Abstract
Currently, the Golden Syrian hamster is widely considered an important model of Clostridium difficile disease, as oral infection of this animal pretreated with antibiotics reproduces many of the symptoms observed in humans. Two C. difficile strains, B1 and 630, showed significant differences in the progression and severity of disease in this model. B1-infected hamsters exhibited more severe pathology and a shorter time to death than hamsters infected with 630. Histological changes in the gut did not correlate with absolute numbers of C. difficile bacteria, but there were clear differences in the distribution of bacteria within gut tissues. Light, scanning, and transmission electron microscopy revealed high numbers of B1 bacteria at the mucosal surface of the tissue, whereas 630 bacteria were more frequently associated with the crypt regions. Both B1 and 630 bacteria were frequently observed within polymorphonuclear leukocytes, although, interestingly, a space frequently separated B1 bacteria from the phagosome wall, a phenomenon not observed with 630. However, pilus-like structures were detected on 630 located in the crypts of the gut tissue. Furthermore, B1 bacteria, but not 630 bacteria, were found within nonphagocytic cells, including enterocytes and muscle cells.
Clostridium difficile is a human pathogen whose importance is increasing. It causes gastrointestinal infections with symptoms that range from asymptomatic colonization to severe diarrhea, pseudomembranous colitis, or death (1, 2). C. difficile-associated diarrhea characteristically occurs following treatment with broad-spectrum antibiotics and is a significant problem in patients over the age of 65 years (29), for which the fatality rate has been calculated to be between 1 and 5% (17). In addition, the number of reported cases is increasing rapidly, making C. difficile one of the most common causes of nosocomial infections worldwide. Recent studies provided evidence of the emergence of ribotype 027 C. difficile with enhanced virulence, particularly with regard to its capacity to induce pseudomembranous colitis in patients (32). The factors favoring the emergence of this derivative are not clear, although changes in toxin production (33) and resistance to fluoroquinolones have been proposed (8). The 027 outbreak strain, which is dominant in North America, has now spread to Europe (16).
Until recently, the manifestations of C. difficile disease have been attributed largely to two exotoxins, toxins A and B, and antibodies that neutralize the effects of these toxins provide some protection in animal studies (11, 15). However, the frequent isolation of toxin A-negative strains that produce disease in humans suggests that other factors produced by the bacteria contribute to disease pathogenesis. While in vitro systems have been used to characterize specific aspects of the disease process (5, 22), in vivo studies with animals or in clinics provide the best opportunity to understand the dynamic interaction between the pathogen and host. The hamster is considered an important model for investigating C. difficile (2, 18, 27). This is in part because many of the clinical symptoms in humans, including diarrhea, histological damage, and relapse following removal of treatment, can be observed in this model.
In this study, a detailed examination of the hamster model was performed using a combination of telemetry, bacteriology, microscopy, and immunocytochemistry to monitor disease progression following challenge with the hamster-virulent strain C. difficile B1 (20) or the fully genome-sequenced strain 630 (28). Inclusion of the B1 strain, which has been used extensively elsewhere (26, 27), allowed direct comparison with data generated by other groups. Significant differences in the time course and severity of the disease caused by the two strains were observed, along with important differences in the distribution of the bacteria associated with and found within the gut tissues.
MATERIALS AND METHODS
Bacterial strains.
C. difficile strain B1 (which is distinct from the 027/BI/NAP1 lineage) was obtained from Dale Gerding (Hines Veterans Affairs Hospital, IL), and strain 630 was obtained from Brendan Wren (London School of Hygiene and Tropical Medicine, United Kingdom). Bacteria were routinely grown on CCFA agar supplemented with 7% horse blood under anaerobic conditions. Table 1 shows a summary of the properties of these strains.
TABLE 1.
Properties of the C. difficile strains used in this study
| Strain | Toxinotypea | Toxin Ab |
Toxin Bc |
CDTd |
Generation time in vitro (min)e | |||
|---|---|---|---|---|---|---|---|---|
| In vitro | In vivo | In vitro | In vivo | cdtA | cdtB | |||
| B1 | 0 | +++ | ++ | + | + | − | − | 70 |
| 630 | 0 | +++ | ++ | + | + | − | − | 40 |
Toxinotyping was kindly performed by M. Rupnik, Institute for Public Health, Slovenia.
Toxin A was measured by immunodot blotting using samples from 24-h cultures grown in YT media in vitro or from gut contents taken from hamsters at the endpoint of infection. The strength of the color change after addition of the substrate is indicated by the number of plus signs.
Toxin B was measured by immunodot blotting using samples from 24-h cultures grown in YT media in vitro or from gut contents taken from hamsters at the endpoint of infection. Uninfected YT media or uninfected gut contents were used as negative controls.
Determined by PCR using cdtA and cdtB primers described by Stubbs et al. (31), who confirmed by PCR, immunodot blotting, and an ADP-ribosyltransferase assay that toxintoype 0 strains do not generate the binary toxin.
Generation times for the two strains were calculated using cultures of the strains grown with shaking in prereduced brain heart infusion at 37°C.
Preparation of spores.
Spores were prepared from confluent bacterial cultures grown anaerobically on agar plates for 5 to 7 days. The cultures were then removed from the anaerobic environment and exposed to the atmosphere for 12 h to promote spore formation. The spores were then harvested from each plate into 1 to 2 ml of ethanol. The spores were collected by centrifugation and resuspended in phosphate-buffered saline (PBS), and 100-μl aliquots were stored at −80°C. To calculate the infectious dose administered, one aliquot was thawed, and the number of spores capable of germinating was calculated by plating serial dilutions on CCFA agar supplemented with blood.
Hamsters.
Female Golden Syrian hamsters that weighed approximately 100 g were purchased from Harlan Olac UK. These animals were housed individually and fed ad libitum. Telemetry chips (Vitalview Emitter) were inserted by laparotomy into the body cavities of the animals at least 3 weeks before infection with C. difficile. Once the wounds healed, the animals were placed on receiver pads, and the body temperature and activity were monitored (Vital View software).
Administration of clindamycin.
Each animal received orogastrically 30 mg/kg of clindamycin phosphate approximately 12 h before infection. Following administration of the antibiotic, all animals were placed in individual sterilized isolator cages with disposable filter top lids and were provided with sterile food, water, and bedding.
Oral infection with C. difficile spores.
A minimum of five animals per group each received approximately 100 spores of either strain B1 or 630. Following infection, each animal was placed in a second sterile cage with appropriate food, water, and bedding. The animals were subsequently transferred into new sterile cages every 24 h after infection until the end of the experiment. To monitor environmental contamination, animals treated with clindamycin alone were subjected to the same treatment and housed in individual cages alongside infected animals.
Quantification of bacterial load.
To estimate colonization, hamsters were sacrificed, and the gut region from the stomach to the anus of each animal was removed. The gut was divided into four sections, the upper small bowel, the lower small bowel, the cecum, and the colon. To enumerate the total bacterial load (spores and vegetative cells), each section was opened longitudinally, and the contents were removed by gentle washing in two changes of 10 ml PBS. The tissues were homogenized in 5 ml of PBS for 1 min using a Stomacher, and viable counts were determined for the homogenates. Serial 10-fold dilutions were plated on CCFA blood agar plates containing 20 μg/ml amphotericin B to suppress yeast growth. To estimate the numbers of spores present in the samples, the samples were heated for 10 min at 56°C, and the numbers of spores present were determined by the viable count method as described above.
Detection of toxin production in vitro and in vivo.
Production of toxins A and B by B1 and 630 was detected in vitro using filtered culture supernatants obtained from cultures grown anaerobically for 96 h and in vivo using filtered cecal contents from animals taken at the endpoint of the infection experiment. Portions (50 μl) of samples were inoculated onto nitrocellulose membranes and allowed to dry. Negative controls consisting of brain heart infusion alone or filtrate from an uninfected animal were also included. Membranes were probed with either monoclonal antibody PCG-4 (Novus Biologicals), which was used for detection of toxin A, or monoclonal antibody 5A8-E11 (Novus Biologicals), which was used for detection of toxin B. The specificity of these antibodies for detection of the toxins was confirmed by inclusion of purified toxins A and B (VWR International) as positive controls. In addition, uninfected YT media or gut contents from an uninfected animal were used as negative controls.
Confirmation of infecting strains.
To confirm that the strain isolated from a hamster was the strain that was originally used for infection, bacteria were grown, and DNA was isolated and sent to Brendan Wren (London School of Hygiene and Tropical Medicine), who compared the genomic content as determined by microarray analysis to the genomic contents previously determined for the infecting strains (30) (data not shown).
Histological changes in the gut.
Samples from different regions of the gut were prepared for simple histology by using 5% paraformaldehyde. As the infected tissue was extremely friable, this was done by gentle instillation of the fixative by insertion of a fine-tip pastette into a 2- to 3-cm section of tissue, which was then immediately placed into a bath of the fixative. Samples were fixed for at least 24 h before they were sectioned and stained with hematoxylin and eosin.
Electron microscopy.
Rings of gut tissue that were approximately 5 mm thick were placed in a primary fixative (2% paraformaldehyde, 2% glutaraldehyde, 0.1 M sodium cacodylate buffer [pH 7.4], 0.1% MgCl2, 0.05% CaCl2) for 2 h on ice immediately after dissection. The specimens were then washed in ice-cold sodium cacodylate buffer with added chlorides, before they were placed in 1% osmium tetroxide in sodium cacodylate buffer for 1 h at room temperature. The samples were then treated with mordant (1% tannic acid) before dehydration with an ethanol series (20%, 30%, 50%, 70%, 90%, and 95% ethanol; 30 min each), staining with 2% uranyl acetate at the 30% ethanol stage, and finally submersion three times for 20 min in 100% ethanol. The samples were treated twice for 15 min with propylene oxide (PO), and then the solution was changed to a 1:1 mixture of PO and TAAB812 resin (with treatment for at least 1 h) and finally to neat TAAB812 resin with a few drops of PO (with treatment overnight). The specimens were then embedded in a flat molded tray and cured in an oven at 60°C. Thin sections (50 nm) were cut with a Leica EMUC6 ultramicrotome, contrasted with uranyl acetate and lead citrate, and imaged with an FEI 120-kV Spirit Biotwin transmission electron microscope (TEM) with a Tietz F415 digital Temcam.
For scanning electron microscopy (SEM) the processing procedure was the same as the procedure described above, except that after the initial osmification tissues were further impregnated with 1% aqueous thiocarbohydrate and osmium tetroxide layers using the protocol for OTOTO described by Malick and Wilson (18a). Dehydration was followed by critical point drying in a Bal-tec CPD030, and tissue was mounted on aluminum stubs with silver dag, sputter coated with 2-nm gold in a Bal-tec SCD050, and examined with an Hitachi S-4800 SEM.
Immunogold labeling of infected sections.
Tissues were fixed in 4% paraformaldehyde, 0.2% glutaraldehyde, 0.1 M phosphate buffer (pH 7.4) at 37°C for 10 min and then at room temperature for a combined total fixation time of 2 h. Tissues were then rinsed in PBS and processed to Lowicryl HM20 by progressively lowering the temperature in a Leica FS unit. Briefly, samples were dehydrated using an ethanol series (30% ethanol at 4°C, 50% ethanol at 1°C, 70% and 90% ethanol at −20°C, and 100% ethanol at −30°C and −50°C; 30 min each) and impregnated with Lowicryl-ethanol (1:3, 1:1, and 3:1; 1 h each) and then with neat Lowicryl overnight before embedding and UV polymerization at −50°C. Ultrathin sections (50 nm) were cut and collected on Formar-carbon-coated grids for labeling using 50-μl droplets on Parafilm. Sections were blocked with 0.02 M glycine in PBS for 10 min, followed by 10% fetal calf serum in blocking diluent for all following steps for 1 h, labeled with serum raised to heat-killed C. difficile 630 diluted 1:200, and rinsed in PBS three times over a 15-min period. For detection of pili, antibody to PilA (CD3507), raised to the recombinant protein in rabbits and absorbed against Escherichia coli, was used. Sections were then incubated on 10-nm protein A gold for 20 min, washed thoroughly with distilled water for 10 min, contrasted with uranyl acetate and lead citrate, and imaged with the TEM.
Individual counts of C. difficile B1 and 630 were obtained for immunogold-localized bacteria present in the infected gut tissue. Eight transverse 3-mm lengths of cecum between the surface of the mucosa and the base of the lamina propria were analyzed for each infection, and the precise location of either C. difficile or the background flora was recorded.
Statistical analysis.
All statistical analyses were carried out using GraphPad Instat 3. Time-to-death data were analyzed using the parametric Student t test, while the levels of colonization of different regions of the gut were analyzed using the nonparametric two-tailed Mann-Whitney test. A P value of <0.05 was considered statistically significant.
RESULTS
Monitoring infection using clinical observation and telemetry.
Clindamycin-pretreated hamsters were orogastrically challenged with spores of either C. difficile B1 or 630 and observed until they succumbed to infection. Previously implanted telemetry chips provided a continual stream of information related to the core body temperature and activity of each hamster during the course of infection. Control readings taken before and after clindamycin treatment showed that antibiotic treatment had no detectable influence on these telemetric measurements (results not shown). Despite the fact that these two strains share several key phenotypic characteristics, such as being toxinotype 0 and producers of both toxin A and B in vitro and in vivo, and despite the fact that they are PCR negative for the actin-specific ADP-ribosyltransferase (binary) toxin (C. difficile binary toxin) (Table 1), they displayed different disease profiles in the hamster model. Following challenge with B1, hamsters routinely developed diarrhea approximately 28 h postinfection. At the onset of diarrhea, the hamsters appeared to be clinically well; however, during the next 2 to 3 h, as the diarrhea continued, a reduction in hamster physical activity was noted. Animals were culled when the body temperature dropped to ethically unacceptable levels (below 35°C), normally at around 33 h. This infection profile has been observed in over 40 infected animals to date. In contrast to hamsters infected with B1, hamsters infected with 630 remained well for at least 36 h before developing diarrhea. The onset of diarrhea coincided with a significant drop in temperature, and the hamsters became rapidly moribund and were sacrificed when the body temperature dropped to 35°C, which occurred between 45 and 48 h postinfection. Interestingly, approximately 90% of the hamsters infected with either strain exhibited a spike in temperature (approximately 1°C) prior to the onset of diarrhea.
To determine the reproducibility of infection, the time taken to reach a core body temperature that was 2°C below the normal temperature when the two C. difficile strains were used was calculated, and the average time to death was 2,001 min (standard deviation, 64 min; n = 5) for B1 and 2,686 min (standard deviation, 90 min; n = 5) for 630. Comparison of data from individual animals using the parametric Student t test indicated that animals infected with B1 had a statistically significantly different (P = 0.001) time to the “endpoint” of the infection (approximately 33 h postinfection) than animals infected with 630 (45 h postinfection). Thus, ongoing monitoring of the core body temperature provides a useful indirect measurement of the time course of infection.
Quantification of C. difficile recovered from the gut of infected hamsters.
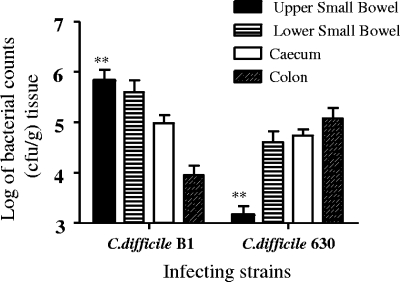
The numbers of viable C. difficile bacteria (vegetative cells and spores) associated with tissues from different regions of the gut were determined at the time of sacrifice by monitoring bacterial viable counts using both luminal washes (data not shown) and homogenized tissue (Fig. 1). While statistically higher numbers of C. difficile B1 bacteria were found in homogenized tissue taken from the upper small bowel (P = 0.0079, Mann-Whitney test), strain 630 showed almost the opposite pattern of colonization and the highest number of bacteria was recovered from homogenized tissue from the colon. Although the difference in the levels of colon colonization between the strains is not statistically significant (P = 0.150, Mann-Whitney test), the trend was consistent in repeated experiments (n = 10) (data not shown). At present it is unclear whether this reflects a real difference in the preferred location of colonization of the strains or is a result of the additional time (12 h) that 630 was located in and able to multiply in the colon. Subsequent analysis of the tissue by electron microscopy (see below) revealed that the distribution of the two C. difficile strains within the gut tissue could at least partially contribute to the differences, as viable counting was performed using material recovered from washed, homogenized tissue. As a consequence, the high levels of 630 bacteria associated with the crypt regions of the mucosal surface may have been recovered more effectively than the B1 bacteria, which were found more frequently within the mucosal tissue inside neutrophils (Table 2).
FIG. 1.
Log bacterial counts recovered from different regions of the bowel following infection with approximately 100 spores of C. difficile B1 or 630. Tissue was obtained from the upper small bowel, the lower small bowel, the cecum, and the colon. The error bars indicate the standard deviations from the means for five animals. Asterisks indicate differences in colonization that are statistically significant.
TABLE 2.
Counts of B1 and 630 bacteria detected in hamster gut tissues by electron microscopy
| Location | No. of bacteriaa |
|||
|---|---|---|---|---|
| Infection 1 |
Infection 2 |
|||
| B1 | 630 | B1 | 630 | |
| On mucosa surface | 120 (5.3) | 178 (4.5) | 166 (14.4) | 131 (11.6) |
| Inside surface neutrophils | 560 (9.4) | 120 (6.7) | 687 (29.5) | 85 (6) |
| Inside surface eosinophils | 252 (8.0) | 0 | 207 (18.7) | 0 |
| Inside epithelial cell | 20 (2.4) | 0 | 26 (3.8) | 0 |
| Inside submucosal neutrophils | 252 (7.7) | 120 (7.9) | 316 (20.5) | 164 (15.5) |
| Inside submucosal eosinophils | 192 (8.8) | 0 | 122 (10.7) | 0 |
| Inside nongranulocytes | 35 (4.4) | 0 | 52 (4.1) | 0 |
| Inside crypts | 0 | 1,996 (78.2) | 0 | 3,040 (213.6) |
| Extracellular in submucosa | 60 (5.7) | 32 (6.4) | 108 (9.7) | 23 (5.6) |
Counts were obtained for immunogold-localized bacteria present in the infected gut tissue. Eight 3-mm transverse lengths of cecum were analyzed for each independent infection, and the standard deviations of the means of the counts are indicated in parentheses. There are no significant differences between the two data sets.
In a more detailed study using strain B1, the numbers of heat-resistant bacteria (spores) present at different locations within the gut at different times after challenge were determined (Table 3). This analysis revealed that spores were present at the late stage of infection, during the onset of diarrhea (28 h) and at 33 h when a higher proportion of heat-resistant organisms was detected.
TABLE 3.
Numbers of vegetative bacteria and heat-resistant bacteria (spores) intimately associated with tissues over a time course of infection with C. difficile B1
| Time postinfection (h) | No. of viable bacteria recovered |
|||||||
|---|---|---|---|---|---|---|---|---|
| Upper small bowel |
Lower small bowel |
Cecum |
Colon |
|||||
| Vegetative cells | Spores | Vegetative cells | Spores | Vegetative cells | Spores | Vegetative cells | Spores | |
| 28 | 8.41 × 104 | 8.6 × 101 | 1.19 × 104 | 9.8 × 102 | 7.83 × 104 | NDa | 1.50 × 103 | ND |
| 33 | 1.35 × 105 | 1.04 × 104 | 1.4 × 106 | 9.3 × 104 | 3.18 × 106 | 1.7 × 105 | 3.57 × 105 | 3.22 × 104 |
ND, not determined.
Histological changes.
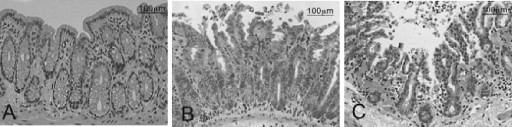
In order to assess the impact of C. difficile infection on gut pathology, sections of gut tissues were sampled after infection at 12 h, at 24 h, at the onset of diarrhea (28 h), and at the time of death (approximately 33 h and 45 h for B1 and 630, respectively). The tissue samples were stained with hematoxylin and eosin and were compared to similar tissue samples from noninfected hamsters (Fig. 2). While samples were prepared for the entire length of the gut, only the sections from the cecum and colon showed an influx of polymorphonuclear leukocytes (PMNs) associated with the infection (3). This was despite the fact that C. difficile strains could be cultured from the other regions at the times examined. In the ceca and colons from both B1- and 630-infected hamsters obvious pathology was apparent by 28 h and 45 h, respectively, corresponding to the terminal stage of the disease in each case. Importantly, despite the fact that fewer C. difficile bacteria were recovered from the homogenized cecal and colonic tissue, hamsters infected with B1 consistently exhibited greater inflammatory damage than hamsters infected with 630. At these times the integrity of the infected epithelial surface in hamsters infected with either strain was almost completely destroyed, and inflammatory exudate was released into the lumen (Fig. 2B and C). Extensive submucosal edema was also observed, together with severe vascular congestion in the submucosa (Fig. 2C). Further, PMNs (predominantly neutrophils) could be observed entering the tissue via infiltrating blood vessels and also extruded through the damaged epithelium.
FIG. 2.
Hematoxylin- and eosin-stained, paraffin-fixed sections of the upper colon of (A) uninfected hamsters, (B) 630-infected hamsters, and (C) B1-infected hamsters. In contrast to the results for the uninfected tissue, increasing numbers of polymorphs are present basally and at the surface of the mucosal of tissue infected with C. difficile 630, and the highest numbers are in tissue infected with C. difficile B1.
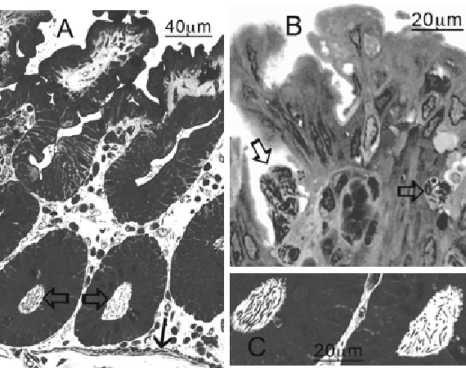
Further analysis using 1-μm semithin Lowicryl sections stained with toluidine blue (Fig. 3) confirmed that there were striking differences in both the associated pathologies of the infected tissue and the distributions of the two strains of C. difficile within the mucosa and submucosa of the cecum (Table 2). C. difficile 630 heavily colonized the crypts, and the numbers of bacteria increased basally without disrupting the integrity of the local mucosa close to the infected crypts (Fig. 3A). In stark contrast, B1 was almost absent from the crypt regions and was observed mainly at or just below the surface epithelium at the neck of the crypts. B1 bacteria were consistently associated with both heavy disruption of the mucosa and infiltration of higher numbers of PMNs to the mucosal surface and to the submucosa (Fig. 3B).
FIG. 3.
Semithin (1 μm) Lowicryl sections of tissue infected with C. difficile B1 and 630 and stained with toluidine blue. (A) In tissue infected with 630, bacteria are at the mucosal surface at the base of the crypts (open arrows) in close proximity to the basal lamina (solid arrow). (C) Higher magnification of bacteria in panel A. (B) In contrast, B1 bacteria were most often associated with PMNs (arrows).
SEM and TEM.
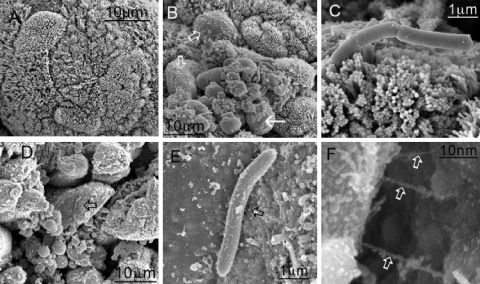
The detailed analysis of the distribution of C. difficile and the characterization of the infiltrate were supported and enhanced by electron and immunoelectron microscopy of adjacent sections of gut tissue. SEM images of infected cecum reinforced the evidence that infection with B1 resulted in damage to the surface of the gut that was greater than that observed with 630 (Fig. 4). Gut tissue from a B1-infected colon was denuded of microvilli (Fig. 4A and D), and the organisms were apparently in contact with the surface and associated with thin structures (Fig. 4E and F). These structures resembled bacterial pili, but they could potentially have been host derived. In direct contrast, 630 bacteria did not appear to cause significant damage to the microvilli of the gut and were more often observed interacting with the gut microvillus via one pole of the organism (Fig. 4C).
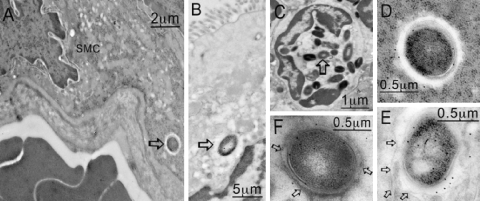
FIG. 4.
SEM images of cecal tissue infected with C. difficile. Tissues were obtained 1 h after the onset of diarrhea (approximately 29 h postinfection for B1 and approximately 44 h postinfection for 630). (A) Control uninfected tissue. (B and D) Infection of the tissue with both 630 (B) and B1 (D) resulted in denuded patches of microvilli (open arrows), and organisms appeared to anchor themselves to the mucosal surface. (C and E) For 630, attachment was mediated largely at the pole of the organism (C), while for B1, the organisms appeared to be interacting with the surface by means of pilus-like structures between 200 and 300 nm long (E) (arrow). (F) Higher magnification of pilus-like structures in panel E (arrows).
Analysis of TEM images identified a significant number of both B1 and 630 bacteria interacting with the mucosal surface, although much higher numbers of B1 bacteria were found within vacuoles inside PMNs attracted to the surface or to the submucosa of the gut (Fig. 5A). Many of the bacteria detected at the surface of the gut and within the underlying submucosa were confirmed to be C. difficile by immunogold labeling using specific polyclonal sera raised to whole C. difficile bacteria (Fig. 5C and 6D). In contrast to B1 and in agreement with the previous observations, high numbers of 630 bacteria were found at the base of the crypts, where they expressed structures that resembled both flagella and pili (Fig. 5D). Interestingly, C. difficile bacteria expressing one of these two structures were recognized by antisera raised to PilA (CD3507), one of five putative type IV pilus proteins encoded by genes identified in the genome (28), suggesting that such pili may be expressed in vivo (Fig. 7F).
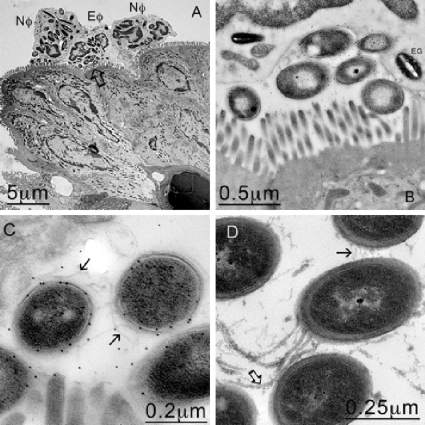
FIG. 5.
PMNs engulfing C. difficile B1 at the mucosal membrane. (A) TEM image showing neutrophils (Nφ) and eosinophils (Eφ) at the mucosal surface that appear to have engulfed several bacteria. (B) Higher magnification of the area indicated by the arrow in panel A, confirming that C. difficile was trapped against the mucosal membrane by the host cells. (C) Higher magnification of panel B. The bacteria were confirmed to be C. difficile by immunogold labeling. (D) In contrast, 630 bacteria found in the crypts appear to express both flagella (open arrow) and pili (solid arrow).
FIG. 6.
(A) C. difficile B1 bacteria were most commonly found in association with PMNs within vacuoles (arrow). (B) Higher magnification of panel A. (C) When images of the vacuoles were enlarged (area indicated by open arrows in panels A and B), filamentous structures between 200 and 400 nm long were discerned (arrowheads). These structures appeared to pin back (solid arrows in panel B and solid arrow in panel C) the contents released from fusing lysosomes (open arrows in panels B and C), suggesting a possible mechanism for the cell to avoid direct contact with the digestive elements. (D) Immunogold labeling confirmed that the organisms inside the vacuole were C. difficile.
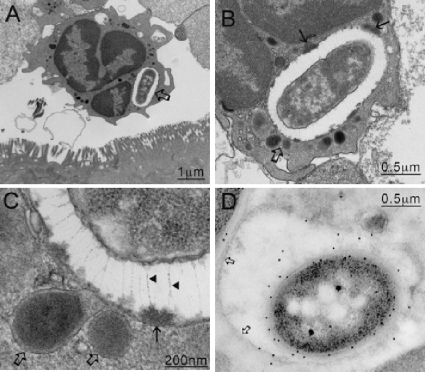
FIG. 7.
TEM images of tissue infected with C. difficile 630. Unlike B1 (Fig. 6), 630 seems to be unable to prevent killing within the neutrophils (A), and organisms located within phagosomes undergo degradation (B). In an enhanced image (D), the phagosomal membrane (open arrow) is located tightly around the bacteria, lysosomal contents are intimately associated with the bacteria, and a second dark vesicle is undergoing fusion (solid arrow) The bacteria within the phagosomal vesicles (C) and the flagellated cells within the crypt regions (E) were confirmed to be C. difficile by immunogold labeling with anti-630 antibody. In addition, antibody raised to PilA (CD3507) recognized the larger of the two structures expressed by free 630 bacteria located in the crypt regions, suggesting that the pili may be expressed in vivo (F).
Intracellular C. difficile bacteria are detected by electron microscopy.
Examination of TEM images of infected gut tissue routinely revealed that the bacteria were within both phagocytic (both 630 and B1) and nonphagocytic cells (only B1). PMNs were regularly detected at the infected mucosa either within tissues or free at the luminal surface (Fig. 5A). Closer examination of TEM images revealed C. difficile within neutrophils (Fig. 6A). When the cells from B1-infected tissues were examined, the intracellular bacteria could be clearly visualized because an electron-translucent space was often visible between the bacteria and the vacuolar membrane (Fig. 6B). Closer examination of these vacuolar spaces revealed the presence of unique filamentous elements that visually resembled capsular elements of a fibrillar glycocalyx array (7). These structures appeared to inhibit direct contact between the phagolysosomal membrane and the bacteria (Fig. 6C). Further study of the images also identified fusing lysosomes that appeared to be releasing their contents into the vacuolar space. In contrast, although 630 could be observed within vacuoles of neutrophils, vacuolar spaces or filaments were never detected (Fig. 7A to D). Visually, the destruction of 630 was more apparent than the destruction of B1 (Fig. 7A to D), and the phagosomal membrane was consistently found to be tightly associated with 630 bacteria and lysosomal contents were found to be in intimate contact with the bacterial membrane. This suggests that the two C. difficile strains interact differently with neutrophils in vivo.
Interestingly, small but significant numbers of B1 bacteria were found inside vacuoles apparently located in a number of different types of nonphagocytic cells, including smooth muscle cells (Fig. 8A), enterocytes at the epithelium of the mucosa (Fig. 8B), and eosinophils (Fig. 8C). In the smooth muscle, lack of disruption of adjacent muscle fibers suggested that this was not an artifact of tissue preparation or damage as a result of infection. The bacteria in nonphagocytic cells were sometimes within open vacuoles similar to those observed in the neutrophils, but on other images there was a close association between the bacteria and vacuolar membranes. Similar observations were not made when 630-infected tissues were examined, and the intracellular bacteria were restricted to phagocytes.
FIG. 8.
C. difficile B1 was in several other locations within the gut, as determined by TEM. These locations included smooth muscle cells (A [arrow] and D), the mucosa (B [arrow] and E), and eosinophils (C [arrow] and F). In panels E and F the membrane around the bacteria is viable (arrows), but a membrane is not present in panel D (arrows).
DISCUSSION
Since the early 1970s, the hamster has been recognized as a valuable model of C. difficile disease, and in particular, it has provided important information about the role of the toxins in disease and potential experimental therapies. However, relatively little work has been done to carefully characterize the infection process (15, 20, 23) in terms of pathology. This may in part reflect the difficulty of housing and managing hamsters, which makes the use of statistically relevant numbers of animals more difficult than the use of other rodents, such as the mouse. However, with the recent developments in Clostridium genetics and the availability of molecular tools for analysis (12, 24), well-characterized models for this type of infection are likely to become more important.
In contrast to the results with hamsters, attempts to successfully infect inbred mice have in the past resulted in mild disease with limited pathology (21). More recently, it has been reported (6) that treatment of mice with a cocktail of antibiotics prior to infection with the highly toxigenic strain VPI10463 can allow development of many of the histopathological features seen in humans, including diarrhea. While this development may provide an alternative and useful model, it does not negate data generated with the hamster, in which treatment with clindamycin can be followed by administration of variety of different clinically isolated C. difficile strains, resulting in fatal infections in the animals.
Here, we describe differences in pathology that were observed when two C. difficile strains, strains 630 (28) and B1 (27), were investigated using the hamster model. Although these strains are clearly distinct, they both produce toxins A and B in vitro and in vivo and are in fact the same toxinotype. However, we observed that 630 caused an extended disease that was less severe than the disease caused by strain B1. This observation is consistent with a previous report comparing 630 to the epidemic BI strains (distinct from B1) that also found that 630 causes less severe disease (20). In our study, the difference in disease severity did not appear to correlate with the total C. difficile burden in the gut, but it did correlate with the distribution of bacteria within tissues. Microscopic analysis revealed that strain 630 was associated predominantly with the deep crypts of infected tissues, whereas B1 was found primarily within the mucosal tissue associated with inflammatory cells, such as neutrophils. Such differences in the tissue distribution of C. difficile strains have not been reported previously and may provide clues to the differences in the pathologies observed. In the past C. difficile-associated diarrhea has been attributed largely to the production of toxins A and B, but the infection is clearly complex, and other factors, such as tissue distribution, could play a role in the pathology. This possibility is supported by the emergence of clinically relevant strains with truncated or deleted forms of the toxin genes (9, 14). The observation that strains that produce high levels of toxins, such as NAP1/027, can cause mortality in hamsters without inducing diarrhea adds to the argument that other bacterial and host-derived factors contribute to the disease process (25).
In addition to differences in tissue distribution, we also noted differences between the C. difficile strains in their interactions with host cells in vivo. B1 could be readily identified inside neutrophils, as they were residents in spacious vacuoles in which the bacterial cell surface was clearly separated from the vacuolar membrane by an electron-translucent space. Careful examination of this space revealed the presence of fine filamentous appendages between the bacteria and the host vacuolar membrane that visually resembled a fibrillar glycocalyx array previously observed when bacteria formed microcolonies on mucosal surfaces (7). Similar structures were not found in neutrophil vacuoles harboring 630, where the bacterial surface was apparently immediately adjacent to the vacuolar membrane. In addition, within the crypts, the 630 bacteria appeared to express pilus-like structures which were recognized by an antiserum raised to PilA (CD3507), suggesting that 630 bacteria express type IV pili. These pili, which are associated with twitching motility, are believed to contribute to effective host colonization and other forms of complex colonial behavior (19). Several C. difficile proteins that appear to play a role in adhesion in vitro have already been identified (4, 5), and passively administered antibodies to the dominant surface layer protein were found to prolong the survival of animals following bacterial challenge (23). Electron micrographs generated in our study suggest that bacterial factors other than surface layer proteins may be involved in the adhesion process, but further work is required. Specific gene products may be upregulated in response to different environmental niches, including the neutrophil phagosome. We consistently identified thin structures between the B1 bacteria and the mucosal surface (Fig. 5F). However, at this point it is not clear whether these structures are host or bacterially derived products.
Differences in the behaviors of the strains in the hamster may also provide clues concerning the clinical picture of C. difficile disease. Strain B1 was originally isolated from a patient with diarrhea (13), while strain 630 was isolated from a patient with pseudomembranous colitis (34). The level of damage following infection of hamsters with B1 suggests that bacterial factors produced in vivo induce acute responses by the host characterized by high levels of neutrophils and high volumes of fluid secretion. In contrast, 630 appeared to cause significantly less tissue damage, and the infected hamsters remained relatively well for a longer period of time. Previous data have indicated that the severity of disease associated with the 027 ribotypes is a consequence of a greater capacity of the 027 ribotype strains to remain in the vegetative form for longer periods than other strains, and it has been postulated that production of the toxins over this time period could account for the enhanced virulence of these strains (10). However, it is possible that persistence in the gut for longer periods in the vegetative form in a toxin-sensitized gut would result in the pathologies observed in humans. This may help explain why hamsters infected with 027 ribotypes appear to show various levels of mortality with and without diarrhea (25).
In conclusion, we provide evidence that there are significant differences in the pathogenesis of two C. difficile strains within infected hamster tissues, and our findings highlight differences in virulence between these strains. This study supports the value of the hamster as an in vivo model of C. difficile infection and provides evidence that further investment in this system is essential if we are to benefit from this valuable model during development of new interventional and prophylactic treatments for this disease.
Acknowledgments
We acknowledge the help of M. Rupnik with toxinotyping of the B1 strain.
G.D. and D.G. were supported by The Wellcome Trust. Work in Glasgow was supported by Wellcome Trust grant 080860 to G.R.D. J.E. was supported by BBSRC grant BBS/B/09708 to N.F.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 14 September 2009.
REFERENCES
- 1.Barbut, F., B. Gariazzo, L. Bonne, V. Lalande, B. Burghoffer, R. Luiuz, and J.-C. Petit. 2007. Clinical features of Clostridium difficile—associated infections and molecular characterisation of strains: results of a restrospective study, 2000-2004. Infect. Control Hosp. Epidemiol. 28:131-139. [DOI] [PubMed] [Google Scholar]
- 2.Bartlett, J. G., A. B. Onderdonk, R. L. Cisneros, and D. L. Kasper. 1977. Clindamycin associated colitis due to a toxin producing species of Clostridium in hamsters. J. Infect. Dis. 136:701-705. [DOI] [PubMed] [Google Scholar]
- 3.Borriello, S. P., J. M. Ketley, T. M. Mitchell, F. E. Barclay, A. R. Welch, A. B. Price, and J. Stephen. 1987. Clostridium difficile—a spectrum of virulence and analysis of putative virulence determinants in the hamster model of antibiotic-associated colitis. J. Med. Microbiol. 24:53-64. [DOI] [PubMed] [Google Scholar]
- 4.Calabi, E., F. Calabi, A. D. Phillips, and N. Fairweather. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerquetti, M., A. Serafino, A. Sebastianelli, and P. Mastrantonio. 2002. Binding of Clostridium difficile to Caco-2 epithelial cell line and extracellular matrix proteins. FEMS Immunol. Med. Microbiol. 32:211-218. [DOI] [PubMed] [Google Scholar]
- 6.Chen, X., K. Katchar, J. D. Goldsmith, N. Nanthakumar, A. Cheknis, D. N. Gerding, and C. P. Kelly. 2008. A mouse model of Clostridium difficile-associated disease. Gastroneterology 135:1984-1992. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., R. T. Irvin, and K. J. Cheng. 1981. The bacterial glycocalyx in nature and disease. Annu. Rev. Microbiol. 35:299-324. [DOI] [PubMed] [Google Scholar]
- 8.Despande, A., C. Pant, A. Jain, T. G. Fraser, and D. D. Rolston. 2008. Do fluoroquinolones predispose patients to Clostridium difficile associated disease? A review of the evidence. Curr. Med. Res. Opin. 24:329-333. [DOI] [PubMed] [Google Scholar]
- 9.Drudy, D., N. Harnedy, S. Fanning, M. Hannan, and L. Kyne. 2007. Emergence and control of fluoroquinolone-resistant, toxin A-negative, toxin B-positive Clostridium difficile. Infect. Control Hosp. Epidemiol. 28:932-940. [DOI] [PubMed] [Google Scholar]
- 10.Freeman, J., S. D. Baines, K. Saxton, and M. H. Wilcox. 2007. Effect of metronidazole on growth and toxin production by epidemic Clostridium difficile PCR ribotypes 001 and 027 in the human gut model. J. Antimicrob. Chemother. 60:83-91. [DOI] [PubMed] [Google Scholar]
- 11.Giannasca, P. J., Z.-X. Zhang, W.-D. Lei, J. A. Boden, M. A. Giel, T. P. Monath, and W. D. Thomas. 1999. Serum anti-toxin antibodies mediate systemic and mucosal protection from Clostridium difficile disease in hamsters. Infect. Immun. 67:527-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heap, J. T., O. J. Pennington, S. T. Cartman, G. P. Carter, and N. P. Minton. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452-464. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, S., C. R. Clabots, F. V. Linn, M. M. Olson, L. R. Peterson, and D. N. Gerding. 1990. Nosocomial Clostridium difficile colonisation and disease. Lancet 336:97-100. [DOI] [PubMed] [Google Scholar]
- 14.Kim, H., T. V. Riley, M. Kim, C. K. Kim, D. Yong, K. Lee, Y. Chong, and J. W. Park. 2008. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J. Clin. Microbiol. 46:1116-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kink, J. A., and J. A. Williams. 1998. Antibodies to recombinant Clostridium difficile toxins A and B are effective treatment and prevent relapse of C. difficile-associated disease in a hamster model of infection. Infect. Immun. 66:2018-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuijper, E. J., B. Coignard, J. S. Brazier, C. Suetens, D. Drudy, C. Wiuff, H. Pituch, P. Reichert, F. Schneider, A. F. Widmer, K. E. Olsen, F. Allerberger, D. W. Notermans, F. Barbut, M. Delmee, M. H. Wilcox, A. Pearson, B. C. Patel, D. J. D. Brown, R. Frei, T. Akerlund, I. R. Poxton, and P. Tull. 2007. Update of Clostridium difficile-associated disease due to PCR ribotype 027 in Europe. Eur. Surveill. 12:E1-E2. [DOI] [PubMed] [Google Scholar]
- 17.Kyne, L., M. B. Hamel, R. Polavaram, and C. P. Kelly. 2002. Health care costs and mortality associated with nosocomial diarrhoea due to Clostridium difficile. Clin. Infect. Dis. 34:346-353. [DOI] [PubMed] [Google Scholar]
- 18.Larson, H. E., A. B. Price, and S. P. Boriello. 1980. Epidemiology of experimental enterocecitus due to Clostridium difficile. J. Infect. Dis. 142:408-413. [DOI] [PubMed] [Google Scholar]
- 18a.Malick, L. E., and R. B. Wilson. 1975. Modified thiocarbohydrate procedure for scanning electron microscopy: routine use for normal, pathological, or experimental tissues. Stain Technol. 50:265-269. [DOI] [PubMed] [Google Scholar]
- 19.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 20.Merrigan, M. M., S. Sambol, S. Johnson, and D. N. Gerding. 2003. Prevention of fatal Clostridium difficile-associated disease during continuous administration of clindamycin in hamsters. J. Infect. Dis. 188:1922-1927. [DOI] [PubMed] [Google Scholar]
- 21.Naaber, P., R. H. Mikelsaar, S. Salmoinen, and M. Mikelsaar. 1998. Bacterial translocation, intestinal microflora and morphological changes in experimental models of Clostridium difficile infection. J. Med. Microbiol. 47:591-598. [DOI] [PubMed] [Google Scholar]
- 22.Nusrat, A., C. Von Eichel-Streiber, J. R. Turner, P. Verkade, J. L. Madara, and C. A. Parkos. 2001. Clostridium difficile toxins disrupt epithelial barrier function by altering membrane microdomain localization of tight junctions. Infect. Immun. 69:1329-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Brien, J. B., M. S. McCabe, V. Athie-Morales, G. S. A. McDonald, D. B. Eidhin, and D. P. Kelleher. 2005. Passive immunisation of hamsters against Clostridium difficile infection using antibodies to surface layer proteins. FEMS Microbiol. Lett. 246:199-205. [DOI] [PubMed] [Google Scholar]
- 24.O'Connor, J. R., D. Lyras, K. A. Farrow, V. Adams, D. R. Powell, J. Hinds, J. K. Cheung, and J. I. Rood. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 61:1335-1351. [DOI] [PubMed] [Google Scholar]
- 25.Razaq, N., S. Sambol, K. Nagaro, W. Zukowski, A. Cheknis, S. Johnson, and D. N. Gerding. 2007. Infection of hamsters with historic and epidemic BI types of Clostridium difficile. J. Infect. Dis. 196:1813-1817. [DOI] [PubMed] [Google Scholar]
- 26.Sambol, S., M. M. Merrigan, J. K. Tang, S. Johnson, and D. N. Gerding. 2002. Colonisation for the prevention of Clostridium difficile disease in hamsters. J. Infect. Dis. 186:1781-1789. [DOI] [PubMed] [Google Scholar]
- 27.Sambol, S. P., J. K. Tang, M. M. Merrigan, S. Johnson, and D. N. Gerding. 2001. Infection of hamsters with epidemiologically important strains of Clostridium difficile. J. Infect. Dis. 183:1760-1766. [DOI] [PubMed] [Google Scholar]
- 28.Sebaihia, M., B. Wren, P. Mullany, N. Fairweather, N. Minton, R. A. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeño-Tárraga, H. Wang, M. T. G. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 29.Spencer, R. C. 1998. Clinical impact and associated costs of Clostridium difficile associated disease. J. Antimicrob. Chemother. 41:5-12. [DOI] [PubMed] [Google Scholar]
- 30.Stabler, R. A., D. N. Gerding, J. G. Songer, D. Drudy, J. Brazier, H. T. Trinh, A. A. Witney, J. Hinds, and B. W. Wren. 2006. Comparative phylogenomics of Clostridium difficile reveals clade specificity and microevolution of hypervirulent strains. J. Bacteriol. 188:7297-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stubbs, S. L., M. Rupnick, M. Gilbert, J. Brazier, B. I. Duerden, and M. Popoff. 2000. Production of actin specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 32.Valiquette, L., D. E. Low, J. Pepin, and A. Mcgeer. 2004. Clostridium difficile infection in hospitals: a brewing storm. Can. Med. Assoc. J. 171:27-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
- 34.Wust, J., N. M. Sullivan, U. Hardegger, and T. D. Wilkins. 1982. Investigation of an outbreak of antibiotic-associated colitis by various typing methods. J. Clin. Microbiol. 16:1096-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]