Abstract
The transcriptional regulator Rgg of Streptococcus pyogenes is essential for expression of the secreted cysteine protease SpeB. Although all isolates of S. pyogenes possess the speB gene, not all of them produce the protein in vitro. In a murine model of infection, the absence of SpeB production is associated with invasive disease. We speculated that naturally occurring mutations in rgg, which would also abrogate SpeB production, may be present in invasive isolates of S. pyogenes. Examination of the inferred Rgg sequences available in public databases revealed that the rgg gene in strain MGAS315 (a serotype M3 strain associated with invasive disease) encodes a proline at amino acid position 103 (Rgg103P); in contrast, all other strains encode a serine at this position (Rgg103S). A caseinolytic assay and Western blotting indicated that strain MGAS315 does not produce SpeB in vitro. Gene-swapping experiments showed that the rgg gene of MGAS315 is solely responsible for the lack of SpeB expression. In contrast to Rgg103S, Rgg103P does not bind to the speB promoter in gel shift assays, which correlates with a lack of speB expression. Despite its inability to activate speB expression, Rgg103P retains the ability to bind to DNA upstream of norA and to influence its expression. Overall, this study illustrates how variation at the rgg locus may contribute to the phenotypic diversity of S. pyogenes.
Streptococcus pyogenes causes non-life-threatening diseases, such as pharyngitis and impetigo, as well as life-threatening diseases, such as streptococcal toxic shock syndrome (STSS) and necrotizing fasciitis. Infection may also precipitate postinfection sequelae, including acute rheumatic fever and poststreptococcal acute glomerulonephritis (12). Worldwide, S. pyogenes is responsible for the death of approximately 500,000 people each year, primarily due to complications associated with acute rheumatic fever (6, 26).
All isolates of S. pyogenes possess a highly conserved speB gene, which encodes a secreted cysteine protease (10). In a mouse model, invasive S. pyogenes variants arising in vivo have mutations in covS, which encodes the histidine kinase component of the CovRS regulatory system (15, 29, 32). The mutations result in the loss of SpeB expression, which can attenuate the invasive potential of the pathogen by degrading secreted virulence factors, including streptokinase (SKA) and a bacteriophage-encoded DNase (Sda1) (1, 10, 11, 32). SKA converts human plasminogen to plasmin, which accumulates on the surface of the bacteria due to the presence of plasmin-binding proteins (11, 32). The plasmin activity mediates host tissue degradation and invasion. Similarly, in the absence of SpeB-mediated degradation of Sda1, the nuclease promotes dissemination of the pathogen by degrading neutrophil extracellular traps (32). The invasive SpeB-negative phenotype is associated only with mutations in covRS (16, 29) and not with mutations in speB, suggesting that additional changes in CovRS-mediated expression contribute to the phenotype. A variety of studies have revealed that SpeB is not produced in vitro by 58 to 93% of the S. pyogenes isolates tested, depending on the source and type of strains collected (8, 19, 27, 30). One possible explanation for the lack of SpeB production by these isolates is the accumulation of mutations in regulatory genes essential for speB expression.
The regulatory protein Rgg, also known as RopB, is essential for speB expression (7, 18). Rgg belongs to a family of transcriptional regulators (TIGR01716) encoded by genes in some low-G+C-content gram-positive bacteria, including Streptococcus gordonii (28) and by gadR in Lactococcus lactis (25), by mutR in Streptococcus mutans (22), by lasX in Lactobacillus sakei (23), and by rovS in Streptococcus agalactiae (24). Staphylococci, bacilli, and gram-negative bacteria do not encode Rgg-like orthologs. Rgg is characterized by a helix-turn-helix motif at the amino terminus, which is presumably involved in DNA binding. In addition to controlling speB expression, rgg inactivation is associated with changes in the expression of 706 genes in strain NZ131 during both the exponential and postexponential phases of growth (14). Finally, the Rgg regulon varies considerably in size and composition in different isolates of S. pyogenes (13, 16).
To determine if variation of rgg might be responsible for the lack of speB expression in certain clinical isolates, we performed rgg gene-swapping experiments. The results showed that the Rgg variant in strain MGAS315 is solely responsible for the lack of speB expression in this invasive isolate.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bacterial strains and plasmids used in this study are described in Table 1. S. pyogenes was grown at 37°C in a 5% CO2 atmosphere without agitation in either Todd-Hewitt broth (Becton Dickinson, Sparks, MD) containing 0.2% (wt/vol) yeast extract (THY broth) or chemically defined medium (CDM). When appropriate, erythromycin and kanamycin were added to media at final concentrations of 2.5 and 500 μg/ml, respectively. The composition of CDM was described previously (14), and this medium contained glucose at a final concentration of 2.0% (wt/vol).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source and/or reference |
|---|---|---|
| S. pyogenes strains | ||
| MGAS315 | M3 serotype | ATCC |
| MGAS315rgg | MGAS315 transformed with p40 to create an rgg mutant, Emr | This study |
| MGAS315rgg (Rgg103S) | MGAS315rgg complemented with pKK131, Emr Kanr | This study |
| MGAS315rgg (Rgg103P) | MGAS315rgg complemented with pKK315, Emr Kanr | This study |
| NZ131 | M49 serotype | D. R. Martin, New Zealand |
| NZ131rgg | rgg mutant, Emr | M. S. Chaussee (7) |
| NZ131speB | speB mutant, Emr | M. S. Chaussee (7) |
| NZ131rgg (Rgg103S) | NZ131rgg complemented with pKK131, Emr Kanr | This study |
| NZ131rgg (Rgg103P) | NZ131rgg complemented with pKK315, Emr Kanr | This study |
| E. faecalis UV202 | recA deficient | K. E. Weaver, University of South Dakota |
| E. coli DH10B | Invitrogen | |
| Plasmids | ||
| pVA891-2 | Cloning vector, Emr | H. Malke, Jena, Germany |
| p40 | pVA981-2 containing a 592-bp internal fragment of the rgg gene, Emr | 13 |
| pST-Blue1 | Cloning vector | EMD Chemicals Inc., Germany |
| pMSP3535Va | pVA380-1 replicon, nisRK pnisA Kanr | G. M. Dunny, University of Minnesota (5) |
| pKK1 | rgg ORF encoding a serine at amino acid 103 cloned into pMSP3535Va, Kanr | This study |
| pKK2 | rgg ORF encoding a proline at amino acid 103 cloned into pMSP3535Va, Kanr | This study |
| pKK131 | Derivative of pKK1 that is defective for replication in S. pyogenes, Kanr | This study |
| pKK315 | Derivative of pKK2 that is defective for replication in S. pyogenes, Kanr | This study |
Emr, resistance to erythromycin; Kanr, resistance to kanamycin.
Inactivation of the rgg gene in S. pyogenes MGAS315.
The rgg gene in MGAS315 was insertionally inactivated as previously described (13). The recombinant suicide vector designated p40 contained a 592-bp internal fragment of the rgg gene (Table 1). p40 was used to transform wild-type strain MGAS315, and transformants were selected on agar plates containing erythromycin. Inactivation of rgg was confirmed by nucleotide sequencing as previously described (13).
Construction of plasmids containing the NZ131 and MGAS315 rgg genes.
The oligonucleotide primers KpnF (5′-GGG GTA CCC CAT ATG GAA ATT GGT GAA-3′) and XbaR (5′-GCT CTA GAG CCT CAG GAC AGT TTA TGT-3′), which contained KpnI and XbaI restriction sites (underlined), were used to amplify the entire rgg open reading frame (ORF) from strains NZ131 and MGAS315. The PCR products were gel purified using a QIAquick PCR purification kit (Qiagen Inc., Valencia, CA) and cloned into pSTBlue-1 (EMD Chemicals Inc., Darmstadt, Germany). The rgg genes were excised using KpnI and XbaI and ligated to the cognate sites in pMSP3535Va (5). The ligation mixtures were used to electroporate Enterococcus faecalis UV202, and transformants were selected on agar plates containing kanamycin. E. faecalis was used to maintain the gram-positive origin of replication for use as a shuttle vector (5). The recombinant shuttle plasmids were designated pKK1, which contained the NZ131 rgg gene (rgg103S), and pKK2, which contained the MGAS315 rgg gene (rgg103P). Integration plasmids were derived from pKK1 and pKK2 following transformation of E. coli strain DH10B and selection for loss of a functional gram-positive origin of replication as previously described (5). The resulting integration vectors containing the NZ131 and MGAS315 rgg genes were designated pKK131 and pKK315, respectively. Nucleotide sequencing confirmed that no mutations occurred in the rgg ORFs during passage in E. coli.
rgg allelic replacements in strains NZ131 and MGAS315.
The MGAS315rgg mutant strain was used to express either the Rgg103S variant or the Rgg103P variant under control of the native rgg promoter. To do this, the MGAS315rgg strain was transformed with pKK131 (rgg103S) or pKK315 (rgg103P). Integration of these plasmids into the 5′ fragment of the interrupted rgg gene resulted in an intact rgg ORF at the native locus and a truncated rgg ORF in the integrated vector. Construction of the strains was confirmed by PCR and nucleotide sequencing (data not shown).
Immunoblot analysis of Rgg in MGAS315.
The MGAS315 and MGAS315rgg strains were grown in 40 ml THY broth. Samples were centrifuged at 4,000 rpm (3,200 × g) for 10 min at 4°C. The pellets were suspended in 0.5 ml phosphate-buffered saline (PBS) and transferred to FastPROTEIN Blue tubes (MB Biomedicals, LLC, Solon, OH). Samples were lysed using FastPrep FP120 (Qbiogene, Inc., Carlsbad, CA) twice at a speed of 6 for 20 s. Lysed samples were centrifuged, and supernatant fluids were collected. The MGAS315 and MGAS315rgg proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) independently. Each sample was transferred to a cellulose membrane using a mini whole-gel eluter (Bio-Rad Laboratories, Hercules, CA), and proteins with molecular masses ranging from 28 to 40 kDa were collected according to the manufacturer's instructions. Samples were dialyzed overnight in 0.25 M PBS and then concentrated from 3 ml to 0.2 ml using a Savant SPD131DD SpeedVac concentrator (Thermo Fisher Scientific Inc., Waltham, MA). Ten microliters of each sample was separated by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was blocked in 5% skim milk in PBS (blocking buffer) for 1 h at room temperature. The membrane was washed with PBS. Anti-Rgg rabbit antibody diluted 1:2,500 in blocking buffer was added and incubated for 1 h. Next, the membrane was incubated with conjugated goat anti-rabbit immunoglobulin G(H+L)-horseradish peroxidase (Bio-Rad Laboratories, Hercules, CA) diluted 1:5,000 in blocking buffer. SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific, Rockford, IL) was added to the membrane according to the manufacturer's instructions, and the signal was detected by exposure to Amersham Hyperfilm ECL (GE Healthcare Limited, Buckinghamshire, United Kingdom).
Casein agar assay.
Proteolytic activity of SpeB was assessed using agar plates containing casein as previously described (7).
Supernatant protein isolation, SDS-PAGE, and immunoblot analysis.
Culture supernatant fluids from S. pyogenes strains were passed through a 0.22-μm filter (Millipore Corp., Bedford, MA), and proteins were precipitated with 100% cold trichloroacetic acid (final concentration, 10%) and acetone (final concentration, 5%). Proteins were analyzed by immunoblotting with SpeB sera from rabbits as previously described (7).
Purification of Rgg and gel shift assays.
Fusion proteins containing the maltose-binding protein and either the Rgg103S variant from strain NZ131 or the Rgg103P variant from strain MGAS315 were constructed and purified as previously described (21). A digoxigenin gel shift kit (second generation; Roche Applied Sciences, Indianapolis, IN) was used to study protein-DNA interactions. DNA fragments were generated by PCR using genomic DNA isolated from NZ131 as the template DNA. The 225-bp upstream promoter region of speB containing the Rgg binding site (21) was generated using primers U3fwd (5′-GCG GGC ATA GTT TTA TCA ACT GTC ATA T-3′) and U1rev345 (5′-CGC TGA TGC TTT TAT TGA CTT CTT ATT-3′). The 227-bp norA putative upstream Rgg binding site was generated using primers norAIRfwd (5′-CGC TTT TCT CCA AAT ATT CTT TTA CCA AT-3′) and norAIRrev (5′-AGA CTA TAA AGT TTA TTA CTG TGA TGG CCA C-3′). The PCR products were gel purified using a SpinPrep gel DNA kit (EMD Chemicals, Inc., Gibbstown, NJ). The speB and norA DNA fragments were end labeled and used in gel shift reactions according to the manufacturer's instructions (Roche). Briefly, different amounts of variant Rgg protein (13.0 pmol, 26 pmol, and 75 pmol) were added to labeled speB (5.0 fmol) or norA (3.3 fmol) DNA, and 1.0 pmol of unlabeled DNA was added as a competitor in a control reaction. The complexes were separated on a 6% native polyacrylamide gel in 0.5× Tris-borate-EDTA buffer and transferred to a Hybond-N+ membrane (Amersham Pharmacia Biotech, Buckinghamshire, England). The membrane was cross-linked by baking it at 120°C for 30 min. The digoxigenin-labeled DNA fragments were detected by immunoblotting with anti-digoxigenin-alkaline phosphatase fragment (1:10,000) and the chloro-5-substituted adamantyl-1,2-dioxetane phosphate (CSPD) chemiluminescent substrate (1:100). The signal was detected by exposure to Amersham Hyperfilm ECL (GE Healthcare Limited, Buckinghamshire, United Kingdom). The gel shift experiments were repeated four times, and representative images are shown below.
Mass spectrometric analysis of E. coli-expressed Rgg variants.
One microgram of purified protein (either Rgg103S or Rgg103P) in a 50% mixture of solutions A (water in 0.1% formic acid) and B (acetonitrile in 0.1% formic acid) was injected into a mass spectrometer (ESI Q-Tof micro; Micromass, United Kingdom). Data were acquired in positive-ion mode using the following parameters: capillary voltage, 3,200 V; sample cone voltage, 30 V; and molecular mass range, 700 to 1,500 Da. The flow rate was 20 μl/min. Data were acquired for 4 min, and the spectrum was deconvoluted using the MaxEnt 1 tool from MassLynx 4.1 software (Micromass, United Kingdom) and the following parameters: output mass range, 75,800 to 78,000 Da; resolution, 0.6 Da/channel; damage model; uniform Gaussian width at half-height, 0.75 Da; left minimum intensity ratio, 33%; right minimum intensity ratio, 33%; and completion options, iterate to convergence.
RNA isolation.
S. pyogenes was grown in 10 ml of THY broth in 15-ml tubes (Fisher Scientific, Pittsburgh, PA) until the exponential phase of growth (A600, 0.3) and the postexponential phase of growth (defined using the first hourly measurement resulting in a change in the A600 of less than 0.05). Cultures were centrifuged at 4,000 rpm (3,200 × g) for 5 min, and the pellets were suspended in 300 μl of RNAlater (Ambion, Austin, TX) and frozen at −80°C. RNA was isolated using an RNeasy mini kit (Qiagen, Valencia, CA) according to the procedure recommended by the manufacturer. The concentration and quality of the RNA were determined with an Agilent 2100 bioanalyzer (Agilent, Palo Alto, CA) using an RNA 6000 Nano LabChip kit (Agilent, Palo Alto, CA).
Quantitative RT-PCR.
The norA probe and norA primers have been described previously (14). Amplification and detection were done with the ABI Prism 7700 sequence detection system (PE Applied Biosystems, Foster City, CA) using TaqMan One-Step reverse transcription (RT)-PCR master mixture reagents (Roche Applied Sciences, Indianapolis, IN) as previously described (14).
RESULTS
rgg variation in sequenced S. pyogenes strains.
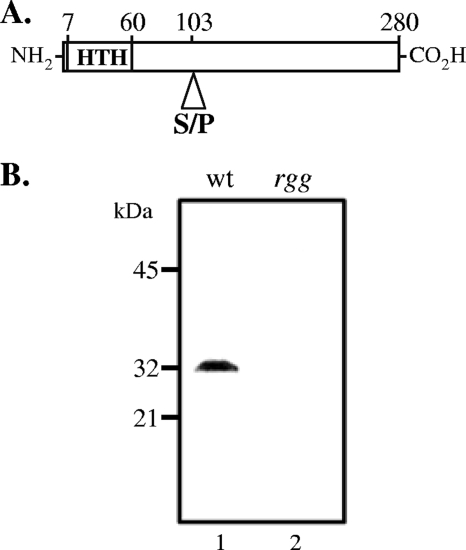
We speculated that some non-SpeB-producing isolates of S. pyogenes may be associated with changes in rgg. To examine this idea further, the nucleotide sequences of rgg in public databases were compared. Eight rgg alleles from 15 different clinical isolates were identified. Serotype M3 strains had the greatest variation, and five alleles were present in six strains. Of particular interest was a serotype M3 strain, designated MGAS315, that was isolated from an invasive disease episode involving STSS (4). Strain MGAS315 encodes the only Rgg variant with a proline residue at amino acid position 103 (Fig. 1A), in contrast to all other sequenced variants, which have a serine at this position. In addition, SpeB was not detected in strain MGAS315 by immunoblotting (2), suggesting that the variation at the rgg locus may be responsible for the absence of SpeB production.
FIG. 1.
Rgg is expressed in strain MGAS315. (A) Diagram of Rgg indicating the position of the N-terminal helix-turn-helix (HTH) motif and the amino acid variation at position 103. (B) Cytoplasmic proteins isolated from wild-type MGAS315 (wt) (lane 1) and an isogenic rgg mutant derivative (lane 2) analyzed by immunoblotting using antibodies to Rgg.
Strain MGAS315 expresses an Rgg variant that does not activate speB expression.
The nucleotide sequence of the Rgg binding site in the promoter region of speB (21) in strain MGAS315 was 97% (90 of 92 bp) identical to that in SpeB-producing strains SF370 and NZ131, suggesting that sequence variation in the upstream promoter region was probably not responsible for the lack of speB expression in strain MGAS315. In addition, the presumptive −35 and −10 sequences of the speB promoter are identical in strain MGAS315 and SpeB-producing strains. To determine if the lack of speB expression in MGAS315 was due to a failure to express rgg, Western blotting using antiserum to Rgg was used. The results showed that Rgg is expressed in strain MGAS315 (Fig. 1B).
To determine if the single amino acid difference between the Rgg proteins of strains NZ131 and MGAS315 (designated Rgg103S and Rgg103P, respectively) was responsible for the difference in speB expression, the two rgg alleles were used to complement an MGAS315 rgg mutant strain. The resulting strains contained identical heterologous DNA, and the variants were under transcriptional control of the native promoter. Thus, the strains differed only in the codon for the amino acid at position 103. PCR and nucleotide sequence analysis confirmed the construction of the strains (data not shown).
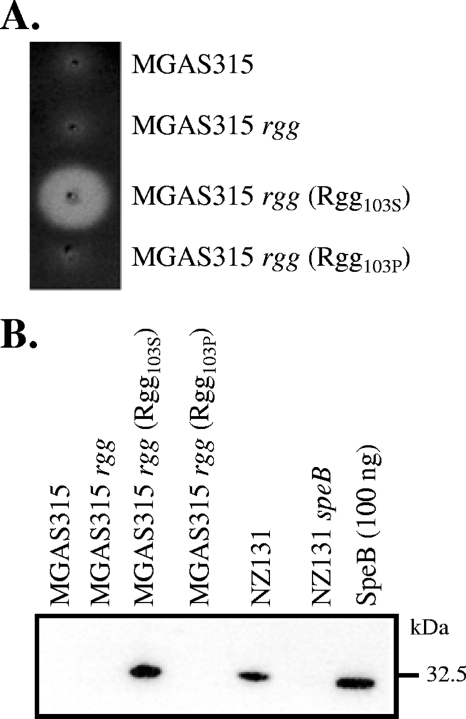
A casein agar assay was used to assess the influence of the two Rgg variants on speB expression. Wild-type strain MGAS315, an rgg mutant of MGAS315, and the mutant strain expressing Rgg103P did not produce caseinolytic activity (Fig. 2A), which is primarily due to SpeB activity (7). In contrast, expression of Rgg103S in strain MGAS315 was associated with caseinolytic activity (Fig. 2A). To confirm the results, culture supernatant fluids were analyzed by immunoblotting with polyclonal sera to SpeB as previously described (7). As predicted from the enzymatic assay, SpeB was detected in the MGAS315 rgg mutant expressing Rgg103S and the positive control strain NZ131. In contrast, strains MGAS315, MGAS315rgg, and MGAS315rgg (Rgg103P) did not produce SpeB (Fig. 2B). Finally, in strains expressing Rgg103P, speB transcripts were not detected by real-time RT-PCR; however, the level of speB transcripts was 16-fold greater than the level of gyrA transcripts in the MGAS315 rgg mutant strain expressing Rgg103S, which confirmed that the effect was at the level of transcription.
FIG. 2.
Rgg103S variant activates SpeB production in strain MGAS315. (A) Strains were stab inoculated into casein agar plates and incubated for 48 h anaerobically. Translucent zones surrounding the stab sites indicate SpeB caseinolytic activity. (B) Culture supernatant proteins analyzed by immunoblotting using antisera to SpeB.
To determine if the results were strain specific, an NZ131 rgg mutant was complemented with either the Rgg103S or Rgg103P variant. The strain transformed with Rgg103P did not express speB; in contrast, complementation with Rgg103S did restore speB expression (data not shown). Together, the results show that the naturally occurring Rgg variant possessing a proline residue instead of a serine residue at position 103 is solely responsible for the absence of speB expression in strain MGAS315.
The Rgg103P variant does not bind to the speB promoter but does bind to a motif upstream of norA.
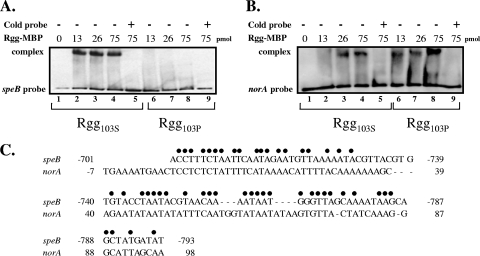
Previously, Rgg (RopB) binding to the speB promoter was characterized in strain HSC5, which expresses speB in the postexponential phase of growth (21). Gel shift assays were used to determine if the inability of Rgg103P to activate speB expression was due to a lack of binding to the speB promoter. To do this, the Rgg103S and Rgg103P variants were overexpressed in E. coli as proteins fused to the maltose-binding protein and were purified using amylose. The purity of the Rgg proteins was assessed by SDS-PAGE using Coomassie blue protein staining (data not shown). To determine if posttranslational modifications occurred during expression in E. coli, the proteins were also analyzed by mass spectrometry (data not shown), which confirmed that the proteins were not modified. Two DNA fragments were prepared for gel shift analysis. The first DNA fragment consisted of a 225-bp PCR product that corresponded to DNA upstream of speB (positions −596 to −820), which was previously shown to contain an Rgg binding site essential for speB expression (21). The second DNA fragment consisted of a 228-bp PCR product that corresponded to DNA upstream of the norA gene (positions −68 to 160). The norA gene was selected because RT-PCR indicated that its expression was influenced by inactivation of rgg in strain NZ131 and because norA expression was partially complemented in strain NZ131 with the Rgg103P variant (complementation increased norA expression twofold compared to the expression in the mutant strain). In addition, the nucleotide sequence of the putative promoter region was similar, but not identical, to the binding site of the speB gene (Fig. 3C). As expected, the Rgg103S variant retarded the speB DNA fragment in a concentration-dependent manner (Fig. 3A, lanes 2 to 4), and the retardation was relieved by addition of unlabeled DNA (Fig. 3A, lane 5). In contrast, the Rgg103P variant failed to retard the speB DNA fragment, even at the highest concentration of protein (Fig. 3A, lanes 6 to 8), indicating that the presence of a proline at position 103 abrogates binding to the speB promoter region. The absence of binding correlated with the lack of speB expression in strains expressing the Rgg103P variant. Both variants, however, specifically retarded DNA fragments designed to the putative Rgg binding site upstream of norA, indicating that the Rgg103P variant could still interact with DNA in a sequence-dependent manner (Fig. 3B, lanes 6 to 8); however, it remains to be determined if binding to this site specifically influences expression of norA. Overall, the results show that the single amino acid difference at position 103 of the Rgg protein influences the specificity of DNA binding.
FIG. 3.
DNA binding ability of the Rgg103S and Rgg103P variants as assessed by electrophoretic mobility shift assays. (A) Lanes 1 to 9 contained 5.0 fmol of speB DNA fragment. Lanes 2 to 5 contained different amounts of Rgg103S, and lanes 6 to 9 contained different amounts of Rgg103P. Lanes 5 and 9 also contained 1.0 pmol of unlabeled speB DNA fragment (Cold probe). (B) Lanes 1 to 9 contained 3.3 fmol of a norA DNA fragment. Lanes 2 to 5 contained different amounts of Rgg103S, and lanes 6 to 9 contained different amounts of Rgg103P. Lanes 5 and 9 also contained 1.0 pmol of an unlabeled norA DNA fragment. (C) Alignment of the Rgg binding site for speB with the predicted binding site in the putative promoter region of norA.
Inactivation of rgg in MGAS315 alters growth.
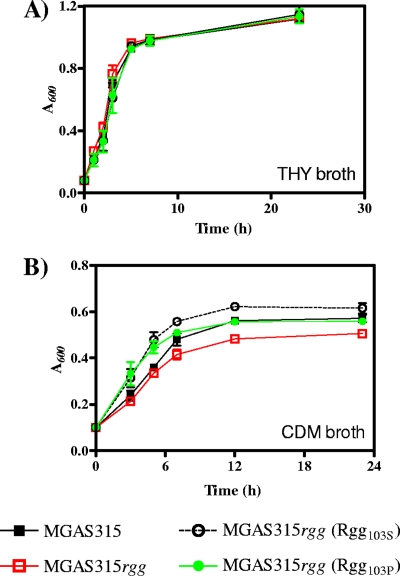
The Rgg regulon is variable in strains (13). For example, inactivation of rgg in S. pyogenes strains MGAS5005, SF370, CS101, and NZ131 alters the transcription of between 3 and 706 genes (13, 14). Although the Rgg protein in strain MGAS315 did not activate speB expression, it was expressed (Fig. 1B) and bound to DNA upstream of the norA gene (Fig. 3B). Therefore, it was of interest to determine if an rgg mutant of strain MGAS315 was phenotypically different from the parental strain. To test this, the growth of wild-type strain MGAS315, the growth of the isogenic rgg mutant derivative strain, and the growth of the mutant strain complemented with the variants encoding either Rgg103S or Rgg103P were analyzed. No significant differences in the growth rate or yield between the strains containing the different rgg genes and the rgg mutant strain were observed when peptide-rich THY medium was used (Fig. 4A).
FIG. 4.
Rgg103P variant influences growth in CDM. Strains were cultured with (A) THY broth or (B) CDM containing 2% glucose as the primary energy source. Growth was measured by determining the absorbance at 600 nm at various time points. The data are the means and standard errors of the means from three independent experiments.
In contrast, when cultured in CDM containing 2% glucose, the MGAS315 rgg mutant had a lower growth rate and lower growth yield than the wild-type MGAS315 strain (Fig. 4B), similar to results previously obtained with an rgg mutant of strain NZ131 (9). In addition, the changes in growth were abrogated by addition of 1% neopeptone to the media (data not shown), a result which is also similar to previous results obtained with strain NZ131 (9). Complementation with the Rgg103P variant restored the growth yield to that of the wild-type strain, while the Rgg103S variant had an increased growth rate and yield compared to the wild-type strain (Fig. 4B). In each case, there were minor differences in the exponential phase of growth compared to the wild-type strain. Nonetheless, the results suggest that the decrease in the growth yield associated with rgg inactivation is specific to loss of the gene, since the effect can be restored by complementation with either allele.
DISCUSSION
The transcriptional regulator Rgg is essential for expression of the extracellular SpeB protease, which is thought to promote a localized infection by degrading streptococcal invasive factors such as SKA and DNase I (1, 11, 29, 32). Serotype M3 strains are more likely than strains of many other serotypes to be associated with current episodes of invasive disease (3, 4). A previous study reported that a serotype M3 strain, designated MGAS315, did not produce SpeB in vitro (2). The rgg gene in this strain encodes a proline residue at position 103 instead of the serine residue that is encoded by all other alleles in current databases. We analyzed strain MGAS315 to determine if the variation was responsible for the lack of SpeB production. The results show that the Rgg variant of strain MGAS315 is unable to activate speB expression both in strain MGAS315 and in a serotype M49 strain background (strain NZ131). Gel shift assays showed that the variant does not bind to the speB promoter region, revealing the molecular basis for the lack of speB expression in MGAS315. Interestingly, the Rgg103P variant binds to DNA upstream of norA, indicating that variation at amino acid 103 does not abrogate binding but rather alters the specificity of binding.
Previous reports suggested that only a subset of S. pyogenes isolates produce SpeB in vitro (8, 19, 27, 30); however, it was not clear if the isolates could produce the protein in vivo during human infection. Identifying the molecular basis for the lack of SpeB production in MGAS315 suggests that the genetic change is stable and thus the isolate probably does not produce SpeB during human infection. Similarly, a naturally occurring truncated variant of Rgg was identified in serotype M1 strain 5628, which did not express speB (16). The results are consistent with the idea that invasive disease and SpeB production are inversely correlated (17); however, it is noteworthy that most isolates of S. pyogenes, including those obtained from invasive infections and other serotype M3 isolates, do produce SpeB in vitro, indicating that there is a role for multiple host and pathogen factors in determining the clinical fate of S. pyogenes infection.
The genome sequences of two serotype M3 isolates from patients with STSS, designated MGAS315 and SSI-1, have been determined (4, 20). Comparison of the sequences identified four nonsynonymous differences in chromosomally encoded proteins, including Rgg (20). In contrast to the MGAS315 protein, Rgg encoded by SSI-1 contains a serine at position 103 of Rgg and a cysteine at position 31 instead of a tyrosine. SSI-1 produces SpeB (20). The extent of Rgg variation in serotype M3 isolates and in other serotypes remains to be determined.
Rgg of S. gordonii is a positive transcriptional regulator of the adjacent gene gtfG, which encodes a glucosyltransferase involved in biofilm formation (28). An S. gordonii strain deficient in gtfG expression encoded an Rgg with a single amino acid difference compared to Rgg in strains expressing normal amounts of gtfG (31); specifically, the variant encoded a histidine at position 271 instead of an aspartate. Further analysis showed that the single amino acid difference was responsible for diminished gtfG expression. In contrast to results reported here, the variant maintained the ability to bind to the gtfG promoter region, indicating that additional factors were likely involved (31).
The SpeB protease can potentially alter host-pathogen interactions by modifying a number of host and streptococcal proteins (10). The changes include (i) alteration of the extracellular matrix by direct degradation of fibronectin and vitronectin and by activation of human matrix metalloproteases, (ii) degradation of complement factors, including C3b and factor H, and (iii) degradation or proteolytic truncation of bacterial surface proteins, including adhesins. Thus, although the occurrence and selection in mice of SpeB-negative invasive variants are associated with mutations in covRS (16, 29), naturally occurring rgg alleles that do not activate speB expression may influence several aspects of the infectious process, including adherence, host immune recognition, and tissue invasion.
Acknowledgments
We thank K. Weaver and A. Manna for critical reviews of the manuscript.
This work was supported by NIH NIAID grant RO1AI052147 to M.S.C. and by NIH grant 2 P20 RR016479 from the INBRE Program of the National Center for Research Resources.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 14 September 2009.
REFERENCES
- 1.Aziz, R. K., M. J. Pabst, A. Jeng, R. Kansal, D. E. Low, V. Nizet, and M. Kotb. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 51:123-134. [DOI] [PubMed] [Google Scholar]
- 2.Banks, D. J., B. Lei, and J. M. Musser. 2003. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 71:7079-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beres, S. B., E. W. Richter, M. J. Nagiec, P. Sumby, S. F. Porcella, F. R. DeLeo, and J. M. Musser. 2006. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 103:7059-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 6.Carapetis, J. R., A. C. Steer, E. K. Mulholland, and M. Weber. 2005. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5:685-694. [DOI] [PubMed] [Google Scholar]
- 7.Chaussee, M. S., D. Ajdic, and J. J. Ferretti. 1999. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect. Immun. 67:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaussee, M. S., J. Liu, D. L. Stevens, and J. J. Ferretti. 1996. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infections. J. Infect. Dis. 173:901-908. [DOI] [PubMed] [Google Scholar]
- 9.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang-Ni, C., and J. J. Wu. 2008. Effects of streptococcal pyrogenic exotoxin B on pathogenesis of Streptococcus pyogenes. J. Formos. Med. Assoc. 107:677-685. [DOI] [PubMed] [Google Scholar]
- 11.Cole, J. N., J. D. McArthur, F. C. McKay, M. L. Sanderson-Smith, A. J. Cork, M. Ranson, M. Rohde, A. Itzek, H. Sun, D. Ginsburg, M. Kotb, V. Nizet, G. S. Chhatwal, and M. J. Walker. 2006. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 20:1745-1755. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dmitriev, A. V., E. J. McDowell, and M. S. Chaussee. 2008. Inter- and intraserotypic variation in the Streptococcus pyogenes Rgg regulon. FEMS Microbiol. Lett. 284:43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dmitriev, A. V., E. J. McDowell, K. V. Kappeler, M. A. Chaussee, L. D. Rieck, and M. S. Chaussee. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J. Bacteriol. 188:7230-7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engleberg, N. C., A. Heath, A. Miller, C. Rivera, and V. J. DiRita. 2001. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183:1043-1054. [DOI] [PubMed] [Google Scholar]
- 16.Hollands, A., R. K. Aziz, R. Kansal, M. Kotb, V. Nizet, and M. J. Walker. 2008. A naturally occurring mutation in ropB suppresses SpeB expression and reduces M1T1 group A streptococcal systemic virulence. PLoS ONE 3:e4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kansal, R. G., A. McGeer, D. E. Low, A. Norrby-Teglund, and M. Kotb. 2000. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect. Immun. 68:6362-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion, and processing of the cysteine proteinase of Streptococcus pyogenes. J. EMBO. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa, I., K. Kurokawa, A. Yamashita, M. Nakata, Y. Tomivasu, N. Okahashi, S. Kawabata, K. Yamazaki, T. Shiba, T. Yasunaga, H. Hayashi, M. Hattori, and S. Hamada. 2003. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 13:1042-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neely, M. N., W. R. Lyon, D. L. Runft, and M. Caparon. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J. Bacteriol. 185:5166-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi, F., P. Chen, and P. W. Caufield. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rawlinson, E. L., I. F. Nes, and M. Skaugen. 2002. LasX, a transcriptional regulator of the lactocin S biosynthetic genes in Lactobacillus sakei L45, acts both as an activator and a repressor. Biochimie 84:559-567. [DOI] [PubMed] [Google Scholar]
- 24.Samen, U. M., B. J. Eikmanns, and D. J. Reinscheid. 2006. The transcriptional regulator RovS controls the attachment of Streptococcus agalactiae to human epithelial cells and the expression of virulence genes. Infect. Immun. 74:5625-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. Kok. 1998. A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 26.Stevens, D. L., M. H. Tanner, J. Winship, R. Swarts, K. M. Ries, P. M. Schlievert, and E. Kaplan. 1989. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N. Engl. J. Med. 321:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Stockbauer, K. E., L. Magoun, M. Liu, E. H. Burns, Jr., S. Gubba, S. Renish, X. Pan, S. C. Bodary, E. Baker, J. Coburn, J. M. Leong, and J. M. Musser. 1999. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αvβ3 and αIIb β3. Proc. Natl. Acad. Sci. USA 96:242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumby, P., A. R. Whitney, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS. Pathog. 2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talkington, D. F., B. Schwartz, C. M. Black, J. K. Todd, J. Elliott, R. F. Breiman, and R. R. Facklam. 1993. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect. Immun. 61:3369-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vickerman, M. M., M. Wang, and L. J. Baker. 2003. An amino acid change near the carboxyl terminus of the Streptococcus gordonii regulatory protein Rgg affects its abilities to bind DNA and influence expression of the glucosyltransferase gene gtfG. Microbiol. 149:399-406. [DOI] [PubMed] [Google Scholar]
- 32.Walker, M. J., A. Hollands, M. L. Sanderson-Smith, J. N. Cole, J. K. Kirk, A. Henningham, J. D. McArthur, K. Dinkla, R. K. Aziz, R. G. Kansal, A. J. Simpson, J. T. Buchanan, G. S. Chhatwal, M. Kotb, and V. Nizet. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 13:981-985. [DOI] [PubMed] [Google Scholar]