Abstract
Rhodococcus equi is a gram-positive facultative intracellular pathogen that can cause severe bronchopneumonia in foals and AIDS patients. Virulence is plasmid regulated and is accompanied by phagosome maturation arrest and host cell necrosis. A replacement mutant in the gene for VapA (virulence-associated protein A), a major virulence factor of R. equi, was tested for its activities during macrophage infection. Early in infection, phagosomes containing the vapA mutant did not fuse with lysosomes and did not stain with the acidotropic fluor LysoTracker similar to those containing virulent wild-type R. equi. However, vapA mutant phagosomes had a lower average pH. Late in infection, phagosomes containing the vapA mutant were as frequently positive for LysoTracker as phagosomes containing plasmid-cured, avirulent bacteria, whereas those with virulent wild-type R. equi were still negative for the fluor. Macrophage necrosis after prolonged infection with virulent bacteria was accompanied by a loss of organelle staining with LysoTracker, suggesting that lysosome proton gradients had collapsed. The vapA mutant still killed the macrophages and yet did not affect the pH of host cell lysosomes. Hence, VapA is not required for host cell necrosis but is required for neutralization of phagosomes and lysosomes or their disruption. This is the first report of an R. equi mutant with altered phagosome biogenesis.
Rhodococcus equi is a nocardioform gram-positive coccobacillus, closely related to mycobacteria, and an important foal pathogen producing severe pyogranulomatous pneumonia in very young horses and tuberculosislike symptoms in AIDS patients (4, 14, 16, 27). R. equi also belongs to the group of facultative intracellular bacteria (9) and interferes with the maturation of its phagosomes in macrophages (5). Phagosomes containing virulent R. equi pass the early phagocytic stages normally, but they neither acidify nor develop into phagolysosomes, making them early-to-late phagosome intermediates (5, 27).
Both abilities to change phagosome maturation and to eventually kill the host cell by necrosis (12) are regulated by a virulence-associated plasmid (VAP) of 80 to 90 kbp that is typical for clinical isolates from foals. This plasmid carries the gene for the immunodominant virulence-associated protein A (VapA), a cell surface protein (20) that is indispensable for multiplication in mouse macrophages and persistence in mouse organs (10). vapA expression is regulated by at least two transcriptional modulators encoded on the VAP: VirR, a LysR-like transcription regulator (2, 17), and Orf8, a two-component response regulator (17). VapA has no known homologues and no specific activity of the protein has been identified.
VapA is an important virulence factor, as judged by the failure of R. equi vapA knockout mutants to multiply in mice or in isolated primary mouse macrophages (10). However, the role of VapA in phagosome development or cytotoxicity is unknown. It was expected that R. equi would, in the absence of only VapA, retain some virulence-related phenotypes, although this mutant is overall avirulent. Knowing virulence-related phenotypes that are present in a vapA mutant would help to assign VapA a role in pathogenesis. To define the role of VapA in intramacrophage multiplication, we sought to determine which of the intramacrophage features of wild-type virulent R. equi would be altered in a vapA deletion mutant, in particular, the degree of phagosome acidification or phagosome-lysosome fusion, and whether the cytotoxic potential of the bacteria would be affected.
MATERIALS AND METHODS
Bacterial strains and mammalian cells, growth conditions, and transformation.
R. equi 103+, an isolate from a pneumonic foal (Department of Pathobiology, University of Guelph, Guelph, Ontario, Canada), its cured derivative 103− (5), and R. equi 103+ with a gene replacement mutation in Orf8 (17) have been described. R. equi was grown overnight in brain heart infusion broth at 37°C with vigorous shaking to an absorbance (600 nm) of 1.5 to 2.5, Escherichia coli was grown at 37°C in Luria-Bertani (LB) broth. Bacteria were heat killed by incubating 100-μl aliquots in 1.5-ml microfuge tubes for 15 min at 85°C. Antibiotics were used at the following concentrations: apramycin, 50 μg/ml; hygromycin, 150 μg/ml; and kanamycin, 200 μg/ml. Expression of vapA was analyzed by immunoblotting with monoclonal antibodies to VapA (Mab10G5, kindly provided by Shinji Takai, Kitasato University, Aomori, Japan [21], or Mab103 [24]) using the same numbers of rhodococci heated to 95°C for 10 min in denaturing sample buffer, followed by the removal of debris by centrifugation before applying the samples to electrophoresis. Electroporation was performed as described previously (19). The macrophagelike cell line J774E was used as described previously (5).
Plasmids and DNA manipulations.
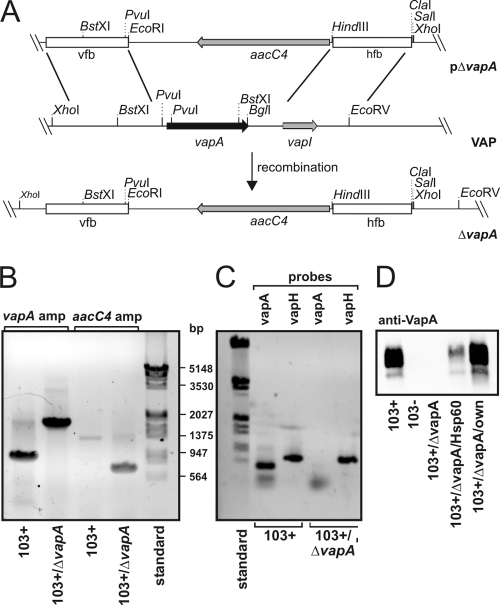
Construction of the vapA gene replacement vector is described in Fig. 1. The DNA primers used are listed in Table 1. Endogenous virulence-associated plasmid was purified as described previously (23). Deletion of the vapA gene was done by following a published strategy (17). Flanking regions of vapA were amplified by PCR for the construction of a gene replacement plasmid. Primer pairs vfB-1/vfB-2 were used to synthesize a 612-bp upstream flanking region, primer pairs hfB-1/hfB-2 to amplify a 586-bp downstream flanking region. The upstream flanking region was cloned through the primers' XbaI and EcoRI sites into pBluescript. The resulting plasmid pBlue-vfB was cut with HindIII and ClaI and ligated with the likewise-cut downstream flanking region to create pBlue-vfB-hfB. A 1.5-kbp PstI fragment from pVK173T (apramycin and hygromycin resistance, shuttle plasmid [15]) containing an apramycin resistance gene was blunt ended with Klenow polymerase and cloned into the EcoRV site of pBlue-vfB-hfB, yielding pBlue-vfB-hfB-Apr. Finally, the 2.7-kbp ClaI/XbaI fragment from pBlue-vfB-hfB-Apr containing the flanking regions and the apramycin resistance marker was blunt end cloned into the SmaI site of pAPVLacZ (E. coli vector; apramycin and hygromycin resistance [17]) to construct the suicide vector pΔvapA (Fig. 1). Competent R. equi 103+ was electroporated with heat-denatured suicide vector DNA and plated for blue or white selection. Five blue transformants were recovered. PCR-based analysis of the clones confirmed a single crossover integration of the vector. A single clone was cultivated further to promote a second crossover event. After the fifth passage, three white colonies were identified of which one proved to be a double-crossover mutant. The primer pair vapA probes 1 and 2 (Table 1) was used for integration analysis.
FIG. 1.
Genetic strategy to create R. equi 103+/ΔvapA. (A) Schematic representation of the creation of R. equi 103+/ΔvapA. Briefly, linearized plasmid pΔvapA was electroporated into 103+ and, after double crossover at the vfb and hfb sites, the entire vapA gene region on the virulence plasmid was replaced by the aacC4 apramycin resistance gene. “VAP” denotes the situation as is found with the relevant section of the virulence-associated plasmid, and ΔvapA describes the situation in the replacement mutant. (B) Agarose gel stained with ethidium bromide. Plasmid DNA isolated from 103+ and from 103+/ΔvapA was used as a template for PCRs with primers to up- and downstream regions of the vapA gene (vapA probes 1 and 2) yielding fragments of 900 bp in the presence of vapA and 1,800 bp in its absence (corresponding to the aacC4 gene with flanking regions of vapA). Primer pair aacC4-F/aacC4-R was used to amplify a fragment of 743 bp in the open reading frame of the apramycin resistance gene with the nucleotide sequence analyzed from pVKT173. A molecular weight marker standard (bp) is shown on the right. (C) The virulence plasmids from 103+ or 103+/ΔvapA were used in PCRs with primers that amplify either the vapA gene as described above or the vapH gene (oligonucleotides vapH-F/-R), which is also located on the VAP. Although vapA is present only in 103+, vapH is present in the wild type and mutant, demonstrating the presence of the VAP in either strain. (D) Immunoblot of R. equi proteins developed with a monoclonal antibody to VapA. Equivalent cell numbers of R. equi overnight cultures were used.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) | Restriction site | Positiona |
|---|---|---|---|
| vapA-pc-1 | CTAGCTAGCTAGCCGCGAAGGCGATCGAAGGGC | NheI | 12503-12523 |
| vapA-pc-2 | CGGGTACCCCTCGCAGCCTGCATGTTTCTGG | KpnI | 13225-13247 |
| vapA probe-1 | GCGATCGCAGCCACAGCCGT | 12576-12595 | |
| vapA probe-2 | GCGTTGTGCCAGCTACCAGAG | 13091-13111 | |
| vapA-endo-fw | CTAGTCTAGACTAGTGCGTCGCGCAGTACGCCGC | XbaI | 12001-12020 |
| vapA-endo-rv | CCATCGATGGCCTCTTCCTCGACCGTCGTCATCAATC | ClaI | 13597-13623 |
| vapH-F | GGACGAGACGAGAGGGGTGC | 7343-7362 | |
| vapH-R | GGATTCGAAAGCAGGTCGAGCG | 8016-8037 | |
| aacC4-F | CGGTGGAGTGCAATGTCGTGC | NA | |
| aacC4-R | GCGGATGCAGGAAGATCAACGG | NA | |
| vfB-1 | GCTCTAGAGGCCGTCGAAGTTGCAAGCG | XbaI | 11924-11943 |
| vfB-2 | CGGAATTCCCTTTCGGACGTCGCCCTTCG | EcoRI | 12516-12536 |
| hfB-1 | CCCAAGCTTCGCGGTCCAGAAACATGCAGG | HindIII | 13219-13239 |
| hfB-2 | CCATCGATGCGCCGGATTGGACGTGCC | ClaI | 13787-13805 |
pSMT3-vapA, expressing vapA from a constitutive mycobacterial hsp60 promoter (shuttle vector; hygromycin resistance [7]), was constructed by PCR of the 103+ virulence plasmid DNA with the primers vapA-pc-1 and vapA-pc-2 (Table 1) containing additional sites for NheI and KpnI, respectively, resulting in a fragment that contained only the vapA gene without promoter regions. The PCR product was digested with NheI and KpnI, respectively, and was cloned into the corresponding pSMT3 sites, yielding pSMT3-vapA.
To express VapA, cloned in pSMT3, from its own promoter, a plasmid was constructed by PCR amplification of a 1.6-kb region from the 103+ virulence plasmid containing the vapA gene using the primers vapA-endo-fw and vapA-endo-rv (Table 1). This region has previously been used for complementation of a vapA deletion strain (10). The PCR product was ClaI/XbaI digested and cloned into the corresponding pSMT3 restriction sites. Propagated in 103+/ΔvapA, this plasmid yielded the “103+/ΔvapA endog. VapA,” or “103+/ΔvapA/own,” strain.
All restriction and DNA-modifying enzymes were from MBI Fermentas (St. Leon-Rot, Germany). DNA fragments separated in agarose gels were isolated and purified with a the Qiaquick gel extraction kit or a QIAEX II gel extraction kit (Qiagen, Hilden, Germany). Plasmid DNA was purified from E. coli using Qiagen kits according to the manufacturer's instructions. DNA amplification was performed by using Pfu polymerase (MBI Fermentas) and the Expand Long Template system (Roche, Mannheim, Germany).
Quantification of intracellular growth, cytotoxicity, intraphagosomal pH, and fluorescence resonance energy transfer (FRET) analysis of phagolysosome formation.
This was performed as described previously (19). Phagolysosome formation was additionally determined microscopically. J774E macrophages on coverslips were incubated with 50 μg of bovine serum albumin rhodamine (BSA-rhodamine)/ml prepared as described by Sydor et al. (19) or with 30 μg ovalbumin-Texas Red (Invitrogen, Karlsruhe, Germany)/ml overnight. Cells were rinsed with warm phosphate-buffered saline (PBS) three times, fresh medium was added, and the fluid phase tracer was chased into lysosomes for 2 h. Macrophages were infected with R. equi strains labeled with ATTO488 as described previously (19) at a multiplicity of infection (MOI) of 20 for 20 min at 37°C. Nonphagocytosed bacteria were removed by two rinses with warm PBS, and the cells were chased in fresh medium for 2 h before they were fixed and quenched with 2% formaldehyde-2.5% glutaraldehyde in PBS and 1 mg of sodium borohydride/ml of PBS (BSA-rhodamine) or 3% formaldehyde and 50 mM ammonium chloride in PBS (ovalbumin-Texas Red). Samples were prepared for fluorescence microscopy, and the percentages of bacteria colocalizing with the fluorescent tracers were determined.
Determination of intracellular survival.
For determination of intracellular survival of bacteria, J774E cells were seeded into 24-well plates at 8 × 104 cells per well 2 days before infection. Infection was with R. equi at an MOI of 0.3 for 30 min at 37°C. Samples were rinsed with PBS twice, and fresh medium containing 3 μg of vancomycin/ml was added. At 2 and 24 h postinfection, medium was removed, and cells were lysed in 1 ml of PBS-0.1% Triton X-100. Serial dilutions were plated onto agar plates containing a Luria broth medium diluted 1:1 with water. The plates were incubated for 30 h at 30°C, and the CFU were counted. The mean CFU for each sample and time point were determined. Plates with less than 10 CFU were ignored unless they were the only plates with colonies.
Visualization of acidic compartments with LysoTracker.
For quantification of acidified compartments, J774E macrophages were infected at an MOI of 1 for 30 min, rinsed twice with PBS, and chased in fresh medium containing 3 μg of vancomycin/ml. At 1.5 or 23.5 h postinfection, medium containing LysoTracker Red DND-99 (product number L7528, 1:10,000; Invitrogen) and SYTO13 (1:1,500; Invitrogen) was added with further incubation for 30 min. Samples were rinsed four times, fixed with 3% formaldehyde in PBS, and mounted onto slides. The samples were analyzed immediately, determining the percentage of infected macrophages with (nonphagosome) LysoTracker-positive compartments.
Statistic analysis.
Data are expressed as means and standard deviations and were analyzed by the two-tailed unpaired Student t test.
RESULTS
To address the question of the virulence-related phenotypes that are present in a vapA mutant, we constructed such mutant by double crossover, exchanging vapA for aacC4, an apramycin resistance gene (Fig. 1). The mutant was denoted 103+/ΔvapA to indicate that the strain was positive for the virulence plasmid but that the vapA gene had been completely removed. Conversely, 103+ denotes a wild-type isolate positive for the virulence-associated plasmid, whereas 103− is the otherwise isogenic, cured derivative.
VapA expression is required early for diversion of phagosome maturation.
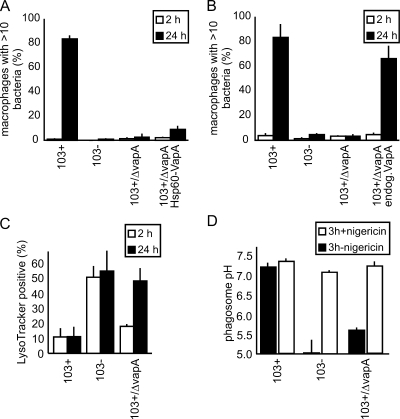
Intramacrophage bacterial numbers were quantified microscopically, and macrophages with more than 10 bacteria were used as a measure for robust intracellular multiplication (3). As expected (10), 103+/ΔvapA did not multiply in macrophages (Fig. 2A). However, expression from the constitutive Hsp60 promoter of pSMT3 led to the production of only approximately one-fifth the amount of VapA produced in 103+, and only a small proportion of the multiplication defect was reversed (Fig. 1D). Expression of vapA from its own promoter in a 103+/ΔvapA background led to approximately wild-type expression of vapA (Fig. 1D) and rescued the multiplication defect in macrophages (Fig. 2B).
FIG. 2.
Characterization of R. equi 103+/ΔvapA virulence phenotypes. (A) Multiplication in J774E of 103+, 103−, 103+/ΔvapA, and 103+/ΔvapA complemented with vapA constitutively expressed from hsp60 promoter (103+/ΔvapA Hsp60-VapA). The percentages of infected macrophages with more than 10 bacteria are indicated. Quantitation was done microscopically, based on staining of bacterial DNA with SYTO13. The data represent means and standard deviations from three independent experiments. (B) Same as in panel A, but complementation was done using the endogenous vapA promoter and the vapA gene (103+/ΔvapA endog.VapA). (C) Qualitative determination of phagosome acidification. J774E were infected as in panel A and, after 2 or 24 h, the samples were stained with red LysoTracker and green fluorescent SYTO13. Colocalization between LysoTracker- and green-labeled bacteria was quantified by using confocal laser scanning microscopy. The percentages of phagosomes positive for LysoTracker are indicated. The data shown are the means and standard deviations of three experiments. In panels A, B, and C, at least 50 infected macrophages were analyzed in each experiment per time and sample type. (D) Quantitative determination of average phagosome pH at 3 h of infection (▪). The addition of nigericin collapses pH gradients between phagosome and cytosol and serves as a control for the calibration process in that the pH should be that of the external buffer (∼7.3) after addition (□). Nigericin was added at 3 h of infection (first pH determination), followed by a 20-min incubation period (redetermination of pH [□]).
Phagosome trafficking was analyzed by quantifying the percentage of phagosomes positive for LysoTracker, an indicator of strongly acidified compartments. At 2 h of infection, most 103+/ΔvapA-containing phagosomes were negative for LysoTracker but, by 24 h, as many phagosomes were positive as those containing 103− (Fig. 2C). To quantify the Rhodococcus-containing vacuole (RCV) pH, we used a calibrated fluorescent microplate assay which quantifies the average pH in a large cohort of phagosomes (19). The pH was 7.18 for 103+, 5.02 for 103−, and 5.61 for 103+/ΔvapA at 3 h postinfection (Fig. 2D). Addition of nigericin, a K+/H+ antiporter at the end of the measurement, immediately brought the phagosomal pH to the level of the externally adjusted pH (Fig. 2D), confirming correct calibration.
Phagolysosome formation was quantified by laser scanning confocal microscopy as the percentage of ATTO488-labeled bacteria colocalizing with lysosomal BSA rhodamine. At 2 h of infection, phagosomes containing 103+/ΔvapA were as frequently lysosomal as those containing wild-type R. equi, whereas almost all phagosomes containing heat-killed bacteria acquired lysosomal contents (Fig. 3A and C). Similar results were obtained with a FRET assay of phagolysosome formation (Fig. 3B).
FIG. 3.
Quantification of phagolysosome formation. (A) J774E macrophage lysosomes were preloaded with BSA-rhodamine (BSA-rhod.) and infected with ATTO488-labeled bacteria for 20 min, and the infection was chased for 2 h before fixation. Colocalization of bacteria with the fluorescent tracer was quantified by using confocal laser scanning microscopy. “103+/ΔT”, phagosomes containing bacteria that were heat-killed (15 min, 85°C) before phagocytosis. The data shown are the means and standard deviations of three experiments with at least 50 infected macrophages analyzed per time and sample type. (B) Phagolysosome formation was determined by using a FRET assay. Phagolyososme formation with 103+ was standardized as “1”, and higher indices indicate more fusion. RFU, relative fluorescence units. (C) Representative microscopic fields of the experiments in panel A. Optical overlays are shown with some transmitted light to visualize the cell outlines. Open and closed arrowheads point to phagosomes without and with lysosome colocalization (yellow color due to red-green overlay), respectively.
Cytotoxicity of R. equi infection does not require VapA but correlates with intracellular viability of bacteria.
Cytotoxicity for J774E macrophages was investigated at 24 h of infection. At this time of infection 103+/ΔvapA was as cytotoxic as 103+ (Fig. 4A), although mutant bacteria did not multiply (Fig. 2). We had previously shown that killing of virulent R. equi by either of five different treatments completely abolishes their cytotoxic potential (12). To analyze whether viability of intracellular bacteria was a critical parameter, we tested their ability to form colonies on nutrient agar after 24 h of infection (Fig. 4B). By this time, 103− bacteria had nearly all been killed, whereas ca. 50% of the initially intracellular 103+/ΔvapA bacteria were alive (Fig. 4B), demonstrating their increased survival.
FIG. 4.
Cytotoxicity for and survival in macrophages. (A) Cytotoxicity of J774E infection with R. equi, quantified by using a lactate dehydrogenase (LDH) release assay. Cytotoxicity is indicated as the percent of LDH release compared to the LDH release by lysing macrophages with the detergent Triton X-100. Infection was done at an MOI of 30 for 1 h, followed by a 24-h chase. (B) J774E macrophages were infected with 103−, 103+/ΔvapA, or 103+/Δorf8. At 2 and 24 h of infection, macrophages were lysed and plated on nutrient agar. The numbers of CFU were quantified after 30 h of incubation at 30°C and were normalized for the 2-h value corresponding to number of ingested bacteria. The data shown are the means and standard deviations of five independent experiments. (C) J774E macrophages were infected at an MOI of 1 for 30 min and chased for 1.5 or 23.5 h before the addition of medium containing LysoTracker and Syto13 for 30 min, rinsing, and fixation. Macrophages that contained at least one nonphagosomal LysoTracker-positive compartment were counted as positive, and all macrophages devoid of any LysoTracker-positive compartment other than phagosomes were counted as negative. Analysis was done by using confocal laser scanning microscopy, and the data shown are the means and standard deviations of three independent experiments with 50 infected macrophages analyzed for each sample type in each experiment. 103+/ΔvapAc, 103+/ΔvapA complemented with vapA in pSMT3 expressed from its own promoter. (D) Representative micrographs of the experiments quantified in panel C. Size bars, 10 μm. Asterisks mark macrophage nuclei. White arrowheads point to bacteria in macrophages without any LysoTracker-positive compartments, and open arrowheads point to bacteria in macrophages that still stain with LysoTracker.
To further analyze this, we tested a gene replacement mutant in orf8. orf8 codes for a two-component response regulator required for vapA expression, and it may also be required for regulation of additional factors on the virulence plasmid or chromosome. The cytotoxicity of 103+/Δorf8 bacteria was also unaltered (Fig. 4A), and ca. 70% survived a 24 h infection of macrophages (Fig. 4B).
Disappearance of functional lysosomes is not a central factor for cytotoxicity.
Hietala and Ardans (8) and Zink et al. (28) concurrently described the lack of fusion of R. equi phagosomes with lysosomes in horse alveolar macrophages. These authors also observed a decreased number of ferritin-labeled lysosomes in infected macrophages, describing it as a degranulation phenomenon. In the present study, we quantified the numbers of acidic compartments (positive for LysoTracker) in macrophages infected with 103+, 103−, 103+/ΔvapA, or 103+/ΔvapA complemented with the vapA gene expressed from its own promoter. After 24 h of infection with virulent R. equi, only a few infected macrophages had LysoTracker-positive compartments, whereas many more macrophages with acidic compartments could be seen after infection with avirulent or mutant bacteria. The complemented strain behaved like wild-type virulent bacteria (Fig. 4C and D).
DISCUSSION
We have investigated the effects of vapA deletion on (i) interference with normal phagosome acidification, (ii) phagolysosome formation in murine macrophages, (iii) intramacrophage bacterial multiplication, and on (iv) cytotoxic effects of the infection, and (v) organelle acidity.
A central determinant and indication of phagosome maturation is phagosome pH: the pH drops from 7.2 to ca. 4.0 to 5.5 between the stage of an early phagosome (immediately after formation) and a mature phagolysosome (∼60 min after formation [6]). At 2 h of infection, most phagosomes containing 103+/ΔvapA were negative for LysoTracker, whereas after 24 h of infection they were positive, which suggested at first that, at 2 h of infection, there was only little difference between the pHs of RCVs containing 103+ or 103+/ΔvapA. However, calibrated phagosome pH measurements revealed that there was indeed some acidification of 103+/ΔvapA containing phagosomes already within the first 3 h of infection, albeit the average pH was higher than in 103− phagosomes. This apparent contradiction could be resolved by analyzing the pH required for the lysosomotropic fluor, LysoTracker, to label a compartment. Literature searches reveal that macrophage phagosomes containing pathogenic mycobacteria (pH ∼6.3) do not stain with LysoTracker (18, 25, 26), whereas more acidic compartments do (1). These data suggest that there is a cutoff pH of ∼6.0 for LysoTracker to stain. We propose that in the case of RCVs containing 103+/ΔvapA, most phagosomes were at pH ∼6.0 at 2 h of infection (i.e., LysoTracker negative) and that some were more acidic than that, whereas wild-type bacteria containing RCVs were mostly at pH ∼7.2. At this time, phagolysosome formation was barely increased with mutant phagosomes compared to the wild type. Together, we interpret these data as indication that, at 2 h of infection, most of the 103+/ΔvapA-containing RCVs were in a transitional state from a late endosome-like to a phagolysosomal compartment. After 24 h, this transition was largely finished and the 103+/ΔvapA-containing RCVs had matured into fully acidic, likely phagolysosomal, compartments. Therefore, VapA is implicated in early phagosome development, and its lack increases phagosome acidification and slightly increases phagolysosome formation. At 24 h of infection, the effects of the mutation became most apparent when 103+/ΔvapA had not multiplied, and its phagosomes were as acidified as those containing the plasmidless strain.
VapA is strongly expressed in 103+ and may actually possess the strongest promoter system in R. equi, as judged by the results of a genetically unbiased promoter search (17). One unexpected finding from the present study was that the expression of vapA on a much lower level did not at all complement the vapA mutant with respect to multiplication in macrophages. Two straightforward interpretations are that either the expression level was too low or that the timing was not correct, i.e., that there are times when the presence of VapA is absolutely required for multiplication and, at others, it may reduce multiplication. We further tested this by expressing vapA in 103+, i.e., a strain containing an endogenous vapA plasmid plus a constitutively expressed copy, but this did not change multiplication (data not shown), suggesting that it is not a timing problem, but that large quantities of VapA are required. VapA is crucially important for virulence in R. equi, possibly as a central scaffold protein. The prolonged half-life of vapA RNA compared to vapC and vapD RNA (2) supports the finding with the mycobacterial Hsp60 promoter construct that the amount of VapA present is important.
We have shown previously (12) that the cytotoxicity of R. equi for macrophages is upregulated in bacteria possessing the virulence plasmid. In agreement with this, 103− caused only minor cytotoxicity. Surprisingly, the cytotoxicity of infection with 103+/ΔvapA or 103+/Δorf8 was as high as with 103+. Based on nutrient agar plating experiments, we propose that intracellular viability of bacteria rather than their actual multiplication is required for cytotoxicity. This view is supported by our previous observations that killing of R. equi by either heat, UV irradiation, antibiotic treatments, chemical membrane permeabilization, or formaldehyde fixation before phagocytosis completely abolished cytotoxicity of an infection, clearly demonstrating that it is not only one defined kind of killing but loss of viability in general that annihilates the cytotoxic potential (12). In addition, some VAP-less isolates of R. equi can be cytotoxic (unpublished data). In summary, these data are most simply explained by chromosomally encoded factors contributing substantially to the cytotoxic effects, whereas VAP genes promote them by increasing intracellular vitality.
LysoTracker-positive compartments (late endosomes and lysosomes) were detected in macrophages infected with 103− or 103+/ΔvapA but not in those infected with 103+. Because infection is followed by macrophage necrosis (12), degranulation of lysosome contents into the macrophage cytoplasm, followed by hydrolysis of cytoplasmic macromolecules could be the major mechanism of cytotoxicity, as proposed by Hietala and Ardans (8).
However, we were surprised to find that 103+/ΔvapA had full cytotoxic potential, and yet did not have an effect on the cellular staining pattern with LysoTracker. Since protons are the smallest “ions” known, loss of a pH gradient across a lysosome membrane would be expected to be the first step in its disintegration. Our data now clearly indicate that disappearance of functional lysosomes is not directly coupled to cytotoxicity and may play a completely different role in pathogenicity of R. equi infection.
The precise molecular role of VapA is still enigmatic, as VapA amino acid sequence analysis did not yield any significant homologues except for the other family members (data not shown [reviewed in references 11 and 13]). Its surface expression on R. equi (22), as well as the cotranscription with other vap genes as part of an operon (2), suggests that it may be important as a central scaffold of a protein complex that may interact with host macrophage structures. A recent comparison of the VapA virulence plasmid from foals with that of the VapB virulence plasmid from swine (11) has identified common elements of the pathogenicity island of the virulence plasmids of R. equi that are likely to be crucial. Speculatively, VapA is likely involved as part of a protein complex in the modulation of the bacterium-phagosome interaction in a host-specific manner. The lack of such effector functions could lead, in a matter of hours to days, to a failure to prevent phagolysosome formation and to normalization of phagosome trafficking. Whether VapA is directly or indirectly involved in maintenance of acidic lysosomes remains to be investigated.
Acknowledgments
We thank Sabine Spürck, Renate Ollig, and Vivian Nicholson for expert technical support; Shinji Takai (Kitasato University) for antibody 10G5; and Julian Davies (University of British Columbia, Vancouver, British Columbia, Canada) for pVK173T.
This study was supported by grants from the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 670, Cell-autonomous Defense) to A.H., by The Natural Sciences and Engineering Research Council of Canada to J.F.P., and by a fellowship of the Studienstiftung des Deutschen Volkes to K.V.B.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 21 September 2009.
REFERENCES
- 1.Anes, E., M. P. Kuhnel, E. Bos, J. Moniz-Pereira, A. Habermann, and G. Griffiths. 2003. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat. Cell Biol. 5:793-802. [DOI] [PubMed] [Google Scholar]
- 2.Byrne, G. A., D. A. Russell, X. Chen, and W. G. Meijer. 2007. Transcriptional regulation of the virR operon of the intracellular pathogen Rhodococcus equi. J. Bacteriol. 189:5082-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darrah, P. A., M. K. Hondalus, Q. Chen, H. Ischiropoulos, and D. M. Mosser. 2000. Cooperation between reactive oxygen and nitrogen intermediates in killing of Rhodococcus equi by activated macrophages. Infect. Immun. 68:3587-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donisi, A., M. G. Suardi, S. Casari, M. Longo, G. P. Cadeo, and G. Carosi. 1996. Rhodococcus equi infection in HIV-infected patients. Aids 10:359-362. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Mora, E., M. Polidori, A. Luhrmann, U. E. Schaible, and A. Haas. 2005. Maturation of Rhodococcus equi-containing vacuoles is arrested after completion of the early endosome stage. Traffic 6:635-653. [DOI] [PubMed] [Google Scholar]
- 6.Haas, A. 2007. The phagosome: compartment with a license to kill. Traffic 8:311-330. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann, J. L., P. O'Gaora, A. Gallagher, J. E. Thole, and D. B. Young. 1996. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19-kDa antigen from Mycobacterium tuberculosis. EMBO J. 15:3547-3554. [PMC free article] [PubMed] [Google Scholar]
- 8.Hietala, S. K., and A. A. Ardans. 1987. Interaction of Rhodococcus equi with phagocytic cells from R. equi-exposed and non-exposed foals. Vet. Microbiol. 14:307-320. [DOI] [PubMed] [Google Scholar]
- 9.Hondalus, M. K., and D. M. Mosser. 1994. Survival and replication of Rhodococcus equi in macrophages. Infect. Immun. 62:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain, S., B. R. Bloom, and M. K. Hondalus. 2003. Deletion of vapA encoding virulence associated protein A attenuates the intracellular actinomycete Rhodococcus equi. Mol. Microbiol. 50:115-128. [DOI] [PubMed] [Google Scholar]
- 11.Letek, M., A. A. Ocampo-Sosa, M. Sanders, U. Fogarty, T. Buckley, D. P. Leadon, P. Gonzalez, M. Scortti, W. G. Meijer, J. Parkhill, S. Bentley, and J. A. Vazquez-Boland. 2008. Evolution of the Rhodococcus equi vap pathogenicity island seen through comparison of host-associated vapA and vapB virulence plasmids. J. Bacteriol. 190:5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luhrmann, A., N. Mauder, T. Sydor, E. Fernandez-Mora, J. Schulze-Luehrmann, S. Takai, and A. Haas. 2004. Necrotic death of Rhodococcus equi-infected macrophages is regulated by virulence-associated plasmids. Infect. Immun. 72:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meijer, W. G., and J. F. Prescott. 2004. Rhodococcus equi. Vet. Res. 35:383-396. [DOI] [PubMed] [Google Scholar]
- 14.Muscatello, G., D. P. Leadon, M. Klayt, A. Ocampo-Sosa, D. A. Lewis, U. Fogarty, T. Buckley, J. R. Gilkerson, W. G. Meijer, and J. A. Vazquez-Boland. 2007. Rhodococcus equi infection in foals: the science of ‘rattles’. Equine Vet. J. 39:470-478. [DOI] [PubMed] [Google Scholar]
- 15.Paget, E., and J. Davies. 1996. Apramycin resistance as a selective marker for gene transfer in mycobacteria. J. Bacteriol. 178:6357-6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prescott, J. F. 1991. Rhodococcus equi: an animal and human pathogen. Clin. Microbiol. Rev. 4:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ren, J., and J. F. Prescott. 2004. The effect of mutation on Rhodococcus equi virulence plasmid gene expression and mouse virulence. Vet. Microbiol. 103:219-230. [DOI] [PubMed] [Google Scholar]
- 18.Schaible, U. E., S. Sturgill-Koszycki, P. H. Schlesinger, and D. G. Russell. 1998. Cytokine activation leads to acidification and increases maturation of Mycobacterium avium-containing phagosomes in murine macrophages. J. Immunol. 160:1290-1296. [PubMed] [Google Scholar]
- 19.Sydor, T., K. von Bargen, U. Becken, S. Spuerck, V. M. Nicholson, J. F. Prescott, and A. Haas. 2008. A mycolyl transferase mutant of Rhodococcus equi lacking capsule integrity is fully virulent. Vet. Microbiol. 128:327-341. [DOI] [PubMed] [Google Scholar]
- 20.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Takamatsu, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuda, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takai, S., M. Iie, C. Kobayashi, T. Morishita, T. Nishio, T. Ishida, T. Fujimura, Y. Sasaki, and S. Tsubaki. 1993. Monoclonal antibody specific to virulence-associated 15- to 17-kilodalton antigens of Rhodococcus equi. J. Clin. Microbiol. 31:2780-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takai, S., M. Iie, Y. Watanabe, S. Tsubaki, and T. Sekizaki. 1992. Virulence-associated 15- to 17-kilodalton antigens in Rhodococcus equi: temperature-dependent expression and location of the antigens. Infect. Immun. 60:2995-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takai, S., T. Sekizaki, T. Ozawa, T. Sugawara, Y. Watanabe, and S. Tsubaki. 1991. Association between a large plasmid and 15- to 17-kilodalton antigens in virulent Rhodococcus equi. Infect. Immun. 59:4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan, C., J. F. Prescott, M. C. Patterson, and V. M. Nicholson. 1995. Molecular characterization of a lipid-modified virulence-associated protein of Rhodococcus equi and its potential in protective immunity. Can. J. Vet. Res. 59:51-59. [PMC free article] [PubMed] [Google Scholar]
- 25.Tan, T., W. L. Lee, D. C. Alexander, S. Grinstein, and J. Liu. 2006. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol. 8:1417-1429. [DOI] [PubMed] [Google Scholar]
- 26.Via, L. E., R. A. Fratti, M. McFalone, E. Pagan-Ramos, D. Deretic, and V. Deretic. 1998. Effects of cytokines on mycobacterial phagosome maturation. J. Cell Sci. 111:897-905. [DOI] [PubMed] [Google Scholar]
- 27.von Bargen, K., and A. Haas. 2009. Molecular and infection biology of the horse pathogen Rhodococcus equi. FEMS Microbiol. Rev. 33:870-891. [DOI] [PubMed] [Google Scholar]
- 28.Zink, M. C., J. A. Yager, J. F. Prescott, and M. A. Fernando. 1987. Electron microscopic investigation of intracellular events after ingestion of Rhodococcus equi by foal alveolar macrophages. Vet. Microbiol. 14:295-305. [DOI] [PubMed] [Google Scholar]