Abstract
Obligate intracellular bacteria of the genus Rickettsia must adhere to and invade the host endothelium in order to establish an infection. These processes require the interaction of rickettsial surface proteins with mammalian host cell receptors. A previous bioinformatic analysis of sequenced rickettsial species identified a family of at least 17 predicted “surface cell antigen” (sca) genes whose products resemble autotransporter proteins. Two members of this family, rOmpA and rOmpB of spotted fever group (SFG) rickettsiae have been identified as adhesion and invasion factors, respectively; however, little is known about the putative functions of the other sca gene products. An intact sca2 gene is found in the majority of pathogenic SFG rickettsiae and, due to its sequence conservation among these species, we predict that Sca2 may play an important function at the rickettsial surface. Here we have shown that sca2 is transcribed and expressed in Rickettsia conorii and have used a heterologous gain-of-function assay in E. coli to determine the putative role of Sca2. Using this system, we have demonstrated that expression of Sca2 at the outer membrane of nonadherent, noninvasive E. coli is sufficient to mediate adherence to and invasion of a panel of mammalian cells, including endothelial cells. Furthermore, soluble Sca2 protein is capable of diminishing R. conorii invasion of cultured mammalian cells. This is the first evidence that Sca2 participates in the interaction between SFG rickettsiae and host cells and suggests that in addition to other surface proteins, Sca2 may play a critical role in rickettsial pathogenesis.
The genus Rickettsia is composed of obligate intracellular gram-negative alphaproteobacteria, many of which are human pathogens. These pathogens are transmitted to the human host by arthropod vectors, such as ticks, which inoculate the host during a blood meal (8). Rickettsial species are divided into two groups, the typhus group (TG) and the spotted fever group (SFG) based on differences in their antigenicity to lipopolysaccharide, the presence of outer membrane proteins, and in part on the diseases that they cause (19). Members of both groups are responsible for severe human disease and two species, Rickettsia prowazekii (TG) and Rickettsia rickettsii (SFG) have been classified as select agents by the Centers for Disease Control and Prevention.
Infection by SFG rickettsiae, such as R. conorii, the causative agent of Mediterranean spotted fever, begins with inoculation of bacteria into the vasculature of the human host during a tick bite (8). Subsequent growth of Rickettsia in proximal endothelial cells can result in a localized dermal and epidermal necrosis called an eschar or tache noire (21). Further damage to the vascular endothelium and infiltration of perivascular mononuclear cells leads to increased fluid leakage into the interstitial space, ultimately resulting in a characteristic dermal rash and acute renal failure with peripheral edema (10, 21). Damage done by R. conorii to target endothelial cells, especially in the lungs, can lead to severe manifestations of disease including pulmonary edema and interstitial pneumonia. Although endothelial cells are the main target cell type for SFG Rickettsia, R. conorii can attach to and invade different cell types in vitro and in vivo and spread via lymphatic vessels to the lymph nodes or via the bloodstream to various tissues (20). Initial clinical manifestations of rickettsial infection include flulike symptoms, often leading to delayed diagnosis or misdiagnosis. There is currently no vaccine available to prevent rickettsial infection but, if properly diagnosed, it can be controlled by broad-spectrum antibiotics. If left untreated, Mediterranean spotted fever and other SFG rickettsial diseases can result in severe morbidity and mortality (22).
Because Rickettsia cannot replicate extracellularly within the mammalian host, establishment of a successful infection relies entirely on the ability of the bacteria to invade a host cell. This process begins with adherence to the host cell through specific interactions, leading to eukaryotic downstream signaling and ultimately bacterial uptake (13). Thus far, two rickettsial surface proteins, rickettsial outer membrane protein A (rOmpA) and rickettsial outer membrane protein B (rOmpB), have been shown to participate in adhesion to mammalian cells in vitro (2, 12, 18) and in eliciting humoral and cellular protective immune responses (5, 6). In addition, rOmpB has been recently demonstrated to be sufficient in mediating bacterial invasion of mammalian cells in vitro through interaction with its mammalian receptor, Ku70 (2, 15). Interestingly, an analysis of rickettsial invasion in cells depleted of endogenous Ku70 by small interfering RNA or in Ku70−/− mouse embryonic fibroblasts revealed that R. conorii invasion is reduced by ca. 50 to 60% (15), suggesting that factors other than Ku70 and rOmpB likely contribute to the invasion process.
rOmpA and rOmpB belong to a family of outer membrane proteins termed autotransporters found in gram-negative bacteria. This class of proteins contains an N-terminal signal sequence to target the protein through the Sec machinery, a central variable passenger domain, and a highly conserved C-terminal β-barrel or translocation domain (11). Once in the periplasm, the β-barrel can insert into the outer membrane and is predicted to form a conduit through which the passenger domain is transported and exposed to the extracellular milieu. A recent bioinformatic analysis of sequenced rickettsial genomes has identified a family of at least 15 genes in addition to rompA (sca0) and rompB (sca5) termed sca (for surface cell antigens) whose predicted products resemble autotransporter proteins (1). Although many genes in the sca family are split, fragmented, or absent in many rickettsial species, five members of this family, namely, ompA (sca0), ompB (sca5), sca1, sca2, and sca4, appear to have evolved under positive selection and are present in the genomes of most rickettsial species (1). Although rOmpA and rOmpB have been shown to participate in adhesion and invasion of mammalian cells, nothing is known about the function of the other sca gene products.
The presence of sca2 in the R. conorii genome and in the genome of virtually all SFG rickettsial species suggests that Sca2 may play a critical role during the Rickettsia life cycle. Due to its similarity to other known autotransporter adhesin proteins in SFG Rickettsia, we hypothesize that Sca2 may play a role in rickettsial adherence to and invasion of target cells. We have adapted a heterologous E. coli expression system, previously used in the study of rOmpB from R. japonica and R. conorii (2, 18), to analyze the function of Sca2. Using this expression system, we demonstrate that Sca2 when expressed at the outer membrane of E. coli is sufficient to mediate adherence to and invasion of a panel of mammalian cell types in the absence of other rickettsial factors. Furthermore, we show that purified soluble Sca2 protein is capable of diminishing the invasion process of R. conorii during infection of cultured mammalian cells. Taken together, these results demonstrate for the first time that Sca2 is expressed in R. conorii and that a conserved rickettsial outer membrane protein other than rOmpA and rOmpB can participate in mediating interactions with target mammalian cells.
MATERIALS AND METHODS
Cell lines and bacterial strains.
HeLa (American Type Culture Collection, Manassas, VA), Vero, and EAHY 926 (obtained from the University of North Carolina Chapel Hill) (4) cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum. HeLa and Vero cell medium was supplemented with 1× nonessential amino acids (Cellgro, Manassas, VA) and 0.5 mM sodium pyruvate (Lonza, Walkersville, MD). Human lung microvascular (HLMV) cells (a gift from J. G. N. Garcia, University of Chicago) were grown in microvascular endothelial cell growth medium 2 (EGM-2 MV; Lonza, Walkersville, MD). All mammalian cells were grown at 37°C/5% CO2. Bacterial genetic manipulations were performed in Escherichia coli TOP 10 (Invitrogen) grown at 37°C in Luria-Bertani (LB) Miller broth supplemented with 100 μg of ampicillin/ml where appropriate. For expression of recombinant Sca2, E. coli BL21(DE3) harboring the plasmid pSca2-200 was grown overnight at 30°C in LB medium with 100 μg of ampicillin/ml. Cultures were diluted 1:10 into fresh media and grown to an optical density at 600 nm (OD600) of 0.5 at 37°C. Protein expression was induced with the indicated concentrations of IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 30°C. R. conorii Malish 7 was propagated by infection of Vero cell monolayers for 5 days at 34°C and 5% CO2. R. conorii were isolated from Vero cells by glass bead disruption of the monolayer, followed by lysis through a syringe. Bacteria were separated from mammalian cell debris by centrifugation through a sucrose gradient as previously described (7). The recovered R. conorii were resuspended in a sucrose phosphate glycine buffer (SPG; 218 mM sucrose, 3.8 mM KH2PO4, 7.2 mM K2HPO4, 4.9 mM l-glutamate [pH 7.4]) and frozen at −80°C. Rickettsia titers were determined by a limiting-dilution infectivity assay, followed by calculation of the Reed and Muench equation (17).
Antibodies and other reagents.
Rabbit anti-Sca234-794 antiserum was prepared by immunization of a White New Zealand rabbit with 700 μg of purified Sca234-794 protein (see the description of protein purification below) mixed 1:1 with complete Freund adjuvant. Three weeks after the primary immunization, a booster of 700 μg of Sca234-794 mixed 1:1 with incomplete Freund adjuvant was administered. One week after the boost, the rabbit was bled, and sera were collected. Anti-His6 rabbit antibody was purchased from Covance, and goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase conjugate was obtained from Sigma. Anti-E. coli antiserum has been previously described (2). Alexa Fluor 488-conjugated goat anti-rabbit IgG, Alexa Fluor 546-conjugated goat anti-rabbit IgG, Texas Red-conjugated phalloidin, and DAPI (4′,6′-diamidino-2-phenylindole) were purchased from Molecular Probes. Rabbit hyperimmune serum against R. conorii (Rc7) was kindly provided by D. Walker (University of Texas Medical Branch, Galveston). R. conorii whole-cell lysate was produced by boiling isolated Rickettsia in sample buffer for 5 min.
Reverse transcription-PCR.
mRNA was isolated from approximately 4 × 105 R. conorii infectious particles by using an RNeasy minikit (Qiagen). RNase OUT (Invitrogen) was added to the samples to increase their stability before being treated with DNase I (Invitrogen) twice. RNA was phenol-chloroform extracted between DNase I treatments, and the concentration was determined by absorbance at 260 nm. A 120-ng portion of DNase I-treated RNA was then reverse transcribed by using an ImProm II kit (Promega) to yield a cDNA/RNA hybrid. Gene-specific primer pairs (see Table 2) were combined with 5 Prime MasterMix (5 Prime, Gaithersburg, MD) to amplify target rickettsial genes from this cDNA/RNA hybrid in a PCR with 30 amplification cycles.
TABLE 2.
RT-PCR primers
| R. conorii gene | Primer sequence (5′-3′) |
|
|---|---|---|
| Forward | Reverse | |
| ompA | AATGACACCGCTACAGGAAGCAGA | AAACGGGCTTATCCACGCACCAAA |
| ompB | CAAACAGGCGTTGTTGATGCGAGT | ATGCGCGAATTGTACTGCACCGTT |
| sca2 | TGGTGCTGATAAAGGTGAGGCAAC | TTGAGAGCGTCCGGTAGATGGTTT |
| sca4 | AGTTCTCAGTCCGGCACAACAACA | AGCTTCAAGCGAGCTTCCTGCAAT |
Plasmid DNA constructs.
The full-length sca2 open reading frame was amplified by PCR from a chromosomal preparation of R. conorii Malish 7 by using the forward and reverse primers 5-ACCATGGATTTACAAAATTCCCAC-3′ and 5′-TTAGTGGTGGTGGTGGTGGTGCAAATTGACTTTTAG-3′, respectively. The resulting PCR product had an NcoI restriction site incorporated 5′ of the sca2 start codon and a His6 epitope tag inserted prior to the stop codon at the 3′ end. The PCR product was initially TOPO TA cloned into pCR2.1 (Invitrogen), resulting in pSca2-100, and then digested with NcoI and XhoI for insertion into the expression vector pET-22b (Novagen), resulting in the plasmid pSca2-200. The N-terminal half of the Sca2 passenger domain encompassing amino acids 34 to 794 was PCR amplified from pSca2-200 by using the forward and reverse primers 5′-AAGGATCCGCAAGCTTTAAAGATTTAGTTAGTAAAA-3′ and 5′-AACTCGAGATTTGTAAGATCAGCTTGTCTTTTTATAAG-3′, respectively. The resulting PCR product was digested with BamHI and XhoI and then ligated into pGEX-2TKP (a gift from T. Kouzarides, Gurdon Institute, United Kingdom), resulting in plasmid pMC024.
Amino acid alignment.
The amino acid sequences of Sca2 from R. conorii Malish 7 (GenBank accession no. AAL02648), R. rickettsii Sheila Smith (ABV75720), R. japonica (AAT79540), R. sibirica (AAT79548), R. australis (AAT79538), and R. typhi Wilmington (AAU03539) were downloaded from GenBank (National Center for Biotechnology Information). The degree of identity and similarity of Sca2 sequences was determined by using the CLUSTAL W alignment application MacVector 9.5.2 (MacVector, Cary, NC) by comparing the translated sca2 open reading frame from each rickettsial species to the predicted R. conorii Malish 7 Sca2 amino acid sequence.
E. coli fractionation.
Ten milliliters of induced E. coli BL21(DE3) cultures was pelleted and resuspended in 1 ml of lysis buffer (phosphate-buffered saline [PBS] plus 1× protease inhibitor [Roche]). Cells were lysed by sonication for 3 s, followed by 10 s of rest on ice until the lysates became translucent. Unbroken cells were pelleted by centrifugation for 10 min at 4°C at 1,000 × g. The supernatant was transferred to a clean tube, and inner membrane protein was extracted with Sarkosyl (final concentration, 0.5%) at room temperature for 5 min. Outer membranes were pelleted by centrifugation at high speed for 30 min at 4°C, resuspended in 50 μl of 2× sample buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and transferred to nitrocellulose. Membranes were analyzed by immunoblotting with anti-His6 rabbit antibody (1:1,000), rabbit anti-Sca234-794 antiserum (1:1,000), and goat anti-rabbit IgG-horseradish peroxidase conjugate (1:3,000) where indicated. Immunoreactive species were revealed with Super Signal West Pico chemiluminescence substrates (Pierce) and exposure of membranes to film.
Protein purification.
Overnight cultures of E. coli BL21(DE3) transformed with pMC024 were diluted 1:10 into 1 liter of fresh LB medium with ampicillin and grown at 30°C until the OD600 reached 0.5. The expression of soluble glutathione S-transferase (GST)-Sca234-794 fusion protein was induced by the addition of 0.5 mM IPTG for 5 h at 30°C. Bacteria were harvested by centrifugation, resuspended in Tris-buffered saline (50 mM Tris [pH 8.0] plus 150 mM NaCl) containing protease inhibitor cocktail, and lysed by sonication (40% amplitude, 5 s on and 10 s off for 20 min; Fisher Scientific sonic dismembrator model 500). Lysates were cleared by centrifugation at 13,000 × g for 30 min at 4°C and applied to a GST-TRAP FF column (GE Healthcare, Piscataway, NJ) by fast protein liquid chromatography on a Åkta fast protein liquid chromatography. Protein was eluted with 30 mM reduced glutathione, and fractions were dialyzed into Tris-buffered saline before storage at −80°C. Protein to be utilized as antigen for antibody production was cleaved with thrombin to remove the GST tag. Thrombin and GST were purified away from Sca234-794 by collection of the flowthrough after passage over a benzamidine Sepharose column (GE Healthcare) and a GST-TRAP FF column.
Cell association and invasion assays.
Cell association and invasion assays were performed as described previously (2, 14). Briefly, mammalian cells were seeded into 24-well plates at 1.2 × 105 cells per well 24 h prior to infection. Cells were washed with serum-free DMEM, infected with 50 μl of induced E. coli (OD600 = 1.0) resuspended in PBS, centrifuged to induce bacterial contact at 200 g for 5 min, and then incubated at 37°C and 5% CO2 for 20 or 60 min to assess the cell association or invasion, respectively. In some experiments, mammalian cells were preincubated with 1 μM GST or GST-Sca234-794 for 30 min at 37°C and 5% CO2 prior to the addition of bacteria. In cell association assays, cells were washed extensively with PBS, lysed with 0.1% Triton X-100 in double-distilled H2O, and plated onto LB agar plates for determination of the CFU. Adherence frequencies were calculated as the percentage of cell-associated bacteria recovered after the PBS washes out of the total bacteria in each well after the 20 min of incubation. To determine bacterial invasion, cells were washed with PBS and then incubated in complete DMEM for an additional 2 h with 100 μg of gentamicin sulfate (MP Biomedicals)/ml. Thereafter, the cells were washed with PBS, lysed with 0.1% Triton X-100 in double-distilled H2O, and plated onto LB agar plates. Invasion frequencies were determined as the number of bacteria surviving the gentamicin challenge out of the total bacterial input after the initial 1 h of incubation. P values were determined by using a two-tailed Student t test.
For R. conorii invasion assays, Vero cells were seeded at 1.5 × 104 cells/well in 96-well plates 24 h prior to infection with R. conorii. Prior to infection, the medium was aspirated from the cells, 50 μl of purified GST or GST-Sca234-794 diluted in cold serum-free DMEM was added, and then Vero cells were incubated for 30 min at 37°C and 5% CO2. R. conorii were diluted to a concentration of 1.6 × 105 bacteria/ml in room temperature serum-free DMEM, and 50 μl was added to each well of Vero cells. The plate was centrifuged at 300 × g for 5 min at room temperature to induce contact, incubated for 30 min at 37°C and 5% CO2, washed with PBS, and fixed in 4% paraformaldehyde (PFA) for 20 min. The infected monolayers were then processed for immunofluorescence and differentially stained to distinguish between extracellular and intracellular R. conorii. Briefly, extracellular R. conorii were stained with Rc7 antiserum (1:1,000), followed by Alexa Fluor 546-conjugated goat anti-rabbit IgG (1:1,000), prior to permeabilization of the mammalian cells with 0.1% Triton X-100. The total R. conorii cells were then stained with Rc7, followed by Alex Fluor 499-conjugated goat anti-rabbit IgG. Images were captured as described for immunofluorescence assays, and the numbers of nuclei and extracellular and total bacteria were enumerated by using the ImageJ software “Analyze Particles” function.
Immunofluorescence.
Cell association assays were performed as described above for the Sca2-expressing E. coli. After the nonadherent bacteria were washed away from the mammalian monolayer with PBS, the cells were fixed in 4% PFA for 20 min at room temperature, washed with PBS, permeabilized with PBS plus 0.1% Triton X-100 for 3 min, and washed again with PBS to removal all Triton X-100. PBS plus 2% bovine serum albumin was used to block the fixed cells, as well as for the antibody dilutions. To assess the association of E. coli with mammalian cells, cells were stained with anti-E. coli antisera (1:1,000), followed by Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:1,000), Texas Red-phalloidin (1:200), and DAPI (1:10,000). Images were digitally captured on a Nikon Eclipse TE2000-u microscope coupled to a charge-coupled device camera using ×200 magnification and processed using Adobe Photoshop.
Electron microscopy.
For scanning electron microscopy, HeLa cells grown on 12-mm glass coverslips in 24-well plates were infected with 100 μl of induced bacteria (OD600 = 1.0) for 20 min and then processed as described previously (2). The samples were visualized by using Fei NovaNano SEM200 at a distance of 5 mm. For transmission electron microscopy, HeLa cells grown in six-well plates (3 × 105 cells per well) were infected with 2 ml of induced bacteria (OD600 = 2.0) for 2 h, washed, and then bathed in fixative (2% glutaraldehyde, 4% PFA, 0.1 M sodium cacodylate buffer.) Cells were carefully scraped from the plate and allowed to fix overnight. The next day, cells were pelleted at 500 × g for 5 min and subsequently processed as previously described (2). The samples were imaged on the FEI Tecnai F30 with a Gatan charge-coupled device digital micrograph.
RESULTS
Conservation of sca2 among spotted fever group rickettsia.
Previous bioinformatic annotations of several rickettsia genomes have demonstrated that a complete sca2 gene is present among most SFG rickettsiae (1, 16). To determine the degree of conservation within the predicted Sca2 protein sequence, an amino acid alignment was performed comparing the sequence of Sca2 from five geographically diverse SFG Rickettsia spp. (R. conorii, R. rickettsii Sheila Smith, R. japonica, R. sibirica, and R. australis) and one TG Rickettsia sp. (R. typhi). As shown in Table 1, the percent identity of Sca2 sequences observed among different SFG rickettsial species is substantial, varying from 83% in R. australis to 94% in R. sibirica to the Sca2 sequence from R. conorii Malish 7. Furthermore, the percent similarity is more striking, ranging from 90 to 95% among the SFG Rickettsia species analyzed. In contrast, the predicted Sca2 amino acid sequence appears to be less conserved among R. typhi possibly due to the degeneration of the sca2 gene within this class of Rickettsia. The high degree of Sca2 sequence identity and conservation among SFG Rickettsia suggests that Sca2 may play a critical function during SFG rickettsial infections.
TABLE 1.
Comparison of Sca2 amino acid sequence among rickettsial species
| Rickettsial species | Size (amino acids) |
R. conorii |
|
|---|---|---|---|
| % Identity | % Similarity | ||
| R. rickettsii Sheila Smith | 1,821 | 90 | 92 |
| R. japonica | 1,813 | 89 | 92 |
| R. sibirica | 1,812 | 94 | 95 |
| R. australis | 1,804 | 83 | 90 |
| R. typhi | 1,483 | 25 | 38 |
Detection of endogenous sca2 protein and expression of recombinant Sca2 protein in E. coli.
We first determined whether sca2 is actively transcribed in R. conorii Malish 7 during association with mammalian cells. We purified mRNA from R. conorii Malish 7, freshly isolated from a Vero cell infection, and removed any contaminating rickettsial or Vero genomic DNA by DNase I treatment. The DNase I-treated rickettsial RNA was then reverse transcribed by using random primers to yield a cDNA/RNA hybrid. The reverse transcription products were used as templates for PCRs amplifying one of four rickettsial sca genes using the indicated primer pairs (Table 2). As shown in Fig. 1A, agarose gel electrophoresis analysis of PCR products reveals the amplification of the rickettsial gene sca2 from R. conorii cDNA, indicating that sca2 mRNA transcript was present prior to reverse transcription. PCR product was also detectable for the genes ompA, ompB, and sca4, which are expressed in R. conorii (data not shown). The PCR products were cloned and sequence verified to confirm the amplification of the correct gene fragment. PCR products were not observed in the control reactions lacking RNA template or reverse transcriptase, indicating that amplification was due solely to the presence of mRNA and not due to contaminating genomic DNA. This result demonstrates that mRNA transcript of sca2 is present in R. conorii isolated from infected Vero cells.
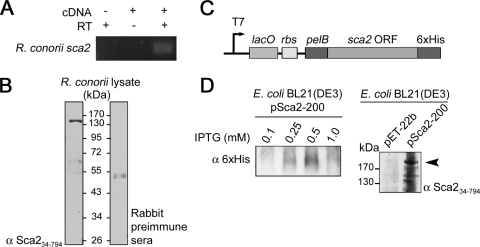
FIG. 1.
Detection of endogenous Sca2 protein in R. conorii and expression of recombinant Sca2 protein in E. coli. (A) Agarose gel electrophoresis analysis of PCR products resulting from amplification of R. conorii cDNA using a sca2-specific primer pair (Table 2). PCR product is 240 bp in size. A no-template control (-cDNA) and no-reverse-transcriptase control (-RT) are included to control for environmental DNA contamination and genomic DNA contamination, respectively. (B) Western immunoblot analysis of R. conorii whole-cell lysate probed with rabbit anti-Sca234-794 hyperimmune sera (left panel) and rabbit preimmune sera (right panel). (C) Schematic of the pET-22b expression vector with the inserted sca2 open reading frame. Relevant features of the 5′ and 3′ ends of pSca2-200, including the in-frame N-terminal pelB leader sequence and the C-terminal His6 tag are shown. (D) Western immunoblot analysis of outer membrane fractions of E. coli BL21(DE3) expressing the empty vector (pET-22b) or full-length R. conorii Sca2 (pSca2-200) probed with anti-His6 (left panel) and anti-Sca234-794 antibodies (right panel). Maximal expression of recombinant Sca2 is observed with 0.5 mM IPTG induction (left panel), resulting in a protein product larger than 170 kDa (arrow).
The sca2 gene is predicted to encode a protein with a molecular mass of ∼200 kDa. To identify endogenous Sca2 protein in R. conorii, Sca2 antiserum was produced by the immunization of a New Zealand White rabbit with a soluble fragment of the predicted R. conorii Sca2 passenger domain encompassing amino acids 34 to 794 (Sca234-794). Western immunoblotting of R. conorii whole-cell lysate reveals the presence of an ∼150-kDa species recognized by anti-Sca234-794 antiserum and not preimmune rabbit sera (Fig. 1B). This result indicates that Sca2 protein is present in R. conorii isolated from a Vero cell infection and suggests, that similar to other rickettsial autotransporter proteins, Sca2 may be processed in vivo.
Previous studies have successfully utilized an E. coli-based heterologous protein expression system to demonstrate that a conserved rickettsial surface cell antigen, rOmpB, expressed at the outer membrane of E. coli is sufficient to mediate adherence and invasion of cultured mammalian cells (2, 18). To study the putative function of Sca2 as a bacterial surface protein, we adapted this system to express Sca2 at the outer membrane of E. coli. The full-length R. conorii sca2 open reading frame was cloned into the E. coli IPTG-inducible expression vector, pET-22b, resulting in plasmid pSca2-200 (Fig. 1C). The plasmid pSca2-200 was transformed into the E. coli expression strain, BL21(DE3), induced for protein expression, and then biochemically fractionated to isolate E. coli outer membrane proteins. As shown in Fig. 1D, increasing concentrations of IPTG leads to the appearance of a high-molecular-weight His6 reactive species as analyzed by Western immunoblotting (left panel) with 0.5 mM IPTG induction, yielding the best protein expression. The identity of this induced His6 reactive protein was confirmed as Sca2 by using anti-Sca234-794 antiserum (right panel). Taken together, these results demonstrate that Sca2 is present at the outer membrane of induced E. coli in this heterologous expression system.
Sca2 expression in E. coli is sufficient to mediate association to epithelial and endothelial cells.
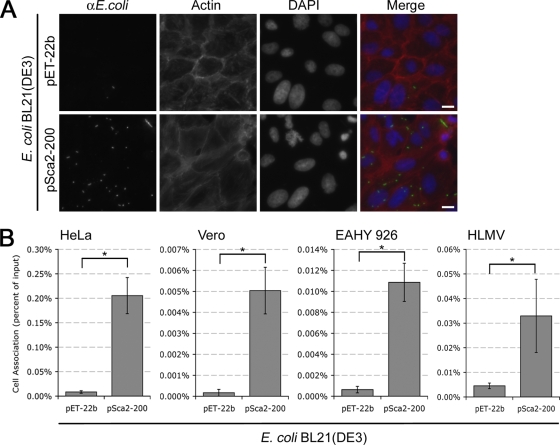
To study the putative contribution of Sca2 in mediating interactions with host cells, E. coli expressing Sca2 at the outer membrane was assessed for the ability to associate with cultured mammalian cells in vitro by immunofluorescence and CFU-based adherence assays (see Materials and Methods). As shown in Fig. 2A, expression of Sca2 was sufficient to mediate cell association of E. coli to cultured Vero cells compared to E. coli harboring the empty vector, pET-22b. A CFU-based assay used to quantify cell association of E. coli expressing Sca2 to epithelial cell lines (HeLa and Vero), transformed human umbilical vein endothelial cells (EAHY 926), and HLMV cells confirmed these results (Fig. 2B). These data illustrate that expression of Sca2 at the E. coli outer membrane is sufficient to mediate adherence to a variety of mammalian cell types.
FIG. 2.
Expression of Sca2 in E. coli is sufficient to mediate adherence to mammalian cells. (A) Fluorescence micrographs of monolayers of Vero cells infected with E. coli BL21(DE3) expressing the empty vector (pET-22b) or the full-length Sca2 protein from R. conorii (pSca2-200). Confluent monolayers of Vero cells were infected for 20 min at 37°C, washed repeatedly with PBS, and then processed for immunofluorescence. Scale bars, 10 μm. (B) CFU-based quantification of bacterial adherence to mammalian host cells. HeLa, Vero, EAHY 926, and HLMV cells were infected with induced cultures as in panel A. Confluent monolayers were infected with induced bacteria for 20 min at 37°C and washed repeatedly with PBS, and then cell-associated bacteria were extracted from host cells by detergent lysis, and plated for CFU quantification. Association was determined as the % CFU of cell-associated bacteria from the initial bacterial inoculums. Actual percentages varied from assay to assay (ranging from 0.001 to 0.2%) depending on the passage number of cells used and the expression of Sca2 at the E. coli outer membrane. *, P < 0.05. The data presented are representative of at least three independent assays for each cell line. Error bars represent the standard deviation of each data set.
Sca2 expression in E. coli is sufficient to mediate invasion of epithelial and endothelial cells.
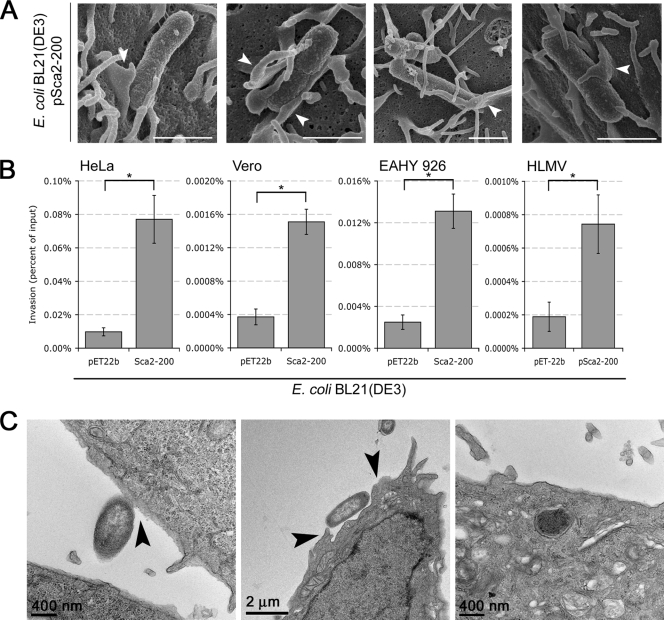
An ultrastructural analysis of Sca2-mediated E. coli association to HeLa cells by scanning electron microscopy (SEM) revealed the induction of subtle membrane perturbations and close contacts between bacteria and host cell (Fig. 3A), an finding reminiscent of those observed during rOmpB-mediated internalization (2, 18). To investigate the ability of Sca2 to mediate invasion of host epithelial and endothelial cells, we utilized a standard gentamicin protection assay. Confluent monolayers of mammalian cells were infected for 60 min with induced E. coli BL21(DE3) harboring either the empty vector, pET-22b, or the Sca2 expression construct, pSca2-200. Extracellular bacteria were killed by gentamicin, and internalized bacteria were plated for CFU enumeration. As shown in Fig. 3B, in the absence of other rickettsial factors, Sca2 is sufficient to mediate uptake by both epithelial and endothelial mammalian cells. These results were confirmed by transmission electron microscopy, demonstrating that Sca2-expressing E. coli induces changes in the plasma membrane of HeLa cells, ultimately leading to internalization (Fig. 3C).
FIG. 3.
Sca2 mediates invasion of mammalian cells. (A) Scanning electron micrographs examining the surface interaction of Sca2-expressing E. coli with HeLa cells. HeLa cell monolayers on glass coverslips were infected with induced bacteria for 20 min and then processed for SEM. White arrowheads highlight possible Sca2-mediated cellular membrane rearrangements. Scale bars, 1 μm. (B) CFU-based quantification of bacterial invasion of HeLa, Vero, EAHY 926, and HLMV cells. Confluent cell monolayers were infected for 1 h with E. coli BL21(DE3) expressing the empty vector (pET-22b) or Sca2 from R. conorii (pSca2-200) and assessed for invasion by gentamicin protection assay. Invasion is presented as the percentage of bacteria recovered after the gentamicin challenge out of the inoculums. Actual percentages varied from assay to assay (ranging from 0.0007 to 0.07%), depending on the passage number of the cells used and the expression of Sca2 at the E. coli outer membrane. *, P < 0.05. The data presented are representative of at least two independent experiments for each individual cell line. Error bars represent the standard deviation of each data set. (C) Transmission electron micrographs of HeLa cells infected with Sca2-expressing E. coli demonstrate different steps of the uptake process, including the initial attachment event (left panel), the induction of changes in the plasma membrane (arrows in middle panel), and the presence of bacteria within membrane bound vacuoles (right panel). Scale bars are indicated in each panel.
Soluble Sca2 protein inhibits rickettsial invasion.
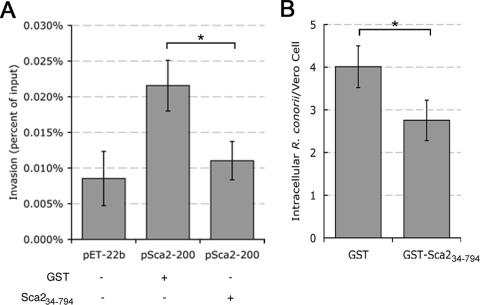
In order to confirm the Sca2-mediated invasive phenotype observed in our heterologous assay, we used purified Sca2 protein to competitively inhibit E. coli BL21(DE3) invasion with mammalian cells when expressing Sca2 at the bacterial surface. We cloned the first half of the Sca2 passenger domain encompassing amino acids 34 to 794 into the E. coli expression vector pGEX-2TKP in order to produce and affinity purify a soluble N-terminal GST tagged Sca2 fragment (GST-Sca234-794). Attempts to express and purify larger soluble portions of Sca2 were unsuccessful due to protein instability. The addition of GST-Sca234-794 to a HeLa cell monolayer prior to infection with E. coli BL21(DE3) expressing full-length R. conorii Sca2 at the surface reduced the ability of these bacteria to invade host cells by 50% (Fig. 4A). This result confirms that bacterial invasion of the host cell in our heterologous system is specifically mediated by Sca2. To assess the role of Sca2 during a rickettsial infection, we similarly used GST-Sca234-794 to competitively inhibit R. conorii invasion of Vero cells. As shown in Fig. 4B, preincubation of Vero cells with GST-Sca234-794 reduced the ability of R. conorii to invade host cells by ca. 30%, suggesting that Sca2 is involved in mediating the rickettsial invasion of host cells.
FIG. 4.
Soluble Sca2 protein inhibits rickettsial invasion. (A) Monolayers of HeLa cells were incubated with 1 μM soluble GST or GST-Sca234-794 protein for 20 min at 37°C and 5%CO2 prior to infection with. E. coli BL21(DE3) expressing pSca2-200. BL21(DE3) harboring pET-22b was used as a negative control. Bacterial invasion was assessed as described in Fig. 3B. (B) Vero cell monolayers seeded at 90% confluence were incubated with 8 μM GST or GST-Sca234-794 protein prior to infection with R. conorii for 30 min. Total rickettsial association and intracellular bacteria were enumerated by differential immunofluorescence microscopy. *, P < 0.05. The data presented are representative of at least two independent experiments. The error bars represent the standard deviation from the average in these experiments.
DISCUSSION
Pathogenesis of a rickettsial infection relies on the ability of the bacterium to adhere to and invade host cells. These interactions are predicted to be governed by rickettsial outer membrane proteins and cognate host cell receptors, whose interactions induce cellular signal transduction events leading to bacterial internalization. A search for genes encoding putative outer membrane proteins in several rickettsia genomes revealed the presence of a family of at least 17 genes, termed sca, whose predicted sequences exhibit similarity to a family of modular proteins, termed autotransporters, found in gram-negative bacteria (1). Overall, the sca gene family exhibits a high degree of degeneration since several genes, including sca6 through sca16, are fragmented, split, or otherwise completely deleted from the genomes of different SFG rickettsia species (1). In contrast, several genes, including sca2, are present in the genomes of a variety of geographically distinct SFG Rickettsia species. The predicted Sca2 amino acid sequence of R. conorii and other SFG Rickettsia spp. exhibits a high degree of identity and similarity among geographically dispersed SFG rickettsial species, including R. japonica (found in Japan and East Asia), R. australis (Australia), and R. rickettsii (North and South America), suggesting that the function of this protein may be conserved. To our knowledge, this is the first demonstration of endogenous sca2 mRNA and Sca2 protein in R. conorii Malish 7 isolated from infected mammalian cells.
Efficient genetic tools to target specific genes in obligate intracellular pathogens have not yet been fully developed. Therefore, to study Sca2 function, we adapted an E. coli heterologous expression system that has been successfully used to characterize rOmpB function of R. conorii and R. japonica (2, 18). By expressing Sca2 at the E. coli outer membrane, we demonstrated that in the absence of other rickettsial virulence factors, Sca2 is sufficient to mediate adherence to and invasion of cultured mammalian epithelial and endothelial cells. Inhibition of these phenotypes with purified GST-Sca2 peptide confirms that invasion of host mammalian cells is specifically mediated by Sca2. Interestingly, the endogenous, native form of Sca2 in isolated R. conorii migrates at a smaller molecular mass (∼150 kDa) than the predicted full-length protein (∼200 kDa) observed in Sca2-expressing E. coli. This suggests that, as has been previously observed for a related Sca protein, rOmpB, (9), Sca2 may also be processed in vivo to release the passenger domain from the translocon domain. The processing machinery is likely absent in E. coli; however, as has been previously demonstrated for rOmpB (2), the processing of a heterologously expressed rickettsial autotransporter on the surface of E. coli is not required to mediate adhesion and invasion of mammalian cells.
Scanning and transmission electron micrographs of HeLa cells infected with Sca2-expressing E. coli revealed the induction of localized changes in the host cell plasma membrane that are likely snapshots of the events leading to invasion. These images revealed cellular membrane protrusions that are reminiscent of those observed in the actin-dependent rOmpB-mediated invasion process (2). Whether Sca2-mediated uptake is mechanistically similar to that mediated by rOmpB is currently under investigation. This is the first demonstration that a Rickettsia gene product other than rOmpA and rOmpB is sufficient to mediate surface interactions with target host cells in vitro.
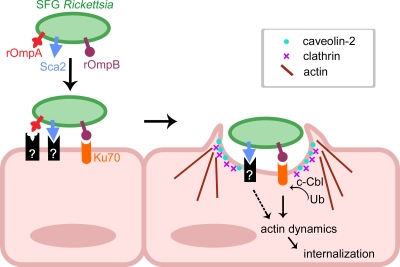
The ability of a pathogen to induce internalization into mammalian cells is likely governed by multiple adhesin-receptor interactions that are coupled to drive changes in the host cell plasma membrane. In the hostile extracellular environment of the host where microorganisms encounter both the innate and adaptive immune systems, it is to the pathogens' advantage to have as many tools as possible to successfully adhere to and in some cases invade host cells. Other invasive pathogens such as Listeria monocytogenes, Yersinia pseudotuberculosis, and uropathogenic E. coli utilize multiple nonfimbrial and fimbrial adhesins that are necessary and sufficient to mediate uptake in nonphagocytic mammalian cells (3, 14). Previous studies have identified rOmpA (Sca0) and rOmpB (Sca5) as proteins involved in adhesion and invasion, respectively, of cultured mammalian cells (2, 12, 18). Whereas rOmpA receptors have yet to be identified, recent studies have identified plasma membrane associated Ku70 as a receptor for rOmpB (2, 15). Since SFG Rickettsia are to an extent still able to invade mammalian cells depleted of Ku70, factors other than rOmpB and Ku70 are likely important. We have demonstrated that recombinant Sca2 protein reduces R. conorii invasion of mammalian cells (∼30%), suggesting that Sca2 plays an important role in R. conorii infection process and may complement invasion mediated by other rickettsial virulence factors (i.e., rOmpB). It is possible that Sca2 may work in concert with other rickettsial proteins to interact with target mammalian cells, especially endothelial cells during the infection process (Fig. 5). The interactions of Sca2 with its cognate receptor(s) may be coordinated with other receptor-ligand pairs to trigger previously observed downstream signaling events, involving the recruitment of host endocytic machinery components (c-Cbl, caveolin-2, and clathrin), and the activation of protein tyrosine and phosphatidylinositol 3-kinases (2, 13) that ultimately lead to localized actin recruitment required for entry. Interestingly, a biochemical analysis of R. conorii proteins that interact with Ku70 identified rOmpB, but not other putative Sca proteins, as a ligand (15), suggesting that Sca2-mediated adherence and entry would be independent of Ku70. The identification of mammalian receptors involved in Sca2-mediated uptake of mammalian cells is of utmost importance and remains the focus of ongoing studies.
FIG. 5.
Model of SFG Rickettsia interactions with mammalian cells. Interactions of SFG Rickettsia with host cells are likely governed by coordinated interactions between rOmpB-Ku70 and Sca2 with an unknown mammalian receptor (black box). Scanning and transmission electron microscopy analyses revealed similarities between rOmpB and Sca2-mediated uptake of mammalian cells, suggesting that while signals may be triggered by different receptors at the plasma membrane, signals ultimately converge to recruit localized actin filaments and components of the endocytic machinery (clathrin, caveolin-2, and c-Cbl) to entry foci.
Clearly, all SFG Rickettsia proteins involved in adherence and invasion of host cells have not been identified. Further characterization of the roles of conserved Sca proteins in vitro and in vivo is crucial to fully understanding the complex host pathogen interactions during SFG rickettsial infections in humans and will hopefully expand our targets for the development of more efficacious therapies against rickettsial diseases.
Acknowledgments
We thank Joe G. N. Garcia (The University of Chicago) for generously providing HLMV cells; Yimei Chen and Qiti Guo for assistance in the transmission and SEM, respectively; Sean P. Riley and Yvonne G. Y. Chan for technical support, suggestions, and critical reading of the manuscript; and Glenn Randall and Todd E. Oakland for discussion regarding the Rickettsia limiting-dilution assay.
This study was in part sponsored by the NIH/NIAID Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program (NIH award 2-U54-AI-057153 to J.J.M.), NIH award RO1-AI072606-01 to J.J.M., and University of Chicago Molecular Cell Biology Training Grant (T32GM007183) to M.M.C. We acknowledge membership within and support from the Region V Great Lakes RCE (GLRCE).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 5 October 2009.
REFERENCES
- 1.Blanc, G., M. Ngwamidiba, H. Ogata, P. E. Fournier, J. M. Claverie, and D. Raoult. 2005. Molecular evolution of Rickettsia surface antigens: evidence of positive selection. Mol. Biol. Evol. 20:2073-2083. [DOI] [PubMed] [Google Scholar]
- 2.Chan, Y. G. Y., M. M. Cardwell, T. M. Hermanas, T. Uchiyama, and J. J. Martinez. 2009. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cell. Microbiol. 11:629-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cossart, P., and P. J. Sansonetti. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242-248. [DOI] [PubMed] [Google Scholar]
- 4.Edgell, C. J., C. C. McDonald, and J. B. Graham. 1983. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. USA 80:3734-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng, H. M., T. Whitworth, J. P. Olano, V. L. Popov, and D. H. Walker. 2004. Fc-dependent polyclonal antibodies and antibodies to outer membrane proteins A and B, but not to lipopolysaccharide, protect SCID mice against fatal Rickettsia conorii infection. Infect. Immun. 72:2222-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng, H. M., T. Whitworth, V. Popov, and D. H. Walker. 2004. Effect of antibody on the Rickettsia-host cell interaction. Infect. Immun. 72:3524-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gouin, E., H. Gantelet, C. Egile, I. Lasa, H. Ohayon, V. Villiers, P. Gounon, P. J. Sansonetti, and P. Cossart. 1999. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Science 112:1697-1708. [DOI] [PubMed] [Google Scholar]
- 8.Hackstadt, T. 1996. The biology of rickettsiae. Infect. Agents Dis. 5:127-143. [PubMed] [Google Scholar]
- 9.Hackstadt, T., R. Messer, W. Cieplak, and M. G. Peacock. 1992. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun. 60:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hand, W. L., J. B. Miller, J. A. Reinarz, and J. P. Sanford. 1970. Rocky Mountain spotted fever. A vascular disease. Arch. Intern. Med. 125:879-882. [PubMed] [Google Scholar]
- 11.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, H., and D. H. Walker. 1998. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microb. Pathog. 24:289-298. [DOI] [PubMed] [Google Scholar]
- 13.Martinez, J. J., and P. Cossart. 2004. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J. Cell Sci. 117:5097-5106. [DOI] [PubMed] [Google Scholar]
- 14.Martinez, J. J., M. A. Mulvey, J. D. Schilling, J. S. Pinkner, and S. J. Hultgren. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 19:2803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez, J. J., S. Seveau, E. Veiga, S. Matsuyama, and P. Cossart. 2005. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell 123:1013-1023. [DOI] [PubMed] [Google Scholar]
- 16.Ngwamidiba, M., G. Blanc, D. Raoult, and P. E. Fournier. 2006. Sca1, a previously undescribed paralog from autotransporter protein-encoding genes in Rickettsia species. BMC Microbiol. 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 18.Uchiyama, T., H. Kawano, and Y. Kusuhara. 2006. The major outer membrane protein rOmpB of spotted fever group rickettsiae functions in the rickettsial adherence to and invasion of Vero cells. Microbes Infect. 8:801-809. [DOI] [PubMed] [Google Scholar]
- 19.Vishwanath, S. 1991. Antigenic relationships among the rickettsiae of the spotted fever and typhus groups. FEMS Microbiol. Lett. 65:341-344. [DOI] [PubMed] [Google Scholar]
- 20.Walker, D. H., and J. H. Gear. 1985. Correlation of the distribution of Rickettsia conorii, microscopic lesions, and clinical features in South African tick bite fever. Am. J. Trop. Med. Hyg. 34:361-371. [DOI] [PubMed] [Google Scholar]
- 21.Walker, D. H., C. Occhino, G. R. Tringali, S. Di Rosa, and S. Mansueto. 1988. Pathogenesis of rickettsial eschars: the tache noire of boutonneuse fever. Hum. Pathol. 19:1449-1454. [DOI] [PubMed] [Google Scholar]
- 22.Yagupsky, P., and B. Wolach. 1993. Fatal Israeli spotted fever in children. Clin. Infect. Dis. 17:850-853. [DOI] [PubMed] [Google Scholar]