Abstract
Leptospirosis is a global public health problem, primarily in the tropical developing world. The pathogenic mechanisms of the causative agents, several members of the genus Leptospira, have been underinvestigated. The exception to this trend has been the demonstration of the binding of pathogenic leptospires to the extracellular matrix (ECM) and its components. In this work, interactions of Leptospira interrogans bacteria with mammalian cells, rather than the ECM, were examined. The bacteria bound more efficiently to the cells than to the ECM, and a portion of this cell-binding activity was attributable to attachment to glycosaminoglycan (GAG) chains of proteoglycans (PGs). Chondroitin sulfate B PGs appeared to be the primary targets of L. interrogans attachment, while heparan sulfate PGs were much less important. Inhibition of GAG/PG-mediated attachment resulted in partial inhibition of bacterial attachment, suggesting that additional receptors for L. interrogans await identification. GAG binding may participate in the pathogenesis of leptospirosis within the host animal. In addition, because GAGs are expressed on the luminal aspects of epithelial cells in the proximal tubules of the kidneys, this activity may play a role in targeting the bacteria to this critical site. Because GAGs are shed in the urine, GAG binding may also be important for transmission to new hosts through the environment.
Leptospirosis is an important zoonotic disease caused by spirochetes of the genus Leptospira. This genus includes free-living nonpathogenic species, as well as pathogenic species, but the essential elements of biology and infection are thought to be similar among most of the pathogens. Leptospirosis has emerged as the most widespread zoonotic disease worldwide (8). The disease in humans varies from a self-limited flu-like illness to an acute life-threatening infection (Weil's disease) with pulmonary hemorrhage, myocarditis, and kidney and liver failure. Virtually every species of mammal, both domesticated and wild, can serve as carriers of the disease, harboring the spirochete in the proximal convoluted tubules of the kidneys and chronically excreting Leptospira through the urine. Humans acquire the disease directly through exposure to urine, indirectly through exposure to fresh water sources contaminated by urine, or through occupational exposure to contaminated tissues and body fluids (15). Leptospira species enter the body through mucous membranes of the eyes, nose, or throat and via cuts or abrasions on the skin. The severity of disease in humans varies with the Leptospira species and serovar involved and the age, health, and immune status of the patient.
Adhesion to host cells and the extracellular matrix (ECM) is a critical feature in the infectious process for virtually all pathogens that require active interaction with the host to cause disease. Adhesion is likely to be critical for Leptospira as well, yet the adhesion mechanisms that Leptospira uses during infection have not been as thoroughly studied as those of many other bacterial pathogens. It is likely that, similar to Borrelia burgdorferi, another pathogenic spirochete that can disseminate widely and chronically infect mammalian hosts, pathogenic Leptospira species bind to multiple receptors on host cells and in the ECM to establish and maintain infection. Several groups have investigated the adhesion of Leptospira interrogans to endothelial, fibroblast, kidney epithelial, and monocyte/macrophage cell lines cultured in vitro (3, 22, 30, 32, 37, 38, 40). The majority of these studies have shown more efficient adherence and/or entry of virulent strains than of avirulent or nonpathogenic (saprophytic) strains. Migration through MDCK cell layers was also associated with an infectious rather than a saprophytic Leptospira strain (5). In one study, adherence was decreased following pretreatment of cell monolayers with proteases but there was no statistically significant change in adherence when cells were pretreated with neuraminidase, sodium metaperiodate, or lipase (37). Most studies of Leptospira adherence, however, have focused on the attachment of the bacteria to ECM components. The adherence of infectious strains of Leptospira to the ECM components collagen type I, fibronectin, and laminin has been documented, and in some cases, one or more bacterial proteins that bind to these substrates have been identified (2, 4, 7, 9, 20, 23, 36). For example, L. interrogans serovar Copenhageni binds to the ECM proteins fibronectin, laminin, and collagen type IV and the plasma protein fibrinogen; adherence to these proteins is mediated largely by LigA and LigB, particularly under physiologically relevant conditions (9).
The Lig proteins are members of the bacterial immunoglobulin (Ig)-like family of proteins and were identified by screening a leptospiral gene expression library with convalescent-phase sera from human leptospirosis patients (27, 33). Physiologic osmolarity has been found to be a powerful inducer of the expression of the Lig proteins, as well as a number of additional proteins (28, 29), and to increase leptospiral adhesion to the ECM proteins to which LigA and LigB bind. A ligB knockout strain of L. interrogans, however, retains infectivity and virulence (11), suggesting that the technically challenging generation of a ligA ligB double mutant may be required to assess the roles of the Lig proteins during infection. Previous studies had identified a 36-kDa fibronectin-binding protein with properties different from the Lig proteins (31). In addition, a leptospiral outer membrane lipoprotein that binds both laminin and factor H has been described elsewhere (4, 39); further work demonstrated that this protein is a member of a larger family that can also bind fibronectin (36). Together, these studies have focused on interactions between Leptospira species and the ECM and have identified several bacterial proteins involved in these interactions.
Interactions of Leptospira species with glycosaminoglycans (GAGs), however, have not yet been reported. GAGs are long, unbranched, disaccharide polymers that may be modified by sulfation and/or epimerization, resulting in considerable functional heterogeneity. Proteoglycans (PGs) consist of GAG chains covalently linked to core proteins. GAGs and PGs are substrates for the attachment of many bacterial pathogens, including another pathogenic spirochete, B. burgdorferi (17-19, 21, 25). Because GAG binding in vitro is a property of many bacterial pathogens (reviewed in reference 35), binding of Leptospira to GAGs and PGs was investigated.
MATERIALS AND METHODS
Bacterial and mammalian cell culture.
Leptospira biflexa serovar Patoc strain 23582 (the reference strain for the serovar [nonpathogenic]) and L. interrogans serovar Canicola (strain 23606, known to be virulent) were obtained from the ATCC. Early experiments were also performed using strains of L. biflexa serovar Patoc from David Haake (UCLA, Los Angeles, CA) and L. interrogans serovar Canicola from Richard Zuerner (USDA, Ames, IA), which behaved similarly to their counterparts from the ATCC. L. interrogans serovar Copenhageni strain Fiocruz L1-130 (pathogenic, with a 50% lethal dose of ≅10 bacteria in hamsters [11]) was provided by David Haake. The bacteria utilized for this study were at low passage numbers (≤8 passages from animals for the virulent strains) or were from the supplier if the passage number was not specified, and they were initially cultured in semisolid Ellinghausen-McCullough-Johnson-Harris medium (2.0 g/liter agarose) supplemented with 100 μg/ml of 5-fluorouracil (Sigma, St. Louis, MO) and 1% heat-inactivated rabbit serum (Sigma) (15) at 30°C. After Dinger zone formation in semisolid medium, bacteria were transferred into liquid, supplemented Ellinghausen-McCullough-Johnson-Harris medium in plastic screw-cap tubes and incubated at 30°C. Radioactive Leptospira cells were prepared by growing the bacteria in liquid medium with 1 mCi of [35S]methionine per 45 ml of culture. The bacteria were then washed to remove excess 35S, suspended in medium supplemented with glycerol to 20%, and stored in aliquots at −80°C, essentially as described previously (10).
The epithelial cell lines HEp-2 (human), HEK293 (human), MDCK (canine), and CHO K1 (and its mutant derivatives) were obtained from the ATCC and were grown at 37°C under 5% CO2 in the media recommended by the ATCC. The human microvascular endothelial cell line HMEC-1 (1) was cultured in endothelial basal medium (Clonetics, San Diego, CA) supplemented with 15% heat-inactivated fetal bovine serum (HyClone, Logan, UT), 1 μg/ml hydrocortisone (Sigma-Aldrich, St. Louis, MO), and 10 ng/ml epidermal growth factor (Sigma-Aldrich). The human macrovascular endothelial cell line EA.hy926 (13, 14) was grown at 37°C under 5% CO2 in Dulbecco's modified Eagle medium with 4.5 g/liter glucose, supplemented with 10% fetal bovine serum, 100 μM hypoxanthine-0.4 μM aminopterin-16 μM thymidine (final concentrations; Sigma), and 2 mM l-glutamine. All cell lines were also grown in the presence of 100 U/ml penicillin and 100 μg/ml streptomycin. The cell layers were washed prior to the addition of the bacteria to remove the antibiotics.
Adhesion assays.
Mammalian cell lines were plated into 96-well tissue culture plates, adjacent to control wells containing cell culture medium only. One hour before the addition of the bacteria, the cell culture medium was removed and the cell layers were washed in phosphate-buffered saline (PBS) and then returned to the incubator in medium supplemented with 3% bovine serum albumin (BSA) but without antibiotics. After 1 h, this medium was removed and radiolabeled bacteria suspended in the cell culture medium without antibiotics were added at a multiplicity of infection of 10 (approximately 3 × 105 bacteria/well) and centrifuged at 800 × g onto the cell layers for 10 min at ambient temperature. The plates were then incubated for 1 h at 37°C under 5% CO2. At the end of the incubation, the motility and integrity of the bacteria in randomly chosen wells were assessed by dark-field microscopy in multiple independent experiments. Unbound bacteria were removed by washing the plates three times with 200 μl/well cell culture medium (without antibiotics). Because one of the strains used (L. interrogans Fiocruz L1-130) has a very low 50% lethal dose in hamsters (11), all wash fluids were treated with bleach to kill the unbound bacteria. The washed wells were solubilized with 1% sodium dodecyl sulfate (SDS), and the contents were transferred onto Luma scintillation plates (Packard, Meriden, CT) and dried before being evaluated by counting with a Packard 96-well-plate scintillation counter. All experiments were performed on at least three independent occasions, with each condition tested in quadruplicate.
For experiments in which cell layers at different degrees of confluence were tested, the cells were plated 4 days in advance to achieve “confluence plus 2 days,” 2 days in advance to achieve confluence, and 6 h prior to the addition of the bacteria for the subconfluent condition. For experiments in which cells and the ECM were compared, confluent layers in 96-well plates were lifted with either 5 mM EDTA in PBS or trypsin-EDTA (Gibco-Invitrogen, Gaithersburg, MD). The lifted cells were transferred into 96-well V-bottom plates containing culture medium without antibiotics and washed twice in the same medium prior to the addition of bacteria and incubation in suspension for 1 h as described above. The ECM (material remaining in the wells after the cells had been lifted) was washed twice with cell culture medium without antibiotics; each well was visually inspected to ensure that no cells remained. The bacteria were then added to the ECM-containing wells and centrifuged or added to the wells containing cells in suspension without a centrifugation step, and the plates were incubated as described above. Washing to remove unbound bacteria and quantification of binding to the cells and the ECM were then performed as described above, with the exception that the cells were washed by centrifugation. In parallel experiments, ECM proteins remaining in the wells were detected by an enzyme-linked immunosorbent assay using polyclonal antisera at the dilutions recommended by the manufacturer (Chemicon, Temecula, CA) after verification of specificity. After the wells were washed, bound primary antibody was detected using anti-rabbit IgG secondary antibody conjugated to alkaline phosphatase (Promega, Madison, WI) at 1:10,000, with development in phosphatase substrate (Sigma Chemical, St. Louis, MO).
The roles of GAGs and PGs in the adhesion of Leptospira strains were tested using the following modifications of the adhesion assay protocol. To inhibit transfer of the GAGs to protein cores, cells were preincubated overnight in medium containing 5 mM p-nitrophenyl-β-d-xyloside (β-xyloside; Sigma-Aldrich, St. Louis, MO), an inhibitor of the xylosyltransferase required for the initial modification of the serine residues that serve as acceptors of GAG chains. Control cells were left untreated or were incubated with an analog, p-nitrophenyl-α-d-galactoside (α-galactoside; Sigma-Aldrich). The cell monolayers were washed and the infection was carried out as described above. To test the competitive effect of exogenous GAGs or GAG analogs on leptospiral attachment to cells, the bacteria were incubated for 30 min at room temperature in cell medium containing 1% BSA supplemented with purified GAGs (Sigma-Aldrich) at concentrations ranging between 0.01 and 1,000 μg/ml. The attachment assay was then performed as described above. The 50% inhibitory concentration (IC50) was determined using the four-parameter logistic model (Hill-Slope model). To determine the effect of the enzymatic removal of different classes of GAGs from cell surface PGs on leptospiral attachment, cell monolayers were incubated at 37°C with a 35-μl/well volume of a lyase (heparinase I, heparitinase, chondroitinase AC, or chondroitinase ABC; Sigma-Aldrich) at 0.5 U/ml for 2 h in cell medium supplemented with 1% BSA, plus the protease inhibitors aprotinin at 10−2 trypsin inhibitory units/ml and phenylmethylsulfonyl fluoride at 150 μg/ml. The monolayers were then washed with PBS three times, and the attachment assay was performed as described above.
Assay of purified-GAG binding.
To screen for GAGs recognized by L. interrogans, 5-mg/ml aliquots of purified GAGs (Sigma-Aldrich) solubilized in water or PBS on the day of plating were added to non-tissue-culture 96-well plates and the plates were incubated at 4°C overnight. The plates were then washed with PBS and blocked for 2 h at room temperature in a solution of 50 mM HEPES, 100 mM NaCl, 1 mM MgCl2, 1 mM MnCl2, 1 mM CaCl2, and 3.5% BSA. Bacteria were added at 3 × 105/well in the same buffer containing 1% BSA. The plates were centrifuged for 15 min and rocked for 45 min at room temperature. The wells were washed with PBS-0.2% BSA and then treated with 1% SDS. The well contents were transferred onto Luma plates and analyzed by liquid scintillation counting as described above.
Gel electrophoresis and immunoblotting.
The ECM proteins associated with mammalian cells lifted with EDTA were compared to those associated with intact monolayers by SDS-10% polyacrylamide gel electrophoresis under nonreducing conditions. Cells were harvested by being lifted with EDTA as described above, or the monolayers were washed three times with PBS and then vigorously scraped off the plastic in the presence of 2× SDS loading buffer without a reducing agent. The cells lifted with EDTA were suspended in the same SDS loading buffer. The viscosities of all samples were reduced by brief sonication on ice. Aliquots of each sample were then diluted further in gel loading buffer, heated to 95°C for 5 min, and loaded immediately onto the gels. After separation by electrophoresis, the proteins were transferred onto Immobilon membranes (Millipore, Bedford, MA). The membranes were either stained with Coomassie blue or blocked in Tris-buffered saline (25 mM Tris, 150 mM NaCl, pH 7.5) containing 5% (wt/vol) nonfat dry milk. The latter membranes were probed with the following rabbit antisera, all from Chemicon (Temecula, CA): antifibronectin (AB1945; 100 ng/ml), antilaminin (AB19012; 200 ng/ml), anti-collagen type I (AB745; 1:200), anti-collagen type IV (AB748; 1 μg/ml), and antivitronectin (AB19014; 1 μg/ml). After development as described below, all membranes were reprobed with antiactin (A-2066; [1:10,000; Sigma Chemical, St. Louis, MO]) as a loading control in addition to the general Coomassie blue stain. The primary antibodies were detected by using goat anti-rabbit IgG (1:10,000; Promega, Madison, WI) and colorimetric development.
Statistical analyses.
The methods used to assess statistical significance varied with the type of experiment and so are described in the figure legends. P values of <0.05 were considered to denote significant differences.
RESULTS
The degree of confluence of the mammalian cells affects Leptospira attachment.
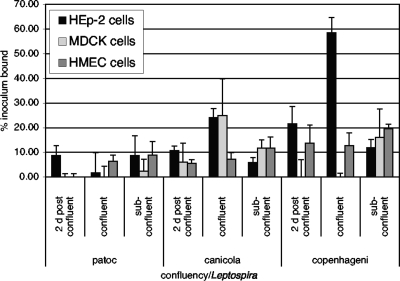
Cells in culture express different receptors at different stages of growth, so we determined how the growth stage of the mammalian cells might influence the adhesion of a few representative Leptospira strains (Fig. 1). The nonpathogenic L. biflexa serovar Patoc showed relatively inefficient cell-specific binding activity under all conditions tested. L. interrogans serovar Canicola bound most efficiently to HEp-2 and MDCK cells just at confluence, while L. interrogans serovar Copenhageni bound very efficiently to HEp-2 cells just at confluence and less efficiently to HEp-2 cells postconfluence. Binding of this strain to MDCK cells was maximal when the cells were subconfluent. For both of the pathogenic strains tested, binding to HEp-2 cells decreased after the cells reached confluence (as demonstrated by comparing binding at 2 days postconfluence to that at confluence); for HMEC-1 cells, this trend was not apparent, and for MDCK cells, it varied among bacterial strains. These results suggest that the HEp-2 cell receptors for pathogenic Leptospira may be masked, become inaccessible, or be downregulated as the cells form completely confluent layers. Some of the variability with the MDCK cells may be due to differential patterns of cell loss from the wells; the same scenario was observed for the human kidney epithelial cell line HEK293 in some experiments (data not shown). HEp-2 and endothelial cells, in contrast, were efficiently retained and so were used for most subsequent studies.
FIG. 1.
The degree of confluence of mammalian cell layers affects the attachment of different Leptospira strains differently. 35S-labeled bacteria were added to wells containing cells that had reached confluence 2 days before the experiment (2 d postconfluent), wells containing cells that were just reaching confluence (with 95 to 100% well area coverage), and wells containing cells expected to reach confluence the day after the experiment (with 45 to 50% well area coverage). Shown are the means of cell-specific attachment levels ± standard deviations, expressed as the percent inoculum bound, from a single experiment representative of multiple experiments. For the HEp-2 cells, the attachment of both L. interrogans strains to postconfluent layers was significantly less efficient than attachment to confluent layers (P ≤ 0.0008); a similar difference (P < 0.0001) in the efficiencies of attachment to confluent versus subconfluent layers was observed, although this result is likely to be affected by the approximately twofold differences in the numbers of cells in the wells. For the other cell lines, confluence did not significantly affect the efficiency of bacterial attachment (P ≥ 0.05). L. interrogans serovar Canicola bound to confluent MDCK cell layers more efficiently than to HEp-2 cell layers (P < 0.016). L. interrogans serovar Copenhageni, in contrast, bound more efficiently to HEp-2 than to MDCK confluent cell layers (P < 0.0001). All comparisons were analyzed by Student's two-tailed t test.
Leptospires bind to mammalian cells as well as to the ECM deposited by the cells.
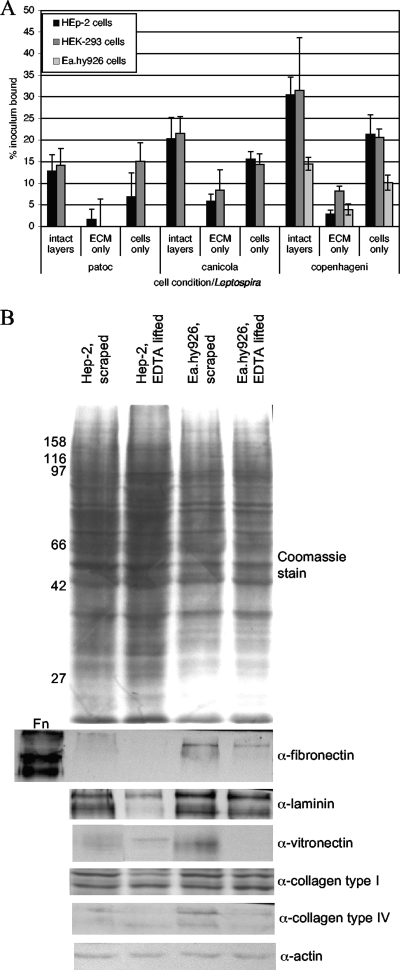
One question arising from the results shown in Fig. 1 and from data in the literature is whether Leptospira bacteria are binding primarily to the ECM or to mammalian cell surface receptors. We therefore lifted cells with EDTA, which disrupts the functions of integrins and other cell attachment molecules, resulting in the detachment of the cells from the ECM, or with trypsin-EDTA, which also degrades proteins that are involved in cell adhesion. We then tested both the ECM remaining in the wells and the cells in suspension for Leptospira attachment. Prior to the addition of the bacteria, both the ECM and the cells were washed with cell culture medium (without antibiotics) to remove the EDTA and restore the divalent cations that might be required for bacterium-cell interactions. We found that binding to intact cell layers, in which both cells and the ECM are present, is most efficient, but that binding to cells lifted with EDTA is more efficient than binding to the remaining ECM alone (Fig. 2A). This finding cannot be attributed simply to the presence of ECM proteins associated with the cells after lifting with EDTA, as most of the ECM proteins examined were diminished or undetectable in analyses of cells lifted with EDTA versus cells scraped off the wells into SDS buffer (Fig. 2B); this trend was more pronounced for the HEp-2 cells than for the Ea.hy926 cells. The increased binding to the cells versus the ECM is particularly notable in light of the lack of centrifugation of the bacteria and cells in suspension, because centrifugation facilitates bacterial interactions with intact cell layers and the ECM. The ECM components detected after removal of the cells varied with the cell line, as determined by an enzyme-linked immunosorbent assay (Table 1). These results suggest that, as has been demonstrated abundantly in studies reported in the literature, Leptospira bacteria do attach to ECM components, but here we demonstrate that these bacteria also bind to mammalian cell surface-specific receptors. At least a portion of the complement of cell surface receptors for Leptospira is proteinaceous in nature, as the level of binding to HEp-2 cells lifted with trypsin-EDTA was less than 10% of the level of binding to cells lifted with EDTA alone (data not shown). This finding is consistent with previous results obtained by other investigators (37). The difference between the effect of trypsin-EDTA and that of EDTA alone on binding to ECM was not statistically significant. This may be due to the overall less efficient attachment to the ECM. Alternatively, the brief trypsin-EDTA treatment required to remove the cells may be insufficient to disrupt interactions between the ECM components, leaving partially digested protein fragments associated with the relatively insoluble matrix available for bacterial attachment. As many ECM proteins have functionally separable domains that retain adhesion activity after proteolytic digestion of the intact protein, these results are consistent with previous data showing that the bacteria recognize specific domains of ECM proteins (9, 36).
FIG. 2.
Attachment of Leptospira to cells versus the ECM. (A) Confluent cell layers were left as is (intact layers) or lifted with EDTA twice to remove all cells from the ECM. The cells were collected by centrifugation, and both the cells and the ECM were washed in medium without antibiotics to remove the EDTA and restore the divalent cations prior to the addition of 35S-labeled Leptospira bacteria at a multiplicity of infection of 10. After incubation for 1 h at 37°C, all wells were washed to remove unbound bacteria, and bound bacteria were quantified by scintillation counting. The results presented were calculated by subtracting the level of background binding to wells without cells or ECM material. The means and standard deviations of results for four replicates from representative experiments (≥4 for each cell line) are shown and are expressed as the percent inoculum bound. By using Student's two-tailed t test, comparisons of binding to lifted cells versus binding to the ECM left behind were as follows: for HEp-2 cells, P > 0.1 for L. biflexa serovar Patoc and P = 0.0002 for L. interrogans serovars Canicola and Copenhageni; for HEK293 cells, P = 0.008 for L. biflexa serovar Patoc, P > 0.06 for L. interrogans serovar Canicola, and P < 0.0001 for L. interrogans serovar Copenhageni; and for Ea.hy926 cells, only serovar Copenhageni was tested, with P < 0.0001 for cell binding versus ECM binding. (B) Immunoblots of ECM proteins associated with cells after either scraping of the cell layers into SDS gel loading buffer or lifting of the cells with EDTA prior to solubilization in gel loading buffer. As a positive control for the antifibronectin (α-fibronectin) antibody, 0.1 μg of purified fibronectin (Fn; soluble form) was loaded. The first lane of the blot probed with antivitronectin (α-vitronectin) was taken from one of the other panels after reprobing with antivitronectin, as the original blot was miscut between lanes. Molecular size markers (in kilodaltons) are shown to the left. α-laminin, α-collagen type I, α-collagen type IV, and α-actin, antilaminin, anti-collagen type I, anti-collagen type IV, and antiactin antibodies.
TABLE 1.
Representative ECM proteins remaining in wells after removal of cells with EDTA
| ECM protein | OD405a value for wells previously containing: |
|
|---|---|---|
| HEp-2 cells | HEK293 cells | |
| Fibronectin | 0.028 ± 0.006 | 0.268 ± 0.008 |
| Laminin | 0.027 ± 0.002 | 0.078 ± 0.004 |
| Vitronectin | NSb | 0.015 ± 0.004 |
| Collagen type IV | NS | NS |
Values were calculated as the difference between readings of optical density at 405 nm (OD405) for wells in which confluent cell layers had been plated 2 days prior to treatment with EDTA, as described in Materials and Methods, and OD405 readings for control wells that had contained medium without cells (background). Shown are means ± standard deviations of results from duplicate experiments, each of which included four replicate wells. The values for HEp-2 cells in these experiments were always low compared to those for other cell lines tested after the same incubation times in a colorimetric substrate.
NS, not significantly above background.
One notable finding from this work is that the different strains of pathogenic leptospires appear to depend on the presence of fibronectin in the ECM to different degrees. While serovar Canicola has lower overall cell-binding activity than serovar Copenhageni and levels of binding to the HEK293 ECM are approximately equal for the two strains, the level of serovar Copenhageni binding to the HEp-2 ECM is lower than that of serovar Canicola binding. Since HEp-2 cells do not express fibronectin (12, 16), one possible explanation is that the serovar Copenhageni bacteria rely more on the presence of fibronectin secreted and deposited by the cells into the ECM, although it is clear that these bacteria must be able to bind to other ECM components or to bind to soluble fibronectin that has been captured from the serum in the growth medium, as suggested by the results shown in Table 1 and by findings from previous work by other groups (9, 36).
PGs account for some, but not all, of L. interrogans attachment to cells.
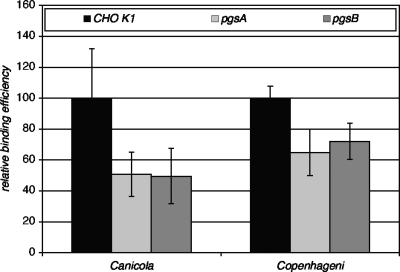
A number of pathogenic bacteria bind to particular GAGs, which may be conjugated to proteins to form PGs. We therefore tested two mutant CHO K1 derivative cell lines that either do not synthesize PGs or that express reduced levels in comparison to wild-type cells (35) in our standard L. interrogans adherence assay. As shown in Fig. 3, both of the pgs mutants tested, which are defective in PG synthesis, showed statistically significant reductions in attachment of both L. interrogans strains tested. However, the reduced binding efficiencies were still high, suggesting that other, non-PG receptors may have a significant role in L. interrogans attachment to cells. Levels of binding of the bacteria to three lec mutants tested (lec1, lec2, and lec8 mutants) were not significantly different for either bacterial strain (data not shown), suggesting that other glycoconjugates play a relatively minor role in L. interrogans attachment to CHO cells.
FIG. 3.
Binding of L. interrogans to wild-type and mutant CHO cells. Cells were plated into 96-well plates, and the attachment assay was performed as described in Materials and Methods. To allow incorporation of all data from multiple experiments, binding to the mutant cell lines in each experiment was normalized to binding to the wild-type CHO K1 cells in the same experiment. Shown are the means and standard deviations of all data points. Binding of both L. interrogans strains to the pgsA and pgsB mutant cells (pgsA and pgsB) was statistically significantly different from that to wild-type CHO K1 cells by Student's two-tailed t test (P ≤ 0.001).
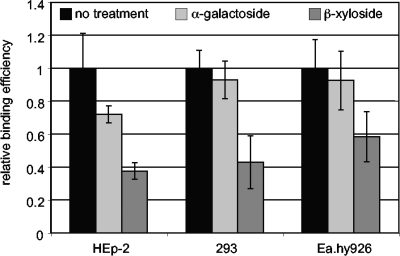
As a second approach to determining whether L. interrogans binds to PGs on mammalian cells, wild-type HEp-2, Ea.hy926, and HEK293 cells were incubated overnight with β-xyloside, an inhibitor of GAG transfer to protein cores. As a control, a different sugar analog, α-galactoside, was tested. As shown in Fig. 4, β-xyloside treatment of all three mammalian cell lines significantly decreased the attachment of L. interrogans, confirming the role of PGs suggested by the results for the CHO pgs mutants.
FIG. 4.
Inhibition of GAG transfer to PGs reduces L. interrogans attachment to mammalian cells. Three cell lines, HEp-2, HEK293 (293), and Ea.hy926, were plated into 96-well dishes and allowed to grow and adhere to the wells. The day before the attachment assay, the media were replaced with fresh media containing no additions (no-treatment control), 5 mM α-galactoside (a control sugar analog), or 5 mM β-xyloside (a sugar analog that competitively inhibits the xylosyltransferase required to transfer the xylose root of GAGs to protein cores). On the day of the attachment assay, the cell layers were washed and bacterial attachment was quantified as described in Materials and Methods. Shown are the means and standard deviations of attachment values normalized with respect to the control; data are from multiple experiments. P values calculated by Student's two-tailed t test for all cell lines were as follows: control versus β-xyloside treatment, P < 0.01; and control versus α-galactoside treatment, P > 0.1.
L. interrogans binds to purified GAGs.
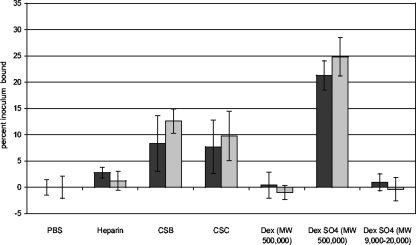
To build on the results suggesting that L. interrogans recognizes PGs expressed by mammalian cells, we determined which GAGs are recognized by L. interrogans. Purified GAGs, the polysaccharides dextran and dextran sulfate, or the buffer control was immobilized in plastic wells and then probed with L. interrogans. The results shown in Fig. 5 demonstrate that L. interrogans binds to high-molecular-weight dextran sulfate but not to high-molecular-weight dextran or to low-molecular-weight dextran sulfate, suggesting that sulfation and polymer size (and consequently, the number of binding sites per molecule) are important determinants of L. interrogans binding to GAGs. This observation is supported by the attachment of the bacteria to heparin, which is more highly sulfated than heparan sulfate, which did not appear to be recognized by the bacteria. The more efficient binding of L. interrogans to chondroitin sulfates B and C than to heparin is indicative of specific Leptospira recognition of particular GAGs.
FIG. 5.
Attachment of L. interrogans serovar Copenhageni strain Fiocruz L1-130 to purified GAGs. GAGs were plated into plastic wells at 1 mg/ml and then probed with 35S-labeled bacteria. After incubation and then washing of the wells to remove unbound bacteria, the percent inoculum bound was calculated. Shown are the means ± standard deviations of results for four replicates from each of two independently grown and radiolabeled batches of bacteria (symbolized by black and gray bars). These data represent results from multiple experiments. CSB, chondroitin sulfate B; CSC, chondroitin sulfate C. The molecular weights (MW) of the dextran (Dex) molecules used are indicated. Binding to heparan sulfate has not been detected in any experiment. The following P values for comparisons to the PBS control were determined by Student's two-tailed t test: for dextran and low-molecular-weight dextran sulfate, P > 0.5; for heparin, P = 0.016; and for CSB, CSC, and high-molecular-weight dextran sulfate, P < 0.001.
Roles of particular GAGs in Leptospira attachment to cells.
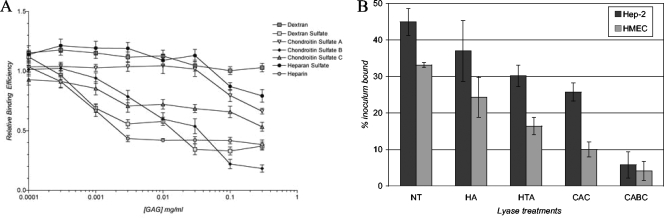
The profile of GAGs recognized by L. interrogans presented in Fig. 5 may be influenced by the efficiency of coating of the plastic wells particular to each GAG. To determine whether the apparent preference of GAGs is consistent with GAGs recognized by L. interrogans when expressed by mammalian cells, two approaches were taken. First, exogenous GAGs in solution were preincubated with L. interrogans to determine whether they could serve as competitive inhibitors of bacterial attachment. As shown in Fig. 6A, dextran sulfate (high molecular weight) competed with cellular GAGs for recognition by L. interrogans (IC50 ≈ 0.007 mg/ml) whereas high-molecular-weight dextran did not. While these results are consistent with the levels of binding to immobilized GAGs shown in Fig. 5, these polysaccharides are model compounds that are not biologically relevant. Chondroitin sulfates B and C, however, are biologically relevant and inhibited L. interrogans attachment to mammalian cells. The IC50 of chondroitin sulfate B was ≈0.023 mg/ml, while that of chondroitin sulfate C was greater than 0.3 mg/ml, the maximum concentration tested, suggesting either that chondroitin sulfate B is the primary GAG recognized by L. interrogans serovar Copenhageni on HEp-2 cells or that this GAG interacts with the leptospiral GAG-binding adhesin(s) with the highest affinity. Heparin, like dextran sulfate, efficiently inhibited leptospiral attachment to HEp-2 cells (IC50 ≈ 0.002), which suggests that sulfation, charge, or both may be important in attachment activity, but heparan sulfate and chondroitin sulfate A did not significantly affect bacterial binding to the mammalian cells. With the exception of the heparin results, these data are also consistent with the levels of binding to purified GAGs shown in Fig. 5. Heparin, however, is the most negatively charged of the naturally occurring GAGs tested, so like dextran sulfate it may affect bacterial attachment for reasons other than specific competition for particular cell surface GAGs.
FIG. 6.
Roles of particular GAGs in the attachment of L. interrogans to cells. (A) GAGs at the concentrations indicated were incubated with bacteria for 30 min prior to the addition of the bacteria to the immobilized mammalian cells. Shown are the means and standard deviations of all data from at least three independent experiments, each of which consisted of four technical replicates. Statistical significance was determined for the entire dose range for each GAG in comparison to dextran by Tukey's multiple-comparison test; P > 0.05 (not significant) for heparan sulfate, P < 0.01 for chondroitin sulfate A, and P < 0.001 for dextran sulfate, heparin, chondroitin sulfate B, and chondroitin sulfate C. Results for chondroitin sulfate B, chondroitin sulfate C, heparin, and dextran sulfate were not significantly different from one another. (B) Mammalian cell layers were treated with 35 μl of 0.5 U/ml of the GAG lyase heparinase (HA), heparatinase (HTA), chondroitinase AC (CAC), or chondroitinase ABC (CABC) for 2 h or were not treated (NT) and were then washed prior to the addition of bacteria. Shown are the means and standard deviations of results, expressed as the percent inoculum bound, for four replicates from a single experiment representative of seven experiments. For each experiment, in comparison to the no-treatment control, the P values for HA were >0.1, for HTA were 0.01 to 0.001, and for CAC and CABC were <0.001 by Student's two-tailed t test for both cell lines.
As a second approach to the identification of cell-associated GAGs that are recognized by L. interrogans, cell layers were treated with GAG lyases, which cleave GAG chains, prior to the addition of the bacteria. As shown in Fig. 6B, heparinase, which degrades heparin, did not affect L. interrogans attachment to epithelial or endothelial cells. In contrast, the two chondroitinases both significantly inhibited subsequent bacterial attachment to the treated cells. The abilities of the chondroitinases to diminish L. interrogans more effectively than heparinase or heparatinase are consistent with the abilities of the chondroitin sulfate GAGs to inhibit L. interrogans attachment to cells (Fig. 6A) and to bind the bacteria (Fig. 5).
DISCUSSION
Pathogenic leptospires have been shown previously to bind to mammalian cell layers in culture and to particular ECM components more efficiently than nonpathogenic leptospires. Here, we report that binding to ECM molecules does not account for all of the Leptospira cell layer-binding activity. Cell surface receptors also serve as targets for Leptospira attachment. In addition, the effects of the confluence of the mammalian cell layers suggest that the availability of cell surface and/or ECM molecules that serve as substrates for the attachment of Leptospira may change as the cells in in vitro monolayers grow. This change may be due to differences in production, modification, and turnover of particular substrates. The identities of these molecules, however, will require further investigation.
Because many bacterial pathogens bind to PGs and, in particular, to the GAG chains that form the major surface-exposed component of PGs, we tested L. interrogans for this binding activity. The data presented here demonstrate that L. interrogans strain L1-130 (serovar Copenhageni), which is a significant pathogen of humans, binds most efficiently to chondroitin sulfate B and C PGs and the corresponding GAGs. GAGs are displayed by diverse cell types and are present in the ECM as well as associated with the mammalian cell surface. They are therefore widely available for pathogen attachment. In fact, preliminary data presented here suggest that a different L. interrogans strain, of serovar Canicola, also binds to PGs. Although our results must be expanded to include additional strains and serovars in the future, the data suggest that L. interrogans utilizes PGs as mammalian substrates for attachment. Our results also suggest that additional, non-GAG or -PG receptors on the mammalian cell surface await identification.
It is apparent from the aggregate of the data presented in Fig. 5 and 6 that L. interrogans L1-130 binds to chondroitin sulfate PGs more efficiently than to heparan sulfate PGs. This pattern is in contrast to that for B. burgdorferi, another pathogenic spirochete that recognizes heparin, heparan sulfate, and chondroitin sulfate B but does not efficiently bind to chondroitin sulfate C (25). It should be noted, however, that although heparin is widely used as a tractable model GAG, it is produced by, and released from, mast cells. It is therefore not typically present in the ECM and available to serve as a target for bacterial attachment to the ECM (34). In contrast, the heparan sulfate and chondroitin sulfate PGs are widely distributed and, accordingly, are substrates for the attachment of a variety of pathogens. In studies such as those described here, the possibility that heparin serves more as a nonspecific inhibitor of interactions, based on the strong negative charge, cannot be dismissed. Additional GAGs that are biologically relevant can, in contrast, assist in the identification of specific receptors for pathogens.
Although both Borrelia and Leptospira species are spirochetes, they are different at the molecular, cellular, and ecologic levels and cause different disease manifestations. For example, the Borrelia species do not contain the genes required for lipopolysaccharide (LPS) synthesis, while the leptospires express LPSs that are variable and determine serovar type. It is unlikely that the LPS, however, contributes to GAG and PG binding, as interactions between carbohydrate polymers would be unlikely due to mutual repulsion by the negative charges. It is far more likely that the GAGs and PGs are recognized by one or more Leptospira proteins that are expressed on the surfaces of the bacteria.
Several proteins, i.e., MSCRAMMs (34), have been identified as Leptospira adhesins mediating attachment to ECM molecules. Like other bacterial proteins that bind to ECM components, these proteins bind to several different mammalian substrates. Since many ECM molecules contain repeated elements, it is possible that the proteins that bind to repeating units may also bind to the repeats of GAGs. This was shown to be the case for the fibronectin-binding protein of B. burgdorferi, which also binds GAGs (16). The identities of leptospiral proteins that mediate interactions with GAGs will be determined in future experiments, but the known adhesins, such as LigA, LigB, LipL32, TlyC, and a family of proteins known by several names (LenA to LenF, Lsa24, and LfhA) constitute a partially characterized subset for prioritized testing (4, 7, 9, 20, 27, 36).
The role of GAG and PG binding in the pathogenesis of Leptospira infections remains to be tested, but several possibilities exist. For example, it is possible that binding to GAGs facilitates leptospiral infection, either initially at mucosal surfaces or later as the bacteria disseminate into diverse tissues. Because these bacteria disseminate from the site of inoculation to virtually any site in the body, tissue tropism may be affected by the GAG-binding preferences of the bacteria and the GAGs available to the bacteria in the tissues. This mechanism has been implicated in the different rates of central nervous system colonization by relapsing-fever Borrelia bacteria that express surface proteins with different GAG preferences (26). The initial encounter of Leptospira species with mammalian hosts is typically at mucous membranes. Epithelia are known to express both heparan sulfate and chondroitin sulfate PGs, including syndecans 1 and 4 and epican. It is therefore possible that the initial host-bacterium interaction is, at least in part, mediated by Leptospira binding to the GAG moieties of chondroitin sulfate PGs.
In the case of Leptospira infection, in particular, a specific site of GAG-bacterium interaction will be interesting to examine. Kidney proximal-tubule epithelial cells produce both heparan sulfate and chondroitin sulfate GAGs in culture (6) and in vivo (24), although different animal species show different GAG profiles in the urine. It is possible that GAG binding facilitates the colonization of kidneys, an epidemiologically critical site. In addition, bacteria bound to GAGs that are released from the epithelial cells of the proximal tubules of the kidneys may provide a mechanism for shedding of the bacteria in the urine. The bacteria making the transition from mammalian host to environmental soil and water may then be associated with host-derived GAGs. In addition, binding to GAGs released into the extracellular environment within the host may allow the bacteria to cloak themselves, which would potentially facilitate the persistent infection established in maintenance hosts. This cloaking may also be a key function for ECM proteins with soluble forms, e.g., fibronectin. Identification of the GAG-binding adhesin(s) of L. interrogans will facilitate further analyses of the roles of attachment to GAGs and PGs in the abilities of Leptospira bacteria to cause infection and disease in mammals.
Acknowledgments
We thank David Haake of UCLA and Richard Zuerner of the USDA for generously providing Leptospira strains and advice on their care and growth. We also thank Edwin Ades of the CDC for providing the HMEC-1 line and Cora-Jean Edgell for providing the Ea.hy926 cell line.
This work was supported by grants R21 AI077560, ROI AI051407, and ROI AI059505 from the NIAID to J.C., by the Advancing a Healthier Wisconsin Initiative at the Medical College of Wisconsin, and by the PREP program at Tufts University School of Medicine, supported by NIH grant R25 GM66567.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 6 October 2009.
REFERENCES
- 1.Ades, E. W., F. J. Candal, R. A. Swerlick, V. G. George, S. Summers, D. C. Bosse, and T. J. Lawley. 1992. HMEC-1: establishment of an immortalized human microvascular endothelial cell line. J. Investig. Dermatol. 99:683-690. [DOI] [PubMed] [Google Scholar]
- 2.Atzingen, M. V., A. S. Barbosa, T. De Brito, S. A. Vasconcellos, Z. M. de Morais, D. M. Lima, P. A. Abreu, and A. L. Nascimento. 2008. Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiol. 8:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballard, S. A., M. Williamson, B. Adler, T. Vinh, and S. Faine. 1986. Interactions of virulent and avirulent leptospires with primary cultures of renal epithelial cells. J. Med. Microbiol. 21:59-67. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa, A. S., P. A. Abreu, F. O. Neves, M. V. Atzingen, M. M. Watanabe, M. L. Vieira, Z. M. Morais, S. A. Vasconcellos, and A. L. Nascimento. 2006. A newly identified leptospiral adhesin mediates attachment to laminin. Infect. Immun. 74:6356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barocchi, M. A., A. I. Ko, M. G. Reis, K. L. McDonald, and L. W. Riley. 2002. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infect. Immun. 70:6926-6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borges, F. T., Y. M. Michelacci, J. A. Aguiar, M. A. Dalboni, A. S. Garofalo, and N. Schor. 2005. Characterization of glycosaminoglycans in tubular epithelial cells: calcium oxalate and oxalate ions effects. Kidney Int. 68:1630-1642. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho, E., A. S. Barbosa, R. M. Gomez, A. M. Cianciarullo, P. Hauk, P. A. Abreu, L. C. Fiorini, M. L. Oliveira, E. C. Romero, A. P. Goncales, Z. M. Morais, S. A. Vasconcellos, and P. L. Ho. 2009. Leptospiral TlyC is an extracellular matrix-binding protein and does not present hemolysin activity. FEBS Lett. 583:1381-1385. [DOI] [PubMed] [Google Scholar]
- 8.CDC. 12 October 2005, revision date. Leptospirosis. CDC, Atlanta, GA. http://www.cdc.gov/ncidod/dbmd/diseaseinfo/leptospirosis_g.htm.
- 9.Choy, H. A., M. M. Kelley, T. L. Chen, A. K. Moller, J. Matsunaga, and D. A. Haake. 2007. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 75:2441-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coburn, J., J. Leong, and J. Erban. 1993. Integrin αIIbβ3 mediates binding of the Lyme disease agent, Borrelia burgdorferi, to human platelets. Proc. Natl. Acad. Sci. USA 90:7058-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croda, J., C. P. Figueira, E. A. Wunder, Jr., C. S. Santos, M. G. Reis, A. I. Ko, and M. Picardeau. 2008. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect. Immun. 76:5826-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dramsi, S., F. Bourdichon, D. Cabanes, M. Lecuit, H. Fsihi, and P. Cossart. 2004. FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol. Microbiol. 53:639-649. [DOI] [PubMed] [Google Scholar]
- 13.Edgell, C. J., J. E. Haizlip, C. R. Bagnell, J. P. Packenham, P. Harrison, B. Wilbourn, and V. J. Madden. 1990. Endothelium specific Weibel-Palade bodies in a continuous human cell line, EA.hy926. In Vitro Cell. Dev. Biol. 26:1167-1172. [DOI] [PubMed] [Google Scholar]
- 14.Edgell, C. J., C. C. McDonald, and J. B. Graham. 1983. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. USA 80:3734-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faine, S., B. Adler, C. Bolin, and P. Perolat. 1999. Leptospira and leptospirosis, 2nd ed. MedSci, Melbourne, Australia.
- 16.Fischer, J. R., K. T. LeBlanc, and J. M. Leong. 2006. Fibronectin binding protein BBK32 of the Lyme disease spirochete promotes bacterial attachment to glycosaminoglycans. Infect. Immun. 74:435-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, J. R., N. Parveen, L. Magoun, and J. M. Leong. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 100:7307-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, B. P., E. L. Brown, D. W. Dorward, L. C. Rosenberg, and M. Hook. 1998. Decorin-binding adhesins from Borrelia burgdorferi. Mol. Microbiol. 30:711-723. [DOI] [PubMed] [Google Scholar]
- 19.Guo, B. P., S. J. Norris, L. C. Rosenberg, and M. Hook. 1995. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect. Immun. 63:3467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoke, D. E., S. Egan, P. A. Cullen, and B. Adler. 2008. LipL32 is an extracellular matrix-interacting protein of Leptospira spp. and Pseudoalteromonas tunicata. Infect. Immun. 76:2063-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaacs, R. 1994. Borrelia burgdorferi bind to epithelial cell proteoglycan. J. Clin. Investig. 93:809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito, T., and R. Yanagawa. 1987. Leptospiral attachment to extracellular matrix of mouse fibroblast (L929) cells. Vet. Microbiol. 15:89-96. [DOI] [PubMed] [Google Scholar]
- 23.Ito, T., and R. Yanagawa. 1987. Leptospiral attachment to four structural components of extracellular matrix. Nippon Juigaku Zasshi 49:875-882. [DOI] [PubMed] [Google Scholar]
- 24.Lensen, J. F., A. L. Rops, T. J. Wijnhoven, T. Hafmans, W. F. Feitz, E. Oosterwijk, B. Banas, R. J. Bindels, L. P. van den Heuvel, J. van der Vlag, J. H. Berden, and T. H. van Kuppevelt. 2005. Localization and functional characterization of glycosaminoglycan domains in the normal human kidney as revealed by phage display-derived single chain antibodies. J. Am. Soc. Nephrol. 16:1279-1288. [DOI] [PubMed] [Google Scholar]
- 25.Leong, J. M., P. E. Morrissey, E. Ortega-Barria, M. E. Pereira, and J. Coburn. 1995. Hemagglutination and proteoglycan binding by the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:874-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magoun, L., W. R. Zuckert, D. Robbins, N. Parveen, K. R. Alugupalli, T. G. Schwan, A. G. Barbour, and J. M. Leong. 2000. Variable small protein (Vsp)-dependent and Vsp-independent pathways for glycosaminoglycan recognition by relapsing fever spirochaetes. Mol. Microbiol. 36:886-897. [DOI] [PubMed] [Google Scholar]
- 27.Matsunaga, J., M. A. Barocchi, J. Croda, T. A. Young, Y. Sanchez, I. Siqueira, C. A. Bolin, M. G. Reis, L. W. Riley, D. A. Haake, and A. I. Ko. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsunaga, J., M. Lo, D. M. Bulach, R. L. Zuerner, B. Adler, and D. A. Haake. 2007. Response of Leptospira interrogans to physiologic osmolarity: relevance in signaling the environment-to-host transition. Infect. Immun. 75:2864-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsunaga, J., Y. Sanchez, X. Xu, and D. A. Haake. 2005. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 73:70-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merien, F., G. Baranton, and P. Perolat. 1997. Invasion of Vero cells and induction of apoptosis in macrophages by pathogenic Leptospira interrogans are correlated with virulence. Infect. Immun. 65:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merien, F., J. Truccolo, G. Baranton, and P. Perolat. 2000. Identification of a 36-kDa fibronectin-binding protein expressed by a virulent variant of Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol. Lett. 185:17-22. [DOI] [PubMed] [Google Scholar]
- 32.Merien, F., J. Truccolo, Y. Rougier, G. Baranton, and P. Perolat. 1998. In vivo apoptosis of hepatocytes in guinea pigs infected with Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol. Lett. 169:95-102. [DOI] [PubMed] [Google Scholar]
- 33.Palaniappan, R. U., Y. F. Chang, S. S. Jusuf, S. Artiushin, J. F. Timoney, S. P. McDonough, S. C. Barr, T. J. Divers, K. W. Simpson, P. L. McDonough, and H. O. Mohammed. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70:5924-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Hook. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 35.Rostand, K. S., and J. D. Esko. 1997. Microbial adherence to and invasion through proteoglycans. Infect. Immun. 65:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stevenson, B., H. A. Choy, M. Pinne, M. L. Rotondi, M. C. Miller, E. Demoll, P. Kraiczy, A. E. Cooley, T. P. Creamer, M. A. Suchard, C. A. Brissette, A. Verma, and D. A. Haake. 2007. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS ONE 2:e1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas, D. D., and L. M. Higbie. 1990. In vitro association of leptospires with host cells. Infect. Immun. 58:581-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuchimoto, M., M. Niikura, E. Ono, H. Kida, and R. Yanagawa. 1984. Leptospiral attachment to cultured cells. Zentralbl. Bakteriol. Mikrobiol. Hyg. Abt. 1 Orig. 258:268-274. [DOI] [PubMed] [Google Scholar]
- 39.Verma, A., J. Hellwage, S. Artiushin, P. F. Zipfel, P. Kraiczy, J. F. Timoney, and B. Stevenson. 2006. LfhA, a novel factor H-binding protein of Leptospira interrogans. Infect. Immun. 74:2659-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vinh, T., S. Faine, and B. Adler. 1984. Adhesion of leptospires to mouse fibroblasts (L929) and its enhancement by specific antibody. J. Med. Microbiol. 18:73-85. [DOI] [PubMed] [Google Scholar]