Abstract
Yersiniae bearing the Yersinia virulence plasmid pYV impact the transcriptome of J774A.1 macrophage-like cells in two distinct ways: (i) by suppressing, in a Yersinia outer protein P (YopP)-dependent manner, the induction of inflammatory response genes and (ii) by mRNA induction of the silencing transcription factor klf2. Here we show that klf2 induction by Yersinia enterocolitica occurs in several cell lines of macrophage and squamous and upper gastrointestinal epithelial origin as well as in bone marrow-derived dendritic cells. Several strains of Pseudomonas aeruginosa and Staphylococcus aureus are equally effective as Y. enterocolitica in inducing klf2 expression. Screening of mutant strains or incubation with recombinant toxins identified the rho-inactivating toxins YopT from Yersinia spp., ExoS from Pseudomonas aeruginosa, EDIN-B from Staphylococcus aureus, and C3bot from Clostridium botulinum as bacterial inducers of klf2 mRNA. klf2 mRNA induction by these toxins does not require de novo protein synthesis. Serum response factor or actin depolymerization does not seem to be involved in regulating klf2 expression in response to bacterial infection. Instead, short hairpin RNA-mediated inactivation of RhoA and its effector rhophilin 1 is sufficient to induce long-term klf2 expression. Thus, bacteria exploit the RhoA-rhophilin signaling cascade to mediate sustained expression of the immunosuppressive transcription factor klf2.
The genus Yersinia comprises three species that are pathogenic to humans and rodents: Yersinia pestis, the etiologic agent of plague, causes systemic and life-threatening disease; and Yersinia enterocolitica and Yersinia pseudotuberculosis are enteropathogens which cause gastrointestinal diseases, including mesenterial lymphadenitis, and, rarely, systemic infections. Yersinia is endowed with a unique capacity to withstand the host attack by injecting antihost effector proteins (Yersinia outer proteins [Yops]) into professional phagocytes via a type III protein secretion system (TTSS) (11) Both the TTSS and Yops are encoded by a 70-kb virulence plasmid (pYV) that is common to all pathogenic Yersinia spp. The six established effector proteins interfere with distinct signaling pathways, resulting in paralysis of phagocyte function. In particular, there are three different effector proteins that interfere with signaling from small GTPases. The protein kinase YopO (YpkA in Y. pseudotuberculosis) physically interacts with RhoA and Rac-1 (7) (although the functional relevance of this interaction has not been shown to date) and inhibits Gαq signaling (42). YopE is a GTPase activating protein (GAP) for Rho, Rac, and Cdc42, accelerating GTP hydrolysis and converting these proteins into the inactive, GDP-bound form (68). YopT, a cysteine protease, inactivates RhoA by cleavage adjacent to a prenylated cysteine located near the carboxy terminus, resulting in membrane release and cytoplasmic redistribution of RhoA (60). However, some studies also showed some effect of YopT on Rac and Cdc42 in biochemical assays performed in vitro (61), but after infection of living cells, YopT seems to act mainly on RhoA (2).
Recent large-scale gene expression studies identified a number of genes induced by the action of Yersinia Yops in macrophages (24, 54) and epithelial cells (8). We and others (43) have identified the transcription factor klf2 as one gene with particularly pronounced induction in host cells in response to, for example, infection with Yersinia enterocolitica (24), Pseudomonas aeruginosa (44), or Staphylococcus aureus (40). However, in most of these cases, the causative bacterial toxins have not been identified, with the exception of Clostridium botulinum C3 toxin (58).
KLF2 (formerly termed LKLF, for lung Kruppel-like factor) belongs to the KLF zinc finger family of transcription factors. KLF family members, such as KLF6, play a role in many cellular processes, including apoptosis, proliferation, differentiation, and development. KLF6, a tumor suppressor, has been shown to be induced by bacterial toxins (43). KLF2 has been identified by virtue of its homology with Eklf and is expressed primarily in the adult lung and, to a much lower extent, in the spleen (5). klf2−/− mice die between 11.5 and 13.5 days postconception, from severe hemorrhage (69) due to abnormal tunica media formation (32). However, experiments employing chimeric mice provided evidence that Klf2 is essential for normal lung development (70). It was shown recently that Klf2 is upregulated upon maturation of single positive T cells and that expression of klf2 is required to program the quiescent state of single positive T cells. klf2−/− T cells have a spontaneously activated phenotype and are rapidly eliminated by Fas ligand-induced apoptosis (32). Consistently, inducible expression of klf2 in Jurkat T cells is sufficient to induce a quiescent phenotype characterized by reduced proliferation, reduced protein synthesis, and decreased surface expression of activation markers (10). Moreover, in endothelial cells, klf2 is induced in response to shear stress (14) and inhibits expression of cell adhesion molecules in response to proinflammatory cytokines (59). Together, these data show that KLF2 has broadly inactivating functions in an ontogenetically diverse collection of cell types.
Here we further investigate bacterial mechanisms to induce mRNA expression of klf2 in host cells. Specifically, we asked if the ability to induce klf2 mRNA is specific for macrophage-like cells, if it is specific to Yersinia, which bacterial proteins are capable of inducing klf2 mRNA, and how exactly bacterial toxins mediate sustained klf2 expression.
MATERIALS AND METHODS
Bacteria and mutants.
Cell lines and bacteria employed are listed in Tables 1 and 2. For generation of P. aeruginosa exoS mutants, the exoS gene of strain PAO1 was subcloned into pCR2.1-Topo (Invitrogen), leading to the plasmid pTexoS (30). The Ω fragment from plasmid pHP45-Ω, conferring resistance to streptomycin and spectinomycin (48), was ligated into the unique HincII restriction site of exoS. The disrupted exoS gene was cloned into the mobilizable suicide vector pEX18Ap, carrying the counterselectable sacB marker (23). The resulting plasmid, pEXexoS::Ω, was conjugated into strains P. aeruginosa PAO1 and PAK by triparental mating with Escherichia coli(pRK2013). Transconjugants were selected on Luria-Bertani (LB) agar plates containing 500 μg/ml carbenicillin. Sucrose-resistant exoS mutant strains with a carbenicillin-sensitive and streptomycin-resistant phenotype were selected on Pseudomonas isolation agar containing 5% sucrose. PAO1 and PAK exoS mutants were confirmed by Southern hybridization (data not shown) and Western blotting using polyclonal ExoS antibody. Recombinant six-His-tagged EDIN-B toxin was a gift from M. Aepfelbacher (University of Hamburg, Germany), and recombinant C. botulinum C3 toxin was a gift from Stefan Linder (Ludwig Maximilians University, Munich, Germany). Recombinant six-His-tagged SycE protein was a gift from G. Wilharm (University of Munich, Germany).
TABLE 1.
Cell lines used in this study
| Cell line | Tissue of origin | Source (strain) or reference |
|---|---|---|
| J774A.1 | Mouse macrophages | ATCC TIB-67 |
| P388D1 | Mouse macrophages | ATCC CCL-46 |
| RAW264.7 | Mouse macrophages | ATCC TIB-71 |
| ANA-1 | Mouse macrophages | 12 |
| U-937 | Human histiocytic lymphoma | ATCC CRL-1593.2 |
| Jurkat | Human T-cell acute lymphoblastic leukemia | 56 |
| Daudi | Human B-cell lymphoma | ATCC CCL-213 |
| HeLa | Human cervix adenocarcinoma | ATCC CCL-2 |
| AGS | Human gastric adenocarcinoma | ATCC CRL-1739 |
| MKN-28 | Human gastric adenocarcinoma | 28 |
TABLE 2.
Bacterial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| Yersinia strains | ||
| WA-C | Plasmidless derivative of WA(pYV) | 21 |
| WA(pYV) | Y. enterocolitica O:8 | 21 |
| 8081 | Y. enterocolitica O:8 | 63 |
| WA(pTTS, pYopT) | WA-C derivative translocating YopT only | 66 |
| WA(pTTS, pYopT C139S) | WA-C derivative translocating a catalytically inactive YopT | 35, 60 |
| WA(pTTS, pYopE) | WA-C derivative translocating YopE only | 66 |
| WA(pTTS, pYopO) | WA-C derivative translocating YopO only | 66 |
| WA(pYVΔYopT) | YopT-deficient WA(pYV) derivative | 67 |
| 534 | Y. pseudotuberculosis clinical isolate | Max von Pettenkofer Institute clinical strain collection |
| 591 | Y. pseudotuberculosis clinical isolate | Max von Pettenkofer Institute clinical strain collection |
| 601 | Y. pseudotuberculosis clinical isolate | Max von Pettenkofer Institute clinical strain collection |
| 686 | Y. pseudotuberculosis clinical isolate | Max von Pettenkofer Institute clinical strain collection |
| Other strains | ||
| Enteropathogenic E. coli E2348/96 | 19 | |
| Enterohemorrhagic E. coli O157:EDL933 | 19 | |
| Enterohemorrhagic E. coli 413/89-1 | 74 | |
| Citrobacter rodentium | 17 | |
| Citrobacter freundii | ATCC 29219 | |
| Listeria monocytogenes EGD | ATCC BAA-697 | |
| Helicobacter pylori P12 | 55 | |
| Helicobacter pylori TIGR | 65 | |
| Campylobacter jejuni C31 | Clinical isolate | R. Haas, Munich, Germany |
| Campylobacter jejuni C63 | 20 | |
| Campylobacter jejuni C64 | 20 | |
| Hafnia alvei 10790 | 29 | |
| Shigella flexneri M90T | 53 | |
| Pseudomonas aeruginosa PAO1 | 64 | |
| Pseudomonas aeruginosa PAK | 16 | |
| Pseudomonas aeruginosa PA103 | 16 | |
| PAO1ΔpcrD | TTSS-deficient derivative of PAO1 | 26 |
| PAO1ΔexoS | exoS-deficient derivative of PAO1 | This study |
| PAKΔpcrD | TTSS-deficient derivative of PAK | 26 |
| PAKΔexoS | exoS-deficient derivative of PAK | This study |
| Staphylococcus aureus 57 | Clinical isolate | M. Aepfelbacher, Hamburg, Germany |
| Staphylococcus aureus Newman | ATCC 25904 | |
| Salmonella enterica serovar Typhimurium SL1344 | 25 |
Cell culture and bacterial infection.
Dendritic cells (DCs) were generated from bone marrows of C57BL/6 mice as described previously (37). Cell lines were cultured in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Unless noted otherwise, stationary-phase overnight bacterial cultures were diluted 1:10 in fresh LB medium and grown to an optical density at 600 nm of 0.5 to 0.6 (about 6 × 108 CFU/ml). Cells were infected at multiplicities of infection of 100:1 and 10:1. Campylobacter jejuni strains were grown on Columbia blood agar plates (Becton Dickinson) in 5% CO2 and restreaked every 48 h. For infections, bacteria were harvested from agar plates and diluted in Dulbecco's modified Eagle's medium. Helicobacter pylori strains were grown on GC agar plates (Doenitz Prolab, Augsburg, Germany) supplemented with 8% horse serum, a complex vitamin mixture, vancomycin (10 mg/liter), trimethoprim (5 mg/liter), and nystatin (1 mg/liter) and were incubated for 1 to 2 days in a microaerobic atmosphere (85% N2, 10% CO2, 5% O2) at 37°C. Stationary-phase Yersinia cultures grown overnight at 27°C were diluted 1:10 in fresh LB medium and shifted to 37°C for 2 h, and infections were performed as described above. Recombinant EDIN-B and SycE were applied at 60 μg/ml overnight, and recombinant C. botulinum C3 toxin was applied at 25 μg/ml overnight. In some experiments, cells were incubated for 2 hours to overnight with 1 to 10 μM Rho kinase inhibitor H1152 (Calbiochem).
Generation of lentivirus and transduction of AGS cells.
The BLOCK-iT lentiviral RNA interference (RNAi) expression system (Invitrogen) was used according to the manufacturer's specifications. Briefly, short hairpin RNA (shRNA) oligonucleotides were selected with BLOCK-iT RNAi Designer software from Invitrogen. To avoid sequence homology with other genes, BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/) was performed. The following target mRNA sequences were used to design shRNAs (starting positions are noted in parentheses): rhoA (687), GCCGGTGAAACCTGAAGAAGG; rhoB (841), GCATCCAAGCCTACGACTACC; rhoC (630), GCCTCCAGGTCCGCAAGAACA; rhpn1 (384), GCTGATCTCAGTGCACTTTGG; cit (685), GCGTTCATCTGATGGGATACG; rok2 (2228), GCAGCTGGAATCTAACAATAG; and ktn1 (1390), GCAGATGAAGTTTCAGCAAGT. A random-sequence shRNA was used as a control. Oligonucleotides were annealed and then cloned into the entry vector pENTR/U6. TOP10 competent E. coli cells were transformed, and colonies were selected on LB plates containing 50 μg/ml kanamycin. After sequence verification, the U6 RNAi cassette was transferred by LR recombination to the pLENTI6/BLOCK-iT-DEST plasmid. A lentiviral stock was produced by cotransfection of 293FT cells with the resulting expression construct and ViraPower packaging mix, using the FuGene 6 transfection reagent (Roche, Mannheim, Germany). At 48 h posttransfection, viral supernatants were harvested and filtered. AGS cells were infected with lentiviruses containing the packaged shRNAs. Gene transfer was detected using a lentiviral vector containing the gfp marker gene. Stable cells were generated by selection with 10 μg/ml blasticidin for 10 days.
Real-time reverse transcription-PCR.
Total cellular RNA was isolated with Trizol RNA isolation reagent (Invitrogen) according to the manufacturer's recommendations. After random-hexamer-primed first-strand cDNA synthesis (Superscript II; Invitrogen), real-time PCR was performed in an ABI Prism 7000 sequence detection system (Applied Biosystems) (with detection of hprt, klf2, rhoA, rhoB, rhoC, rok2, rhpn1, cit, and ktn1). Amplification was done for 40 cycles, using an initial denaturation at 95°C for 10 min followed by cycles of 95°C for 15 s and 60°C for 1 min. Primers and fluorescent probes for murine klf2 and hprt were described previously (24). Human klf2 and hprt PCR assay kits were purchased from Applied Biosystems (Assays-on-Demand). A Universal ProbeLibrary (Roche, Mannheim, Germany) was used in combination with the Universal ProbeLibrary assay design center to design specific primers and probes for rhoA, rhoB, rhoC, rhpn1, rok, cit, and ktn1. Gene expression levels were recorded relative to the hprt housekeeping control as follows: E = 2−ΔCT(E = gene expression value; ΔCT = difference in crossing points between threshold cycles for hprt and the gene of interest). All PCR experiments were performed at least four times, and standard deviations were calculated and displayed as error bars. For graphical display, the maximum or Y. enterocolitica-elicited klf2 mRNA expression value in every graph was given an arbitrary value of 10, and the remaining values and standard deviations were scaled accordingly, graphwise. Statistical analysis was performed with Student's t test, as implemented in Microsoft Excel, considering P values of ≤0.05 statistically significant.
RESULTS
Yersinia enterocolitica, Yersinia pseudotuberculosis, Pseudomonas aeruginosa, and Staphylococcus aureus induce klf2 mRNA in macrophages.
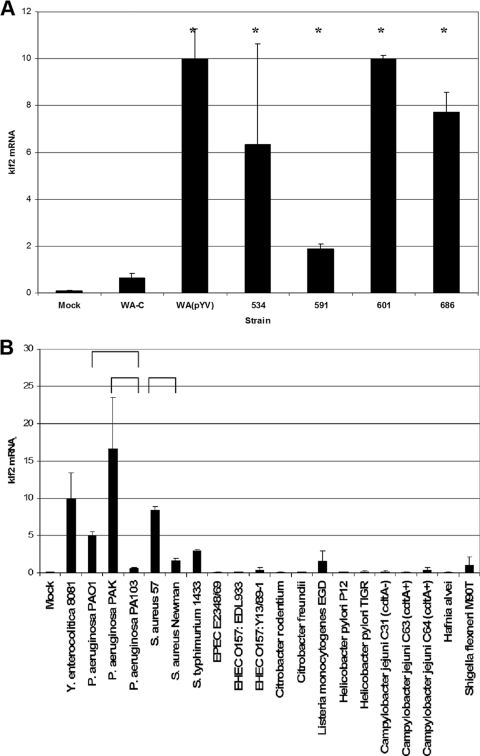
To confirm and extend our previous analyses (24), we first evaluated whether bacteria other than Yersinia enterocolitica were able to induce klf2 mRNA expression in J774A.1 cells (Fig. 1). We first examined a panel of clinical Y. pseudotuberculosis isolates, which demonstrated that these strains are also able to induce klf2 mRNA expression in host cells (Fig. 1A). Next, we screened a panel of phylogenetically diverse bacterial strains, many of whom are known to possess systems for delivery of bacterial proteins into the cytoplasm of host cells (Fig. 1B). In addition to Yersinia, Pseudomonas aeruginosa strains PAO1 and PAK, but not strain PA103, and Staphylococcus aureus strain 57, but not strain Newman, induced klf2 mRNA. Salmonella enterica serovar Typhimurium strain SL1344 induced only low levels of klf2 mRNA in J774A.1 cells. No significant klf2 induction could be demonstrated for strains of enterohemorrhagic Escherichia coli, enteropathogenic Escherichia coli, Citrobacter spp., Listeria monocytogenes, Helicobacter pylori, Campylobacter jejuni, Hafnia alvei, or Shigella flexneri. Similar results were obtained with multiplicities of infection of 10:1 and 100:1 and after 2 and 4 h of infection (not shown).
FIG. 1.
klf2 mRNA induction in J774A.1 cells by different gram-positive and gram-negative bacteria (detailed in Table 2). The Y. enterocolitica-elicited klf2 level was scaled to an arbitrary value of 10. Levels of klf2 mRNA were recorded relative to the HPRT housekeeping control. Error bars represent 1 standard deviation from the mean. Mock, uninfected cells. Horizontal brackets or asterisks indicate statistically significant (t test; P ≤ 0.05) differences in klf2 mRNA expression levels. (A) klf2 induction by different clinical isolates of Y. pseudotuberculosis. (B) klf2 induction by a collection of phylogenetically diverse bacteria.
YopT from Y. enterocolitica, ExoS from P. aeruginosa, EDIN-B from S. aureus, and C3 toxin from C. botulinum induce klf2 mRNA expression.
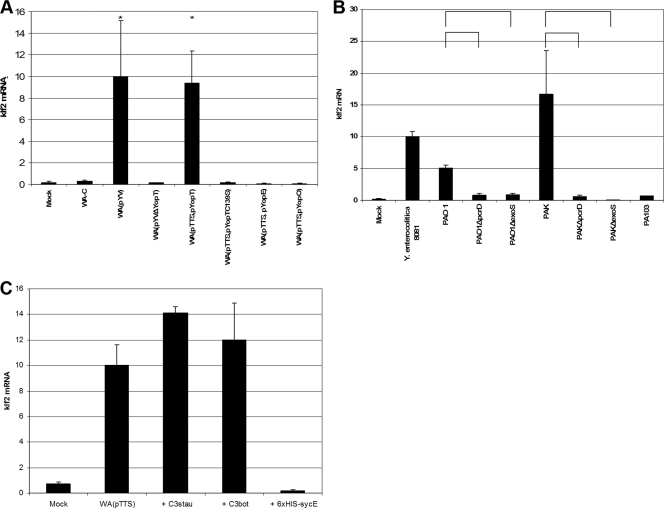
To identify the bacterial protein responsible for klf2 induction, we screened several mutant strains of Y. enterocolitica and P. aeruginosa for klf2 induction in host cells. As displayed in Fig. 2A, translocation of YopT alone induced klf2 mRNA expression, while the yopT deletion mutant WA(pYVΔYopT) did not induce significant klf2 mRNA levels in J774A.1 cells. A catalytically inactive YopT C139S mutant or strains secreting only YopE or YopO proteins were not able to induce klf2 mRNA expression.
FIG. 2.
Identification of bacterial klf2-inducing proteins. (A) klf2 mRNA expression in J774A.1 cells after infection with different Y. enterocolitica mutant strains (detailed in Table 2). The graphical display is as described in the legend to Fig. 1. *, statistically significant (t test; P ≤ 0.05) induction of klf2 mRNA compared to infection with strain WA-C. (B) klf2 mRNA levels in J774A.1 cells after infection with different P. aeruginosa wild-type and mutant strains (see Table 2). The graphical display is as described in the legend to Fig. 1. Horizontal brackets indicate statistically significant (t test; P ≤ 0.05) differences in klf2 mRNA expression levels. (C) klf2 induction by recombinant EDIN-B or C. botulinum C3 toxin in J774A.1 cells. The graphical display is as described in the legend to Fig. 1. sycE, irrelevant protein used as negative control.
For P. aeruginosa, strains PAO1 and PAK, but not strain PA103, induced klf2 mRNA in J774A.1 cells. klf2 induction was dependent on the TTSS, as pcrD mutant strains PAO1ΔpcrD and PAKΔpcrD, which harbor an impaired TTSS, were not able to induce klf2 mRNA anymore (Fig. 2B). Together, these findings suggest ExoS, a type III secreted toxin produced by strains PAO1 and PAK, but not by strain PA103, as a likely candidate mediating klf2 induction. Consistently, exoS deletion mutants PAO1ΔexoS and PAKΔexoS were no longer able to induce klf2 mRNA (Fig. 2B).
For S. aureus, strain 57 (a virulent clinical isolate [13]) induced high levels of klf2 mRNA in J774A.1 cells, while the S. aureus reference strain Newman did not. One important difference is that strain 57, but not Newman, produces EDIN-B, a C3-like ADP-ribosyltransferase which modifies Rho GTPases (M. Aepfelbacher, unpublished observations). We incubated J774A.1 cells overnight with purified recombinant EDIN-B, and this treatment indeed resulted in the induction of high levels of klf2 mRNA compared to treatment with an irrelevant protein (the Y. enterocolitica YopE chaperone SycE) (Fig. 2C). This finding could be confirmed by using the prototypical Rho-specific ADP ribosyltransferase, C3 toxin from C. botulinum (4), which also induced high levels of klf2 mRNA in host cells (Fig. 2C).
YopT induces klf2 mRNA in cell lines of macrophage and epithelial origin.
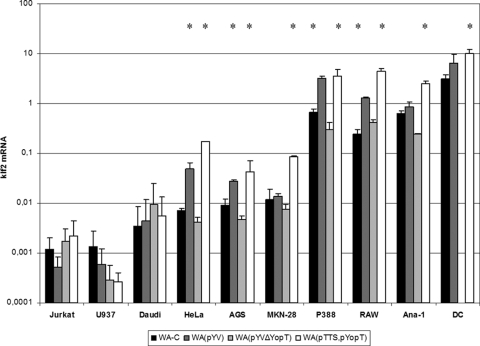
We next wondered whether induction of klf2 mRNA upon bacterial infection could occur in cells other than J774A.1 cells. We tested a panel of mouse and human cell lines of macrophage, lymphocyte, and epithelial origin for klf2 induction upon infection with Y. enterocolitica (Fig. 3). klf2 mRNA was induced in macrophage-like (J774A.1, RAW264.7, P388D.1, and Ana-1), squamous epithelial (HeLa), and gastric epithelial (AGS and MKN-28) cells by strain WA(pTTS, pYopT). We also found klf2 mRNA expression induced in bone marrow-derived DCs (Fig. 3). Thus, YopT is the only effector Yop required to induce klf2 in macrophage/DC and epithelial cell lines, although other bacterial components may also contribute. In contrast, B (Daudi) or T (Jurkat) lymphocytes or histiocytes (U937) did not induce klf2 mRNA in response to Y. enterocolitica (Fig. 3). It was shown earlier, however, that Yops are effectively translocated into these cells (9, 73). We concluded that YopT-mediated induction of klf2 mRNA expression takes place mainly in macrophages, DCs, and epithelial cells.
FIG. 3.
klf2 mRNA levels induced by wild-type Y. enterocolitica and YopT mutants in cell lines derived from different human or mouse tissues. The graphical display is as described in the legend to Fig. 1. Note the logarithmic scale. *, statistically significant (t test; P ≤ 0.05) induction of klf2 mRNA compared to infection of the respective cell line with strain WA-C. DC, mouse bone marrow-derived DCs.
shRNA-mediated knockdown of RhoA induces klf2 mRNA expression.
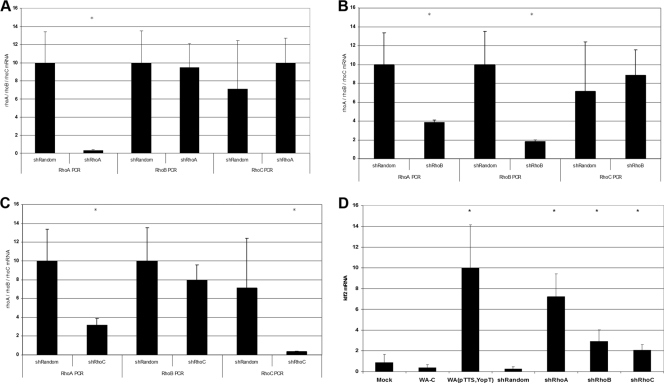
As stated above, all bacterial toxins identified to induce klf2 mRNA expression interfere with small GTPases of the rho family, with the C3 toxins interacting specifically with RhoA, RhoB, and RhoC (4). To identify the Rho protein responsible for klf2 induction, we generated stable shRNA-expressing cell lines for each of the Rho proteins individually. As shown in Fig. 4A to C, rhoA, rhoB, and rhoC shRNA-expressing cells showed substantial downregulation of the corresponding mRNA levels relative to those in control shRNA-expressing cells. However, rhoB and rhoC shRNAs also had modest effects on rhoA mRNA expression levels: rhoB shRNA resulted in a 62% reduction of the rhoA expression level, and rhoC shRNA resulted in a 68% reduction. This problem occurred with several different shRNA constructs. As displayed in Fig. 4D, however, klf2 mRNA expression was highest in rhoA shRNA-transduced cells, while rhoB shRNA- and rhoC shRNA-transduced cells expressed lower levels of klf2 mRNA. This suggests that rhoA is involved in the regulation of klf2 mRNA expression levels.
FIG. 4.
Effect of shRNA-mediated knockdown of Rho GTPases on klf2 expression levels. (A) rhoA, rhoB, and rhoC mRNA expression levels in rhoA shRNA-transduced cells. (B) rhoA, rhoB, and rhoC mRNA expression levels in rhoB shRNA-transduced cells. (C) rhoA, rhoB, and rhoC mRNA expression levels in rhoC shRNA-transduced cells. (D) Effect of Rho GTPase knockdown on klf2 mRNA expression level.
The Rho effector protein rhophilin 1 is involved in klf2 mRNA induction.
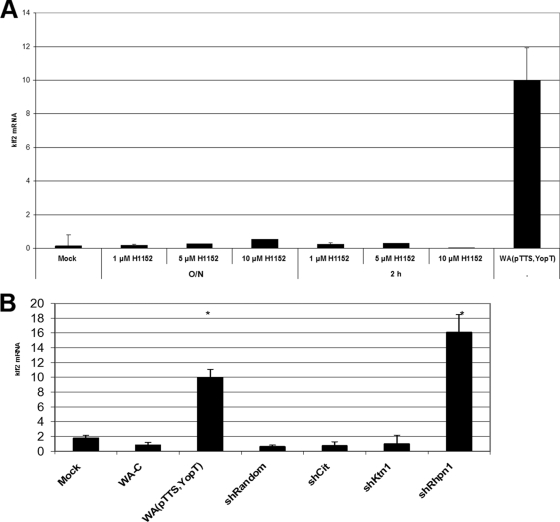
Rho proteins exert their cellular function by binding, in their active GTP-bound state, to Rho effector proteins. To better characterize the RhoA-dependent signaling cascade regulating klf2 expression, we first investigated the role of the well-characterized effector protein Rho kinase. Treatment of J774A.1 cells with different concentrations of the specific, cell-permeating inhibitor H1152 for different times, however, did not result in klf2 induction (Fig. 5A). Similar results were obtained with the less specific inhibitor H1077, which also inhibits the Rho effector proteins PRK1, PRK2, and MSK1 (not shown). Thus, Rho kinase seems not to be involved in regulation of klf2 expression levels.
FIG. 5.
Impact of Rho effector proteins on klf2 expression level. (A) klf2 mRNA levels in J774A.1 cells treated with the ROCK inhibitor H1152 for the indicated times (O/N, overnight) at the indicated concentrations. Similar results were obtained with the inhibitor H1077. (B) Impact of indicated Rho effector protein knockdown by shRNA on klf2 expression level.
We next investigated the role of several other Rho effector proteins by shRNA-mediated knockdown. shRNAs specific for citron, rho kinase 2, kinectin 1, and rhophilin 1 all mediated effective knockdowns of their respective target mRNAs, as measured by real-time reverse transcription-PCR (not shown). Only rhophilin 1 shRNA-transduced cells, however, induced substantial levels of klf2 mRNA (Fig. 5B). Importantly, the specificity of the Rhpn-1 small interfering RNA was controlled by extensive manual BLAST searches, which demonstrated no occurrence of the small interfering RNA sequence in any other mRNA transcript (data not shown). Thus, we suggest that a signaling axis via RhoA and rhophilin 1 regulates klf2 mRNA levels in response to bacterial infection.
DISCUSSION
Recent large-scale gene expression studies identified a number of genes induced by the action of Yersinia Yops in macrophages (24, 54) and epithelial cells (8). We have identified the transcription factor klf2 as one gene with particularly pronounced induction in J774A.1 cells in response to infection with virulent, Yop-translocating yersiniae. Given the immunosuppressive action of klf2 in a variety of cell types, it seems plausible that klf2 induction constitutes a novel immunosuppressive strategy of bacteria.
In the case of infection with Yersinia, the Rho-inactivating cysteine protease YopT mediates sustained klf2 expression in host cells. In the absence of YopT, klf2 is only transiently expressed. Yersinia possesses a second Rho-GTPase-inactivating protein, YopE, which acts as a GAP for Rho, Rac, and Cdc42 in vitro (68). However, after infection of living cells with YopE-translocating yersiniae, YopE seems to interact with a much smaller range of Rho proteins: it acts primarily on Rac rather than on Rho or Cdc42 in vivo, and it inhibits Cdc42-mediated Rac activation induced by bradykinin but not direct activation of Rac by sphingosine-1-phosphate (6). Consistently, YopT and YopE have been shown to differently affect the cytoskeleton and phagocytic capacity of DCs (1). Compared to P. aeruginosa ExoS, it should be noted that Yersinia ExoS contains an additional ADP ribosyltransferase domain inactivating small GTPases (49) and that the GAP domain of ExoS interacts with a much broader range of host proteins than YopE does (31). Our results are consistent with the targeting of RhoA by ExoS ADP ribosyltransferase, which has not been shown previously. These differences in specificity may explain differences in the klf2-inducing capacities of YopT, YopE, and ExoS.
Knowledge about the mechanisms regulating expression of klf2 is scarce. Tumor necrosis factor receptor-associated factor 2 (TRAF2) and signaling via p38 mitogen-activated protein kinase (MAPK), but not tumor necrosis factor signaling, have been suggested to regulate klf2 mRNA expression (34). In both human and mouse cells, critical regions within the klf2 promoter have been identified (27, 57), and heterogeneous nuclear ribonucleoproteins as well as acetyltransferases have been shown to bind to these regions (3). Other results suggest that statin-mediated klf2 induction in endothelial cells is a result of interference with Rho signaling (58), although the precise mechanism remains obscure. Our results show that a signaling cascade from RhoA via rhophilin 1 regulates klf2 expression levels. Since it is the inactivation of RhoA and rhophilin 1 which induces klf2 mRNA expression, this signaling cascade likely suppresses klf2 expression.
To date, several links between Rho and transcriptional regulation have been described. Rho is required for signaling to Srf by several stimuli (22). However, a direct effector protein of Rho acting on Srf or the sre remains to be elucidated. The best-characterized link between Rho and the serum response is via the Srf cofactor Mal. Mal functions as a cytoplasmic sensor for depolymerized actin and is translocated to the nucleus upon actin polymerization, where it serves as a cofactor for Srf (39). Rho has also been shown to indirectly induce the transcriptional activity of NF-κB by phosphorylation of the inhibitor IκBα, enabling active NF-κB to translocate to the nucleus (47). A third link between Rho and transcriptional regulation has been made by demonstrating that Rho is able to stimulate c-jun expression via activation of ERK6 (p38γ), a recently identified MAPK (38). However, all of these findings identify Rho as a potent activator, rather than a suppressor, of gene transcription. As far as we are aware, only one other gene besides klf2 has been described as being suppressed by Rho: expression of the cyclin-dependent kinase inhibitor cdkn1a (encoding p21Waf1/Cip1) is induced by signaling through activated Ras, and this induction can be inhibited by RhoA by action on the cdkn1a promoter (45). Data concerning the rho effector responsible for inhibition of cdkn1a expression are conflicting: expression of cdkn1a protein could be induced by ROCK inhibition in phorbol myristate acetate-treated human erythromyeloblast D2 cells (33) but not in Ras-V12-transformed Swiss 3T3 cells (52). However, we do not think that any of these signaling pathways is responsible for YopT-mediated regulation of klf2 expression, for the following reasons: first, we did not find any influence of YopT on expression of cdkn1a in J774A.1 cells; second, we could not demonstrate an effect of SRF activity on klf2 expression levels (data not shown); third, by performing experiments with the actin-depolymerizing agents latrunculin B and cytochalasin D, we could show that the actin polymerization status is not involved in regulating klf2 expression (data not shown); and fourth, we found rhophilin 1 rather than ROCK to be involved in klf2 regulation.
Rhophilin 1 was first described in 1996, when it was detected in a yeast two-hybrid screen with Rho as bait. It interacts strongly with GTP-bound RhoA, less with RhoB, and little with RhoC (71). Rhpn1 is expressed in various tissues (41) and is highly expressed in the mouse testis, where it interacts with ropporin (18). Other putative binding partners have been identified by yeast two-hybrid screens. Among these are Trim37, Krt15, Cnksr1, Efemp2, and Ndp52 (51). Rhpn1 contains several protein-protein interaction motifs, such as HR1, a central BRO1 domain, and a C-terminal PDZ domain (46). The HR1 domain or Rho binding domain was first described as part of protein kinase PRK1, which binds RhoA (15). It is found in a range of signaling proteins, including rhotekin and PRK2, and is required for GTPase binding (50, 71). The exact molecular functions of BRO1 domains are not known. They are required for cargo protein deubiquitination and play a role in endosome metabolism (36). PDZ domains are protein interaction domains that are often found in multidomain scaffolding proteins that organize intracellular signaling at particular subcellular locations (62). Taken together, the presence of different protein interaction motifs suggest that rhophilin 1 may serve as a signaling protein. Thus, the following sequence of events seems likely in YopT-modified regulation of klf2 expression. YopT cleaves RhoA from its geranylgeranylated membrane anchor, which must not necessarily inactivate RhoA (72) but could also change the subcellular localization of RhoA sufficiently to inhibit effector protein binding. However, the exact subcellular localization of rhophilin 1 in macrophages is unclear, so it remains to be determined whether YopT-mediated RhoA inactivation or compartmentalization is responsible for inhibiting the RhoA-rhophilin 1 interaction. How exactly rhophilin 1 then regulates klf2 mRNA expression remains to be determined.
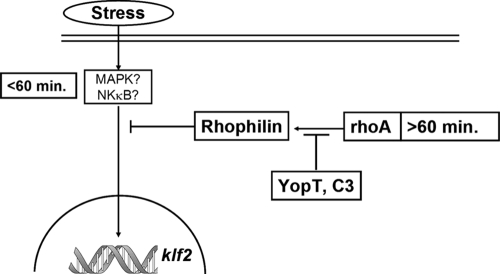
We described earlier that klf2 is also induced transiently (≤1 h after infection) by Yop-devoid yersiniae. Translocation of YopT results in a sustained (≥2 h) expression of klf2 mRNA (24). Together with the data presented here, this suggests the following model of klf2 regulation in the context of bacterial infection (Fig. 6). Immediately after bacterial contact, host cells induce klf2 by an as yet uncharacterized signaling cascade, but MAPK or NF-κB signaling may be involved. This early induction of klf2 may be viewed as a physiological regulatory “loop” to prevent overwhelming inflammatory activation of the host cell. Two hours after infection, signaling via RhoA and rhophilin 1 suppresses klf2 expression. Bacteria mediate long-term expression of klf2 by suppressing this inhibitory action of RhoA via Rho-inactivating enzymes. As noted above, this may constitute a novel immunosuppressive strategy. Future studies will identify the relevance of klf2 induction for infections in vivo as well as the precise role of rhophilin 1 in suppression of transcriptional responses to bacterial infection.
FIG. 6.
Proposed model of klf2 regulation in the context of bacterial infection. Immediately after bacterial contact, host cells induce klf2 by an as yet uncharacterized signaling cascade; however, MAPK or NF-κB signaling may be involved. Two hours after infection, signaling via RhoA and rhophilin 1 suppresses klf2 expression. Bacteria mediate long-term expression of klf2 by suppressing this inhibitory action of RhoA via Rho-inactivating enzymes.
Acknowledgments
This work was supported by the German Federal Ministry of Education and Research under the auspices of the National Genome Research Network, NGFN 2 (grant no. IE-S31T10).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 28 September 2009.
REFERENCES
- 1.Adkins, I., M. Koberle, S. Grobner, E. Bohn, I. B. Autenrieth, and S. Borgmann. 2007. Yersinia outer proteins E, H, P, and T differentially target the cytoskeleton and inhibit phagocytic capacity of dendritic cells. Int. J. Med. Microbiol. 297:235-244. [DOI] [PubMed] [Google Scholar]
- 2.Aepfelbacher, M., C. Trasak, G. Wilharm, A. Wiedemann, K. Trulzsch, K. Krauss, P. Gierschik, and J. Heesemann. 2003. Characterization of YopT effects on Rho GTPases in Yersinia enterocolitica-infected cells. J. Biol. Chem. 278:33217-33223. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad, N., and J. B. Lingrel. 2005. Kruppel-like factor 2 transcriptional regulation involves heterogeneous nuclear ribonucleoproteins and acetyltransferases. Biochemistry 44:6276-6285. [DOI] [PubMed] [Google Scholar]
- 4.Aktories, K., and J. T. Barbieri. 2005. Bacterial cytotoxins: targeting eukaryotic switches. Nat. Rev. Microbiol. 3:397-410. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, K. P., C. B. Kern, S. C. Crable, and J. B. Lingrel. 1995. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Kruppel-like factor: identification of a new multigene family. Mol. Cell. Biol. 15:5957-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andor, A., K. Trulzsch, M. Essler, A. Roggenkamp, A. Wiedemann, J. Heesemann, and M. Aepfelbacher. 2001. YopE of Yersinia, a GAP for Rho GTPases, selectively modulates Rac-dependent actin structures in endothelial cells. Cell. Microbiol. 3:301-310. [DOI] [PubMed] [Google Scholar]
- 7.Barz, C., T. N. Abahji, K. Trulzsch, and J. Heesemann. 2000. The Yersinia Ser/Thr protein kinase YpkA/YopO directly interacts with the small GTPases RhoA and Rac-1. FEBS Lett. 482:139-143. [DOI] [PubMed] [Google Scholar]
- 8.Bohn, E., S. Muller, J. Lauber, R. Geffers, N. Speer, C. Spieth, J. Krejci, B. Manncke, J. Buer, A. Zell, and I. B. Autenrieth. 2004. Gene expression patterns of epithelial cells modulated by pathogenicity factors of Yersinia enterocolitica. Cell. Microbiol. 6:129-141. [DOI] [PubMed] [Google Scholar]
- 9.Boyd, A. P., N. Grosdent, S. Totemeyer, C. Geuijen, S. Bleves, M. Iriarte, I. Lambermont, J. N. Octave, and G. R. Cornelis. 2000. Yersinia enterocolitica can deliver Yop proteins into a wide range of cell types: development of a delivery system for heterologous proteins. Eur. J. Cell Biol. 79:659-671. [DOI] [PubMed] [Google Scholar]
- 10.Buckley, A. F., C. T. Kuo, and J. M. Leiden. 2001. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat. Immunol. 2:698-704. [DOI] [PubMed] [Google Scholar]
- 11.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘type III’ weaponry. Nat. Rev. Mol. Cell Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 12.Cox, G. W., B. J. Mathieson, L. Gandino, E. Blasi, D. Radzioch, and L. Varesio. 1989. Heterogeneity of hematopoietic cells immortalized by v-myc/v-raf recombinant retrovirus infection of bone marrow or fetal liver. J. Natl. Cancer Inst. 81:1492-1496. [DOI] [PubMed] [Google Scholar]
- 13.Czech, A., T. Yamaguchi, L. Bader, S. Linder, K. Kaminski, M. Sugai, and M. Aepfelbacher. 2001. Prevalence of Rho-inactivating epidermal cell differentiation inhibitor toxins in clinical Staphylococcus aureus isolates. J. Infect. Dis. 184:785-788. [DOI] [PubMed] [Google Scholar]
- 14.Dekker, R. J., S. van Soest, R. D. Fontijn, S. Salamanca, P. G. de Groot, E. VanBavel, H. Pannekoek, and A. J. Horrevoets. 2002. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2). Blood 100:1689-1698. [DOI] [PubMed] [Google Scholar]
- 15.Flynn, P., H. Mellor, R. Palmer, G. Panayotou, and P. J. Parker. 1998. Multiple interactions of PRK1 with RhoA. Functional assignment of the Hr1 repeat motif. J. Biol. Chem. 273:2698-2705. [DOI] [PubMed] [Google Scholar]
- 16.Frank, D. W., G. Nair, and H. P. Schweizer. 1994. Construction and characterization of chromosomal insertional mutations of the Pseudomonas aeruginosa exoenzyme S trans-regulatory locus. Infect. Immun. 62:554-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel, G., A. D. Phillips, M. Novakova, H. Field, D. C. Candy, D. B. Schauer, G. Douce, and G. Dougan. 1996. Intimin from enteropathogenic Escherichia coli restores murine virulence to a Citrobacter rodentium eaeA mutant: induction of an immunoglobulin A response to intimin and EspB. Infect. Immun. 64:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujita, A., K. Nakamura, T. Kato, N. Watanabe, T. Ishizaki, K. Kimura, A. Mizoguchi, and S. Narumiya. 2000. Ropporin, a sperm-specific binding protein of rhophilin, that is localized in the fibrous sheath of sperm flagella. J. Cell Sci. 113:103-112. [DOI] [PubMed] [Google Scholar]
- 19.Gansheroff, L. J., M. R. Wachtel, and A. D. O'Brien. 1999. Decreased adherence of enterohemorrhagic Escherichia coli to HEp-2 cells in the presence of antibodies that recognize the C-terminal region of intimin. Infect. Immun. 67:6409-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebert, B., W. Fischer, E. Weiss, R. Hoffmann, and R. Haas. 2003. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 301:1099-1102. [DOI] [PubMed] [Google Scholar]
- 21.Heesemann, J., and R. Laufs. 1983. Construction of a mobilizable Yersinia enterocolitica virulence plasmid. J. Bacteriol. 155:761-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, C. S., J. Wynne, and R. Treisman. 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell 81:1159-1170. [DOI] [PubMed] [Google Scholar]
- 23.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann, R., K. Van Erp, K. Trulzsch, and J. Heesemann. 2004. Transcriptional responses of murine macrophages to infection with Yersinia enterocolitica. Cell. Microbiol. 6:377-390. [DOI] [PubMed] [Google Scholar]
- 25.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 26.Hornef, M. W., A. Roggenkamp, A. M. Geiger, M. Hogardt, C. A. Jacobi, and J. Heesemann. 2000. Triggering the ExoS regulon of Pseudomonas aeruginosa: a GFP-reporter analysis of exoenzyme (Exo) S, ExoT and ExoU synthesis. Microb. Pathog. 29:329-343. [DOI] [PubMed] [Google Scholar]
- 27.Huddleson, J. P., S. Srinivasan, N. Ahmad, and J. B. Lingrel. 2004. Fluid shear stress induces endothelial KLF2 gene expression through a defined promoter region. Biol. Chem. 385:723-729. [DOI] [PubMed] [Google Scholar]
- 28.Imanishi, K., K. Yamaguchi, M. Suzuki, S. Honda, N. Yanaihara, and K. Abe. 1989. Production of transforming growth factor-alpha in human tumour cell lines. Br. J. Cancer 59:761-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janda, J. M., S. L. Abbott, and M. J. Albert. 1999. Prototypal diarrheagenic strains of Hafnia alvei are actually members of the genus Escherichia. J. Clin. Microbiol. 37:2399-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaufman, M. R., J. Jia, L. Zeng, U. Ha, M. Chow, and S. Jin. 2000. Pseudomonas aeruginosa mediated apoptosis requires the ADP-ribosylating activity of exoS. Microbiology 146:2531-2541. [DOI] [PubMed] [Google Scholar]
- 31.Krall, R., J. Sun, K. J. Pederson, and J. T. Barbieri. 2002. In vivo Rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, C. T., M. L. Veselits, K. P. Barton, M. M. Lu, C. Clendenin, and J. M. Leiden. 1997. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 11:2996-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai, J. M., S. Wu, D. Y. Huang, and Z. F. Chang. 2002. Cytosolic retention of phosphorylated extracellular signal-regulated kinase and a Rho-associated kinase-mediated signal impair expression of p21(Cip1/Waf1) in phorbol 12-myristate-13-acetate-induced apoptotic cells. Mol. Cell. Biol. 22:7581-7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Y., J. Ryan, J. Lewis, M. A. Wani, J. B. Lingrel, and Z. G. Liu. 2003. TRAF2 exerts its antiapoptotic effect by regulating the expression of Kruppel-like factor LKLF. Mol. Cell. Biol. 23:5849-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Locher, M., B. Lehnert, K. Krauss, J. Heesemann, M. Groll, and G. Wilharm. 2005. Crystal structure of the Yersinia enterocolitica type III secretion chaperone SycT. J. Biol. Chem. 280:31149-31155. [DOI] [PubMed] [Google Scholar]
- 36.Luhtala, N., and G. Odorizzi. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutz, M. B., N. Kukutsch, A. L. Ogilvie, S. Rossner, F. Koch, N. Romani, and G. Schuler. 1999. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods 223:77-92. [DOI] [PubMed] [Google Scholar]
- 38.Marinissen, M. J., M. Chiariello, and J. S. Gutkind. 2001. Regulation of gene expression by the small GTPase Rho through the ERK6 (p38 gamma) MAP kinase pathway. Genes Dev. 15:535-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miralles, F., G. Posern, A. I. Zaromytidou, and R. Treisman. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329-342. [DOI] [PubMed] [Google Scholar]
- 40.Moreilhon, C., D. Gras, C. Hologne, O. Bajolet, F. Cottrez, V. Magnone, M. Merten, H. Groux, E. Puchelle, and P. Barbry. 2005. Live Staphylococcus aureus and bacterial soluble factors induce different transcriptional responses in human airway cells. Physiol. Genomics 20:244-255. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura, K., A. Fujita, T. Murata, G. Watanabe, C. Mori, J. Fujita, N. Watanabe, T. Ishizaki, O. Yoshida, and S. Narumiya. 1999. Rhophilin, a small GTPase Rho-binding protein, is abundantly expressed in the mouse testis and localized in the principal piece of the sperm tail. FEBS Lett. 445:9-13. [DOI] [PubMed] [Google Scholar]
- 42.Navarro, L., A. Koller, R. Nordfelth, H. Wolf-Watz, S. Taylor, and J. E. Dixon. 2007. Identification of a molecular target for the Yersinia protein kinase A. Mol. Cell 26:465-477. [DOI] [PubMed] [Google Scholar]
- 43.O'Grady, E., H. Mulcahy, C. Adams, J. P. Morrissey, and F. O'Gara. 2007. Manipulation of host Kruppel-like factor (KLF) function by exotoxins from diverse bacterial pathogens. Nat. Rev. Microbiol. 5:337-341. [DOI] [PubMed] [Google Scholar]
- 44.O'Grady, E. P., H. Mulcahy, J. O'Callaghan, C. Adams, and F. O'Gara. 2006. Pseudomonas aeruginosa infection of airway epithelial cells modulates expression of Kruppel-like factors 2 and 6 via RsmA-mediated regulation of type III exoenzymes S and Y. Infect. Immun. 74:5893-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olson, M. F., H. F. Paterson, and C. J. Marshall. 1998. Signals from Ras and Rho GTPases interact to regulate expression of p21Waf1/Cip1. Nature 394:295-299. [DOI] [PubMed] [Google Scholar]
- 46.Peck, J. W., M. Oberst, K. B. Bouker, E. Bowden, and P. D. Burbelo. 2002. The RhoA-binding protein, rhophilin-2, regulates actin cytoskeleton organization. J. Biol. Chem. 277:43924-43932. [DOI] [PubMed] [Google Scholar]
- 47.Perona, R., S. Montaner, L. Saniger, I. Sanchez-Perez, R. Bravo, and J. C. Lacal. 1997. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 11:463-475. [DOI] [PubMed] [Google Scholar]
- 48.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 49.Radke, J., K. J. Pederson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect. Immun. 67:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reid, T., T. Furuyashiki, T. Ishizaki, G. Watanabe, N. Watanabe, K. Fujisawa, N. Morii, P. Madaule, and S. Narumiya. 1996. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J. Biol. Chem. 271:13556-13560. [DOI] [PubMed] [Google Scholar]
- 51.Rual, J. F., K. Venkatesan, T. Hao, T. Hirozane-Kishikawa, A. Dricot, N. Li, G. F. Berriz, F. D. Gibbons, M. Dreze, N. Ayivi-Guedehoussou, N. Klitgord, C. Simon, M. Boxem, S. Milstein, J. Rosenberg, D. S. Goldberg, L. V. Zhang, S. L. Wong, G. Franklin, S. Li, J. S. Albala, J. Lim, C. Fraughton, E. Llamosas, S. Cevik, C. Bex, P. Lamesch, R. S. Sikorski, J. Vandenhaute, H. Y. Zoghbi, A. Smolyar, S. Bosak, R. Sequerra, L. Doucette-Stamm, M. E. Cusick, D. E. Hill, F. P. Roth, and M. Vidal. 2005. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437:1173-1178. [DOI] [PubMed] [Google Scholar]
- 52.Sahai, E., M. F. Olson, and C. J. Marshall. 2001. Cross-talk between Ras and Rho signaling pathways in transformation favours proliferation and increased motility. EMBO J. 20:755-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sansonetti, P. J., D. J. Kopecko, and S. B. Formal. 1982. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect. Immun. 35:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauvonnet, N., B. Pradet-Balade, J. A. Garcia-Sanz, and G. R. Cornelis. 2002. Regulation of mRNA expression in macrophages after Yersinia enterocolitica infection. Role of different Yop effectors. J. Biol. Chem. 277:25133-25142. [DOI] [PubMed] [Google Scholar]
- 55.Schmitt, W., and R. Haas. 1994. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol. Microbiol. 12:307-319. [DOI] [PubMed] [Google Scholar]
- 56.Schneider, U., H. U. Schwenk, and G. Bornkamm. 1977. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer 19:621-626. [DOI] [PubMed] [Google Scholar]
- 57.Schrick, J. J., M. J. Hughes, K. P. Anderson, M. L. Croyle, and J. B. Lingrel. 1999. Characterization of the lung Kruppel-like transcription factor gene and upstream regulatory elements. Gene 236:185-195. [DOI] [PubMed] [Google Scholar]
- 58.Sen-Banerjee, S., S. Mir, Z. Lin, A. Hamik, G. B. Atkins, H. Das, P. Banerjee, A. Kumar, and M. K. Jain. 2005. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation 112:720-726. [DOI] [PubMed] [Google Scholar]
- 59.SenBanerjee, S., Z. Lin, G. B. Atkins, D. M. Greif, R. M. Rao, A. Kumar, M. W. Feinberg, Z. Chen, D. I. Simon, F. W. Luscinskas, T. M. Michel, M. A. Gimbrone, Jr., G. Garcia-Cardena, and M. K. Jain. 2004. KLF2 is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shao, F., P. M. Merritt, Z. Bao, R. W. Innes, and J. E. Dixon. 2002. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109:575-588. [DOI] [PubMed] [Google Scholar]
- 61.Shao, F., P. O. Vacratsis, Z. Bao, K. E. Bowers, C. A. Fierke, and J. E. Dixon. 2003. Biochemical characterization of the Yersinia YopT protease: cleavage site and recognition elements in Rho GTPases. Proc. Natl. Acad. Sci. USA 100:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheng, M., and C. Sala. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24:1-29. [DOI] [PubMed] [Google Scholar]
- 63.Snellings, N. J., M. Popek, and L. E. Lindler. 2001. Complete DNA sequence of Yersinia enterocolitica serotype 0:8 low-calcium-response plasmid reveals a new virulence plasmid-associated replicon. Infect. Immun. 69:4627-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 65.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 66.Trulzsch, K., A. Roggenkamp, M. Aepfelbacher, G. Wilharm, K. Ruckdeschel, and J. Heesemann. 2003. Analysis of chaperone-dependent Yop secretion/translocation and effector function using a mini-virulence plasmid of Yersinia enterocolitica. Int. J. Med. Microbiol. 293:167-177. [DOI] [PubMed] [Google Scholar]
- 67.Trulzsch, K., T. Sporleder, E. I. Igwe, H. Russmann, and J. Heesemann. 2004. Contribution of the major secreted Yops of Yersinia enterocolitica O:8 to pathogenicity in the mouse infection model. Infect. Immun. 72:5227-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Von Pawel-Rammingen, U., M. V. Telepnev, G. Schmidt, K. Aktories, H. Wolf-Watz, and R. Rosqvist. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737-748. [DOI] [PubMed] [Google Scholar]
- 69.Wani, M. A., R. T. Means, Jr., and J. B. Lingrel. 1998. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 7:229-238. [DOI] [PubMed] [Google Scholar]
- 70.Wani, M. A., S. E. Wert, and J. B. Lingrel. 1999. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J. Biol. Chem. 274:21180-21185. [DOI] [PubMed] [Google Scholar]
- 71.Watanabe, G., Y. Saito, P. Madaule, T. Ishizaki, K. Fujisawa, N. Morii, H. Mukai, Y. Ono, A. Kakizuka, and S. Narumiya. 1996. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science 271:645-648. [DOI] [PubMed] [Google Scholar]
- 72.Wong, K. W., and R. R. Isberg. 2005. Yersinia pseudotuberculosis spatially controls activation and misregulation of host cell Rac1. PLoS Pathog. 1:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yao, T., J. Mecsas, J. I. Healy, S. Falkow, and Y. Chien. 1999. Suppression of T and B lymphocyte activation by a Yersinia pseudotuberculosis virulence factor, yopH. J. Exp. Med. 190:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu, C., T. S. Agin, S. J. Elliott, L. A. Johnson, T. E. Thate, J. B. Kaper, and E. C. Boedeker. 2001. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect. Immun. 69:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]