significantly to the binding of CfrA to its substrate, ferric enterobactin. Previous mutagenesis studies with E. coli also showed that two aromatic amino acids (Y260 and F329) (Fig. 4A) in FepA also play a critical role in ferric enterobactin utilization (10, 38). The sequence alignment program did not automatically reveal similar aromatic amino acids at corresponding positions in CfrA. However, glutamine at position 271 in CfrA and the corresponding aromatic amino acid Y260 in E. coli FepA both have uncharged polar side chains (Fig. 4A), suggesting that Q271 in CfrA may also be involved in binding to ferric enterobactin. In addition, manual adjustment aligned the highly conserved F337 residue in CfrA with the F329 residue in E. coli FepA (Fig. 4A).
FIG. 4.
Identification of amino acids in CfrA critical for ferric enterobactin assimilation. (A) Cross-species sequence alignment of ferric enterobactin receptor. The FepA sequences of E. coli (33), S. enterica (4), P. aeruginosa (13), and B. pertussis (5) and the CfrA sequences of 13 C. jejuni and 2 C. coli strains were used for the alignment. The specific amino acids in E. coli FepA that have been demonstrated to contribute to the interaction with ferric enterobactin are indicated by arrows above the alignment. Corresponding conserved amino acids in Campylobacter CfrA are indicated by solid arrows below the alignment. The amino acids conserved across species that might contribute to ferric enterobactin assimilation are enclosed in boxes. (B) Effect of a specific amino acid substitution on CfrA-dependent ferric enterobactin assimilation. The enterobactin growth promotion test was performed using an isogenic cfrA mutant of C. jejuni NCTC 11168 containing a plasmid bearing the wild-type cfrA gene or the cfrA gene with a specific amino acid substitution mutation. The feature of the cfrA gene (wild type or specific amino acid substitution) in the plasmid of each construct is indicated in parentheses below the construct designation.
To test prediction described above, we conducted site- directed amino acid substitution mutagenesis in conjunction with an enterobactin growth promotion assay. As shown in Fig. 4B, a plasmid encoding the R327A mutation in CfrA failed to complement the cfrA mutation, and no growth zone was observed for construct JL446, indicating that R327 in CfrA is essential for ferric enterobactin assimilation in C. jejuni. However, replacement of three other predicted amino acids with alanine appeared to have little effect on the function of CfrA because plasmids pCfrA(Q271A), pCfrA(K297A), and pCfrA(F337A) all successfully complemented the cfrA mutation and resulted in significant growth zones with sizes similar to that observed for control plasmid pCfrA (Fig. 4B).
DISCUSSION
In this study, we obtained compelling evidence demonstrating the feasibility of targeting CfrA for immune protection against Campylobacter. First, CfrA is conserved in Campylobacter strains with levels of amino acid identity ranging from 89% to 98%. The positively charged amino acid R327 that is essential for ferric enterobactin acquisition in C. jejuni is highly conserved in CfrA (Fig. 4A). Second, the CfrA-associated ferric enterobactin uptake system is widely distributed, expressed, and conserved in various C. jejuni strains (Fig. 1B). Third, CfrA plays an important role in the responsiveness of C. jejuni to the catecholamine hormone NE (Fig. 2B), suggesting that CfrA has multiple physiological functions during Campylobacter colonization in the intestine. Finally, specific CfrA antibodies clearly interacted with C. jejuni and inhibited enterobactin-mediated growth promotion in C. jejuni (Fig. 3A and 3B). These findings, plus the fact that CfrA is expressed during the course of Campylobacter colonization of chickens (Fig. 3C), strongly indicate that CfrA is a promising candidate for Campylobacter vaccine development.
Guerry et al. (18) first reported that CfrA is an iron-regulated OMP involved in iron acquisition in Campylobacter. Recently, CfrA was demonstrated to be an outer membrane receptor specific for ferric enterobactin assimilation (39). However, the published whole-genome sequence of C. jejuni 81-176 showed that this strain does not have a typical CfrA, although it has the cognate components of the ferric enterobactin uptake system, such as TonB and the ceu ABC transport system (20). To fill a significant gap in our understanding of the prevalence, sequence, and antigenic homology of CfrA in Campylobacter, we conducted a comprehensive survey in this study by taking advantage of our diverse Campylobacter strain collections. The C. jejuni and C. coli strains tested in this study were obtained from nine different animal hosts and environmental niches and were from 11 United States states and European countries. When sequence homology was examined, our analysis showed that CfrA proteins display a high level of homology. In particular, the R327 amino acid that is critical for ferric enterobactin acquisition is highly conserved in CfrA (Fig. 4A). When the prevalence and expression of CfrA were examined, immunoblotting in conjunction with PCR analysis and an enterobactin growth promotion assay indicated that 26 of 32 Campylobacter strains contain and express a functional CfrA under iron-restricted conditions (Fig. 1B). The cfrA insertional mutation in JL324 was also successfully transferred to a majority of the strains tested by natural transformation, which eliminated the production of CfrA and the ability to use enterobactin for growth under iron-restricted conditions (data not shown). This finding further confirmed that a functional CfrA is present in the majority of the Campylobacter isolates tested. Only a small subset of the Campylobacter strains (5 of 32) could not utilize ferric enterobactin as a sole iron source for growth in this study (Fig. 1B). Thus, the prevalence of a functional CfrA in Campylobacter suggests that enterobactin, which is produced by a wide variety of bacteria belonging to the family Enterobacteriaceae present in intestine, may be a remarkable iron source during Campylobacter colonization in animal and human intestines.
Interestingly, all five C. jejuni strains that showed negative results in the PCR and the enterobactin growth promotion assay also expressed an iron-regulated OMP that reacted to CfrA-specific antibodies (Fig. 1B). Since one of these strains is C. jejuni 81-176 (JL242), which clearly lacks typical CfrA (20), we speculated that C. jejuni 81-176 and the other isolates may contain an iron-regulated OMP homologous to CfrA. Comparative genome sequence analysis indicated that CJJ81176_0471 in 81-176 shares the highest level of homology with CfrA of C. jejuni NCTC 11168 (levels of amino acid similarity and identity, about 54% and 34%, respectively). Our site-directed mutagenesis and complementation experiments demonstrated that CJJ81176_0471 was indeed the iron-regulated OMP that cross-reacted with CfrA-specific antibodies in C. jejuni 81-176. At this time, the function of CJJ81176_0471 is unknown. According to sequence analysis and secondary structure prediction (http://www.ncbi.nlm.nih.gov/protein/121612217), it is likely that CJJ81176_0471 is also a ferric enterobactin receptor, but C. jejuni 81-176 does not have appropriate cognate components of the ferric enterobactin assimilation system, which leads to the inability of C. jejuni 81-176 to utilize ferric enterobactin as a sole iron source (Fig. 1B). The validity of this hypothesis and the physiological significance of CJJ81176_0471 remain to be determined in future studies. Another intriguing finding of our survey is that C. jejuni JL11, which was originally isolated from bovine feces, displayed an exceptional ability to utilize ferric enterobactin, although this strain did not have a typical cfrA gene (Fig. 1B). The lack of a typical cfrA gene in JL11 was confirmed by PCR analysis using different sets of cfrA-specific primers (data not shown). Therefore, it is tempting to speculate that JL11 has a novel ferric enterobactin receptor or system for ferric enterobactin-mediated uptake. This possibility remains to be examined in future studies.
Microbial endocrinology, representing the intersection of microbiology and neurobiology, is an emerging field in microbial pathogenesis (16, 23, 42, 45). Recent findings showed that a family of stress-related neuroendorine hormones (e.g., NE) can influence both the growth and elaboration of virulence-associated properties in a number of bacterial pathogens (16, 23, 42, 45). Notably, although these stress hormones, such as NE, are usually present at nanomolar levels in sera, the level of NE in the intestine is much higher due to the existence of the enteric nervous system, which is rich in adrenergic neurons (16, 23, 42, 45). The enteric nervous system, consisting of about 100 million neurons (19), can secrete NE, the principal autonomic neurotransmitter, into the gastrointestinal lumen at concentrations up to the millimolar level or at even higher concentrations in pathological situations (14). Thus, NE is considered an important in vivo cue to promote the growth and modulate the virulence of enteric pathogens (16, 23, 42, 45). Recently, Cogan et al. (12) reported that NE also increased the growth rate of C. jejuni under iron-restricted conditions and enhanced some virulence-associated properties of Campylobacter, such as motility and invasive ability. In contrast to the significant progress that has been made with other enteric pathogens (16, 23, 42, 45), how NE interacts with C. jejuni is still largely unknown. In this study, we consistently observed that the growth response of the isogenic CfrA mutant to NE under iron-limited conditions was greatly impaired compared to that of the wild-type strain; complementation of the CfrA mutation completely restored NE-mediated growth promotion. Therefore, CfrA was at least partially responsible for the NE-mediated growth promotion, although a mutation in CfrA did not completely eliminate NE-mediated growth promotion in the CfrA mutant. The underlying mechanism of NE-mediated growth promotion in C. jejuni is not clear. The simplest and most straightforward explanation is that NE might facilitate the release of ferric iron from chelators or iron binding proteins to fill a nutritional need of Campylobacter (17). However, given the recent novel findings for E. coli (16, 23, 42, 45), we cannot rule out the possibility that NE also functions as an in vivo cue to modulate physiological and pathogenic properties in C. jejuni. Transcriptome analysis of C. jejuni in response to NE treatment may help us address this issue and understand the molecular basis of the interaction between NE and Campylobacter.
The ferric enterobactin receptor possesses intensively surface-exposed loops and has relatively high aqueous solvent exposure for ligand binding (8, 11). Given this unique structural feature and its inducibility in vivo, the ferric enterobactin receptor has been considered a promising subunit vaccine candidate (30). In our previous studies, we demonstrated that both monoclonal and polyclonal antibodies directed against FepA effectively inhibited the function of FepA and reduced the growth of coliform bacteria under iron-restricted conditions (28, 29). Based on these findings, we speculated that CfrA-specific antibodies may also physically block the binding of Fe-enterobactin to the ligand binding site of CfrA, resulting in the impaired Campylobacter growth with ferric enterobactin as a sole iron source. Our results clearly showed the effectiveness of purified anti-CfrA IgG for inhibiting enterobactin-mediated growth promotion of Campylobacter under iron-limited conditions (Fig. 3A). The inhibitory effect of anti-CfrA IgG was also dose dependent (Fig. 3B), further confirming the intimate interaction between CfrA antibodies and the CfrA receptor. Notably, in contrast to our previous studies (28, 29), the cells used for the growth assay in this study were grown in normal MH broth with no or low-level CfrA expression instead of in iron-depleted MH broth with high-level CfrA expression. We believe that such a modification, in which “low-iron-preadapted” cells are not used, should better mimic the CfrA expression pattern during infection from the initial oral ingestion (low-level CfrA expression due to exposure to an iron-rich environment) to colonization of the intestine (high-level CfrA expression due to iron-restricted conditions in vivo) and thus better evaluate the inhibitory effect of CfrA antibodies.
To enhance the production of specific antibodies directed against the ligand binding site of CfrA, it is important to determine the structure of CfrA and its functional epitopes. A recent report (11) indicated that CfrA contains many of the structural motifs conserved in other siderophore transporters and demonstrated the relatively high aqueous solvent exposure and high thermal stability of CfrA. In this study, we were particularly interested in which amino acids in CfrA may be involved in ferric enterobactin uptake. The mutagenesis study demonstrated that the basic amino acid R327 in CfrA is essential for ferric enterobactin uptake in C. jejuni. R327 is located in the fourth extracellular loop of CfrA, which is highly conserved in Campylobacter. Thus, the conserved protective epitope(s) covering R327 of CfrA may be cloned into appropriate delivery systems, such as an attenuated Salmonella mutant, leading to development of effective, inexpensive, and practical oral vaccines for preventing Campylobacter infections in humans and animal reservoirs.
Acknowledgments
We are grateful to Sandra K. Armstrong (University of Minnesota) for providing E. coli AN102 for purification of enterobactin and to Julian M. Ketley and Richard Haigh (University of Leicester) for providing CJJ81176_0471 mutant genomic DNA. We also thank Qijing Zhang (Iowa State University), Norman J. Stern (USDA), and Stephen P. Oliver (The University of Tennessee) for providing the majority of the clinical and environmental C. jejuni isolates used in this study.
This study was supported by grant 1R21AI07255101 from NIH.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 8 September 2009.
REFERENCES
- 1.Akiba, M., J. Lin, Y. W. Barton, and Q. Zhang. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J. Antimicrob. Chemother. 57:52-60. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M. T., and S. K. Armstrong. 2004. The BfeR regulator mediates enterobactin-inducible expression of Bordetella enterobactin utilization genes. J. Bacteriol. 186:7302-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, M. T., and S. K. Armstrong. 2006. The Bordetella Bfe system: growth and transcriptional response to siderophores, catechols, and neuroendocrine catecholamines. J. Bacteriol. 188:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler, A. J., T. L. Norris, T. Lasco, W. Voight, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 6.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 7.Braun, V., and K. Hantke. 2002. Microbial transport systems, p. 289-311. In G. Winkelmann (ed.), Microbial transport systems. Wiley-VCH Verlag GmBH & Co., Berlin, Germany.
- 8.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 9.Burton, C. L., S. R. Chhabra, S. Swift, T. J. Baldwin, H. Withers, S. J. Hill, and P. Williams. 2002. The growth response of Escherichia coli to neurotransmitters and related catecholamine drugs requires a functional enterobactin biosynthesis and uptake system. Infect. Immun. 70:5913-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao, Z., Z. Qi, C. Sprencel, S. M. Newton, and P. E. Klebba. 2000. Aromatic components of two ferric enterobactin binding sites in Escherichia coli FepA. Mol. Microbiol. 37:1306-1317. [DOI] [PubMed] [Google Scholar]
- 11.Carswell, C. L., M. D. Rigden, and J. E. Baenziger. 2008. Expression, purification, and structural characterization of CfrA, a putative iron transporter from Campylobacter jejuni. J. Bacteriol. 190:5650-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cogan, T. A., A. O. Thomas, L. E. Rees, A. H. Taylor, M. A. Jepson, P. H. Williams, J. Ketley, and T. J. Humphrey. 2007. Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut. 56:1060-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, C. R., and K. Poole. 1993. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J. Bacteriol. 175:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eldrup, E., and E. A. Richter. 2000. DOPA, dopamine, and DOPAC concentrations in the rat gastrointestinal tract decrease during fasting. Am. J. Physiol. Endocrinol. Metab. 279:E815-E822. [DOI] [PubMed] [Google Scholar]
- 15.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freestone, P. P., R. D. Haigh, and M. Lyte. 2007. Blockade of catecholamine-induced growth by adrenergic and dopaminergic receptor antagonists in Escherichia coli O157:H7, Salmonella enterica and Yersinia enterocolitica. BMC Microbiol. 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freestone, P. P., M. Lyte, C. P. Neal, A. F. Maggs, R. D. Haigh, and P. H. Williams. 2000. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol. 182:6091-6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerry, P., J. Perez-Casal, R. Yao, A. McVeigh, and T. J. Trust. 1997. A genetic locus involved in iron utilization unique to some Campylobacter strains. J. Bacteriol. 179:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, M. B. 2003. The enteric nervous system I: organisation and classification. Pharmacol. Toxicol. 92:105-113. [DOI] [PubMed] [Google Scholar]
- 20.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galan. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. M. Ketley, and J. M. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243-257. [DOI] [PubMed] [Google Scholar]
- 22.Huang, S., T. Luangtongkum, T. Y. Morishita, and Q. Zhang. 2005. Molecular typing of Campylobacter strains using the cmp gene encoding the major outer membrane protein. Foodborne Pathog Dis. 2:12-23. [DOI] [PubMed] [Google Scholar]
- 23.Hughes, D. T., and V. Sperandio. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6:111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 143:5-21. [DOI] [PubMed] [Google Scholar]
- 25.Kumar, S., M. Nei, J. Dudley, and K. Tamura. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, J., M. Akiba, O. Sahin, and Q. Zhang. 2005. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob. Agents Chemother. 49:1067-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, J., J. S. Hogan, M. Aslam, and K. L. Smith. 1998. Immunization of cows with ferric enterobactin receptor from coliform bacteria. J. Dairy Sci. 81:2151-2158. [DOI] [PubMed] [Google Scholar]
- 28.Lin, J., J. S. Hogan, and K. L. Smith. 1999. Growth responses of coliform bacteria to purified immunoglobulin G from cows immunized with ferric enterobactin receptor FepA. J. Dairy Sci. 82:86-92. [DOI] [PubMed] [Google Scholar]
- 29.Lin, J., J. S. Hogan, and K. L. Smith. 1998. Inhibition of in vitro growth of coliform bacteria by a monoclonal antibody directed against ferric enterobactin receptor FepA. J. Dairy Sci. 81:1267-1274. [DOI] [PubMed] [Google Scholar]
- 30.Lin, J., S. Huang, and Q. Zhang. 2002. Outer membrane proteins: key players for bacterial adaptation in host niches. Microbes Infect. 4:325-331. [DOI] [PubMed] [Google Scholar]
- 31.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, J., O. Sahin, L. O. Michel, and Q. Zhang. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect. Immun. 71:4250-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundrigan, M. D., and R. J. Kadner. 1986. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J. Biol. Chem. 261:10797-10801. [PubMed] [Google Scholar]
- 34.Lyte, M. 2004. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 12:14-20. [DOI] [PubMed] [Google Scholar]
- 35.Moeck, G. S., and J. W. Coulton. 1998. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol. Microbiol. 28:675-681. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, C. K., V. I. Kalve, and P. E. Klebba. 1990. Surface topology of the Escherichia coli K-12 ferric enterobactin receptor. J. Bacteriol. 172:2736-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newton, S. M., J. S. Allen, Z. Cao, Z. Qi, X. Jiang, C. Sprencel, J. D. Igo, S. B. Foster, M. A. Payne, and P. E. Klebba. 1997. Double mutagenesis of a positive charge cluster in the ligand-binding site of the ferric enterobactin receptor, FepA. Proc. Natl. Acad. Sci. USA 94:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 41.Projan, S. J. 2003. Why is big Pharma getting out of antibacterial drug discovery? Curr. Opin. Microbiol. 6:427-430. [DOI] [PubMed] [Google Scholar]
- 42.Rasko, D. A., C. G. Moreira, R. Li de, N. C. Reading, J. M. Ritchie, M. K. Waldor, N. Williams, R. Taussig, S. Wei, M. Roth, D. T. Hughes, J. F. Huntley, M. W. Fina, J. R. Falck, and V. Sperandio. 2008. Targeting QseC signaling and virulence for antibiotic development. Science 321:1078-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahin, O., N. Luo, S. Huang, and Q. Zhang. 2003. Effect of Campylobacter-specific maternal antibodies on Campylobacter jejuni colonization in young chickens. Appl. Environ. Microbiol. 69:5372-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahin, O., Q. Zhang, J. C. Meitzler, B. S. Harr, T. Y. Morishita, and R. Mohan. 2001. Prevalence, antigenic specificity, and bactericidal activity of poultry anti-Campylobacter maternal antibodies. Appl. Environ. Microbiol. 67:3951-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stintzi, A., A. H. M. V. Vliet, and J. M. Ketley. 2008. Iron metabolism, transport, and regulation, p. 591-610. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 47.van Vliet, A. H., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 180:5291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh, C. 2003. Where will new antibiotics come from? Nat. Rev. Microbiol. 1:65-70. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 50.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, Q., J. C. Meitzler, S. Huang, and T. Morishita. 2000. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68:5679-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, L., U. Baumann, and J. L. Reymond. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]