Abstract
Shigella flexneri is a gram-negative, facultative intracellular pathogen that invades the colonic epithelium and causes bacillary dysentery. We previously demonstrated that S. flexneri inhibits staurosporine-induced apoptosis in infected epithelial cells and that a ΔmxiE mutant is unable to inhibit apoptosis. Therefore, we hypothesized that an MxiE-regulated gene was responsible for protection of epithelial cells from apoptosis. Analysis of all MxiE-regulated genes yielded no mutants that lacked the ability to prevent apoptosis. Spa15, which is defined as a type III secretion system chaperone, was analyzed since it associates with MxiE. A Δspa15 mutant was unable to prevent staurosporine-induced apoptosis. C-terminal hemagglutinin-tagged spa15 was secreted by S. flexneri within 2 h in the Congo red secretion assay, and secretion was dependent on the type III secretion system. Spa15 was also secreted by Shigella in infected epithelial cells, as verified by immunofluorescence analysis. Spa15 secretion was decreased in the ΔmxiE mutant, which demonstrates why this mutant is unable to prevent staurosporine-induced apoptosis. Our data are the first to show that Spa15 is secreted in a type III secretion system-dependent fashion, and the absence of Spa15 in the Δspa15 mutant results in the loss of protection from staurosporine-induced apoptosis in epithelial cells. Thus, Spa15 contributes to the intracellular survival of Shigella by blocking apoptosis in the infected host cell.
Shigella flexneri is a gram-negative, facultative intracellular pathogen that causes bacillary dysentery. Clinical symptoms of disease include watery diarrhea, severe abdominal pain, and bloody stools (39). Disease is a result of the ability of the pathogen to invade the colonic epithelium. Once S. flexneri enters the colon, the bacteria transit through M cells and encounter resident macrophages. The bacteria escape the macrophages by inducing cell death and subsequently invade epithelial cells at the basolateral face (19). Proinflammatory signaling and a subsequent efflux of polymorphonuclear cells (PMNs) into the infected tissue allow the bacteria to invade more epithelial cells at the basolateral pole, while the PMNs contribute to the disease severity by causing local tissue destruction (19). Once inside the cytoplasm of the host cell, S. flexneri induces actin polymerization, which allows the bacteria to move to adjacent cells without the need to enter the extracellular environment (5). The epithelial cells provide the bacteria with an intracellular niche to multiply and spread to adjacent cells.
S. flexneri virulence requires a 220-kb virulence plasmid that encodes a type III secretion system (T3SS), the Ipa proteins essential for entry into the host cells, and other effector proteins. The T3SS is comprised of a needle complex that has a seven-ringed basal body and a protruding needle. The needle complex delivers proteins directly to the host cell from the bacterial cytoplasm (14). Temperature regulation of the genes on the virulence plasmid allows the needle complex to be synthesized and assembled at 37°C. The secretion of proteins is induced upon contact of the bacteria with the host cell. The Ipa proteins mediate entry of the bacteria into the epithelial cell through localized actin depolymerization and membrane engulfment. After engulfment, the bacteria are inside a vacuole that is subsequently lysed, allowing the bacteria to enter the cytoplasm of the host cell (39). Once inside the cytoplasm, the bacteria spread and secrete additional effector proteins. These proteins are encoded by genes that are scattered throughout the 220-kb virulence plasmid and are subsequently secreted through the T3SS for postinvasion virulence (6).
We previously showed that S. flexneri inhibits staurosporine (STS)-induced apoptosis in epithelial cells by preventing the activation of caspase-3, a key protein in apoptotic cell death, and that a ΔmxiE mutant is unable to prevent STS-induced apoptosis (7). MxiE is a transcriptional activator that induces the expression of ∼20 bacterial genes when the bacteria are inside the cytosol of the host cell. The subsequent protein products are secreted through the T3SS (22, 24). We therefore hypothesized that an MxiE-regulated gene is responsible for protection. In this study, we analyzed all of the MxiE-regulated genes and found that none was required to inhibit STS-induced apoptosis. We also analyzed a Δspa15 mutant since Spa15 associates with MxiE (35). This report describes the inability of a Δspa15 mutant to prevent STS-induced apoptosis and demonstrates, for the first time, that Spa15 is secreted through the T3SS. Spa15 was originally described as a T3SS chaperone and a coantiactivator to MxiE (34, 35). We are proposing a new, third function in which Spa15 is involved in apoptosis inhibition in epithelial cells since Spa15 is secreted by the T3SS and the Δspa15 mutant is unable to prevent apoptosis in epithelial cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains of S. flexneri used are listed in Table 1. Bacteria were routinely cultured at 37°C either in Luria-Bertani broth with aeration or on tryptic soy broth plates with 1.5% agar and 0.025% Congo red (CR; Sigma). Antibiotics were used at the following concentrations: kanamycin, 50 μg/ml; streptomycin, 50 μg/ml; chloramphenicol, 5 μg/ml; and ampicillin, 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. flexneri | ||
| 2457T | Wild-type serotype 2a | 13 |
| BS611 | 2457T/ΔmxiE2::aphA-3 | 22 |
| BS613 | BS611/pRRS13 (PlacmxiE+) | 22 |
| BS652 | 2457T/Δspa47::aadA | Lab stock |
| BS766 | 2457T transformed with pKM208 | 7 |
| BS902 | 2457T/Δspa15::aphA-3 | This study |
| BS904 | BS902 transformed with pBSKS-spa15 | This study |
| BS905 | 2457T transformed with pSpa15-2HA | This study |
| BS906 | BS611 transformed with pSpa15-2HA | This study |
| BS907 | BS613 transformed with pSpa15-2HA | This study |
| BS908 | BS652 transformed with pSpa15-2HA | This study |
| BS909 | 2457T transformed with pMxiE-2HA | This study |
| BS914 | BS902 transformed with pSpa15-2HA | This study |
| E. coli | ||
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 (ΔlacIZYA-argF) U169 deoR (φ80dlacΔ[lacZ]M15) | 17 |
| ATM951 | E. coli expressing a plasmid that contains the Shigella entry region | Lab stock (in preparation) |
| Plasmids | ||
| pKD4 | oriR6K; bla aphA-3 | 10 |
| pKM208 | Temperature-sensitive red-, gam-, and lacI-expressing plasmid driven by PTac promoter; bla | 10 |
| pBSKS(−) | pBluescript cloning vector; bla | Stratagene |
| pBSKS-spa15 | spa15 cloned into pBSKS | This study |
| pDZ1 | Cloning intermediate for 2HA fusions; cat | 46 |
| pSpa15-2HA | spa15 cloned into pDZ1; cat | This study |
| pMxiE-2HA | mxiE cloned into pDZ1; cat | This study |
Strain and plasmid construction.
BS902 was constructed using the λ red linear recombination method as previously described (7). PCR was used to amplify a kanamycin resistance cassette gene (kan) from pKD4 (Table 1) with 5′ and 3′ overhangs identical to the 5′ and 3′ regions of spa15 internal to the start and stop codons (Table 2). Kanamycin-resistant recombinants were purified and screened via PCR using primers (Table 2) that annealed ∼70 bp upstream and downstream of the spa15 coding region to detect the size difference due to the insertion of the kanamycin cassette. Next, this mutant was used as the donor strain for transduction of 2457T using P1L4 and selection for kanamycin resistance.
TABLE 2.
Primers used in this study
| Purpose | Forward primer |
Reverse primer |
||
|---|---|---|---|---|
| Name | Sequencea | Name | Sequencea | |
| Amplify aphA-3 cassette for spa15 deletion | S15kanF | 5′-AGTAACATTAATTTAGTTCAATTAGTTAGAGATAGTCTTTTCACGATTGGTGTGTAGGCTGGAGCTGCTTC-3′ | S15kanR | 5′-TAAGACCCCATTTAAGATTTCCATCCTCTGATAAAACTCATGCAGAATCTCATATGAATACCTCCTTAG-3′ |
| Confirm Δspa15 deletion | Spa15F2 | 5′-GTTATATCTATGCTGAGATTG-3′ | Spa15R2 | 5′-CCAATCGAAACATCGCTAAG-3′ |
| Clone spa15 into pBSKS | spaF | 5′-GATCGGTACCATGAGTAACATTAATTTAGTTC-3′ | spaR2 | 5′-GATCGGATCCATTATAAGACCCCATTTAAGATTTC-3′ |
| Sequence spa15 in pBSKS | pBSKF | 5′-AGCGGATAACAATTTCACACAGGAAAC-3′ | pBSKR | 5′-GTTTTCCCAGTCACGACGTTG-3′ |
| Clone spa15 into pDZ1 | spaF | 5′-GATCGGTACCATGAGTAACATTAATTTAGTTC-3′ | spaR | 5′-GATCAGATCTTAAGACCCCATTTAAGATTTC-3′ |
| Clone mxiE into pDZ1 | mxiE2HAF | 5′-CTAGGGTACCATGGAAGGGTTTTTTTTTGTCCG-3′ | mxiE2HAR | 5′-AGATCGGATCCAATTTTTTCATTTATTTTTTTCAC-3′ |
| Sequence spa15 or mxiE in pDZ1 | pDZ1F | 5′-CTGGGTTGAAGGCTCTCAAG-3′ | pDZ1R | 5′-TCAGCCCCATACGATATAAG-3′ |
Restriction enzyme sites are underlined.
Additional primers listed in Table 2 were used to construct the pBSKS-spa15, pSpa15-2HA, and pMxiE-2HA plasmids. spa15 was amplified by colony PCR using Platinum Taq high-fidelity DNA polymerase (Invitrogen) and cloned into pGEM-T (Promega) according to the manufacturers' protocols. Subsequently, spa15 was subcloned into pBSKS (Stratagene) via the Acc65I and BamHI restriction enzyme sites using T4 DNA ligase (New England Biolabs). The ligation reaction was transformed into Escherichia coli DH5α for production of the new plasmid, pBSKS-spa15. The spa15 insert sequence was verified with primers that anneal outside the multiple cloning site of the vector. pBSKS-spa15 was subsequently transformed into BS902 to generate BS904. To create C-terminal hemagglutinin (HA) tags of spa15 and mxiE, vector pDZ1 was used. pDZ1 contains two HA epitopes and is a low-copy vector with a pACYC184 origin of replication (46). spa15 and mxiE were amplified by colony PCR as mentioned above, cloned into pGEM-T, and subcloned into pDZ1 using Acc65I and BglII for spa15 and Acc65I and BamHI for mxiE. The ligation reactions were transformed into E. coli DH5α for production of the new plasmids pSpa15-2HA and pMxiE-2HA. The inserts were sequenced using primers that anneal outside the multiple cloning site of pDZ1. The plasmids were transformed into the various strains listed in Table 1 and selected on medium with the appropriate antibiotic.
Virulence assays.
The invasion, plaque, and apoptosis assays were performed as previously described (7, 18, 32). Briefly for the apoptosis assay, bacterial cultures were grown to mid-log phase, standardized to an optical density at 600 nm of 0.72, washed in 1× phosphate buffered saline (PBS), resuspended in 1× Dulbecco's modified Eagle's medium (DMEM), and applied to a semiconfluent monolayer of HeLa cells. The plates were centrifuged at 3,000 rpm for 10 min at 37°C to facilitate the invasion process by allowing the bacteria to make contact with the HeLa cells. The plates were incubated at 37°C with 5% CO2 for 30 min. Both infected and uninfected controls were then washed with 1× PBS, DMEM plus 50 μg/ml gentamicin was added, and the plates were incubated for 3 h at 37°C with 5% CO2. Infected and uninfected cells were then washed again, and DMEM plus 50 μg/ml gentamicin and 4 μM STS (Calbiochem) was added for an additional 3 h. After the assay, the cells were processed for immunofluorescence or Western blot analysis.
CR secretion assay.
The CR secretion assay was used to identify the proteins secreted by the bacteria through the T3SS and was performed as previously described (4). Briefly, bacteria were grown to mid-log phase, resuspended in 1× PBS, and CR was added to a final concentration of 30 μg/ml. The bacteria were incubated at 37°C for 30 min for secretion of early T3SS proteins or at 1-h intervals for secretion of late T3SS effector proteins (41). After incubation, the bacteria were pelleted by centrifugation, and the supernatant was collected and filtered through a 0.22-μm-pore filter and then stored at −20°C. The proteins in the supernatant represent the secreted proteins and were concentrated by trichloroacetic acid precipitation. Trichloroacetic acid pellets were resuspended in 50 μl sodium dodecyl sulfate (SDS) loading buffer for protein analysis and stored at −20°C. The bacterial pellets, representing the nonsecreted proteins, were resuspended in 500 μl of SDS loading buffer and stored at −20°C.
Immunofluorescence and Western blot analysis.
The same procedures were followed for immunofluorescence or Western blot analysis as previously described (7). For the immunofluorescence analysis, infected monolayers were fixed with 1× PBS with 3% formaldehyde (36% stock; Sigma) and 0.2% glutaraldehyde (25% stock; Sigma). To visualize nuclei, 5 mg/ml of 4′,6-diamido-2-phenylindole (DAPI; Molecular Probes) was diluted 1:1,000 in 1× PBS. Nuclei were monitored for a characteristic apoptotic phenotype, namely DNA fragmentation and chromatin condensation, as demonstrated previously (7). To detect activated caspase-3, a rabbit anti-human cleaved caspase-3 antibody (Cell Signaling Technologies) was used with a secondary goat anti-rabbit immunoglobulin G (IgG) antibody conjugated to Alexa 488 (Invitrogen). To measure secretion of the HA-tagged Spa15, infected cells were stained with a mouse monoclonal anti-HA antibody (Covance) followed by a goat anti-mouse IgG antibody conjugated to Alexa 488 (Invitrogen). After the staining procedure, antifade reagent (Molecular Probes) was applied to all samples, which were then overlaid with coverslips and stored at 4°C in the dark.
For Western blot analyses, each sample was resolved by SDS-polyacrylamide gel electrophoresis, and Coomassie blue staining verified equal loading of total protein for all samples. Caspase-3 was detected with rabbit anti-human caspase-3 or cleaved caspase-3 antibodies (Cell Signaling Technology) followed by a donkey anti-rabbit IgG antibody conjugated to horseradish peroxidase (Amersham Biosciences). The HA-tagged Spa15 was detected using a mouse monoclonal anti-HA antibody in a 1:1,000 dilution with a sheep anti-mouse IgG antibody conjugated to horseradish peroxidase (Amersham Biosciences). IpaB and IpaC were detected with mouse monoclonal anti-IpaB (1:20,000 dilution) and anti-IpaC (1:5,000 dilution) antibodies as previously described (31). The same secondary antibodies were used as with the anti-HA antibody. Western blots were developed using the Visualizer developing system (Upstate Cell Signaling Solutions) according to the protocol provided, and the blots were imaged using a Fuji Intelligent Dark Box with a Fujinon lens and Image Reader LAS-3000 software (Fuji) on the chemiluminescence setting in increment mode at 10-s intervals. Densitometry comparisons were made using the Image Gauge V4.22 software provided.
Statistical analysis.
The densitometry results for the caspase-3 Western blot were analyzed using Tukey's analysis of variance (ANOVA) post hoc test on the SPSS program, version 12.0.1, for Windows. For the strain comparison in the CR secretion assay, the ratios of Spa15 secretion in wild-type bacteria (strain BS905) to the ΔmxiE mutant (strain BS906) and to the ΔmxiE/mxiE+-complemented strain (strain BS907) were compared. A repeated-measures ANOVA was performed using Tukey's post hoc analysis for all time points on the SPSS program version 12.0.1 for Windows. Ratios were analyzed on a log scale to comply with the assumptions of ANOVA, and the summaries are reported as geometric mean ratios relative to BS905, averaging across three repetitions.
RESULTS
Identifying spa15 as the antiapoptosis gene.
We constructed deletion mutants in most of the MxiE-regulated genes (Table 3), and we did not find any genes that were responsible for apoptosis protection since all of the mutants prevented STS-induced apoptosis (data not shown). We next utilized a strain of E. coli expressing the Shigella entry region (SER), which contains all of the genes necessary for epithelial cell invasion (27), in order to rule out the MxiE-regulated genes carried on the chromosome (3). This strain of E. coli also prevented STS-induced apoptosis (data not shown). Thus, Shigella-specific chromosomal genes are not required for the antiapoptotic phenotype. Since this strain of bacteria expresses only Shigella genes carried within the SER, we hypothesized that the protective gene/protein is either mxiE itself or a protein that associates with MxiE, namely IpgC, OspD1, or Spa15. IpgC is a coactivator needed for the induction of MxiE-regulated genes, while OspD1 and Spa15 have been described as antiactivators of MxiE (35, 36). We found that a ΔipgC mutant was not invasive, and it was not further pursued. OspD1 was eliminated as a candidate since the E. coli strain carrying the SER blocks apoptosis and ospD1 is not located within the SER (6). Finally, the spa15 gene was a likely candidate since it is located within the SER and encodes a T3SS chaperone that has the ability to bind to several T3SS effector proteins (6, 34). Therefore, we constructed BS902, a Δspa15 mutant of wild-type S. flexneri strain 2457T. The mutant showed a slightly (10%) reduced entry efficiency, similar to published data on a Δspa15 mutant constructed in S. flexneri strain M90T (34). In addition, the mutant produced plaque sizes about half the size as those of wild-type bacteria in the plaque assay (data not shown).
TABLE 3.
Shigella genes analyzed that had no effect on apoptosis protectiona
| MxiE-regulated deletion mutant gene(s) | MxiE-regulated genes via E. coli strainb | Deletion mutant gene whose product associates with Spa15 | Gene whose product associates with Spa15 (via E. coli strain)b |
|---|---|---|---|
| ospBc | ipaH_1 | ipaA | ospC2 |
| ospC1 | ipaH_2 | ipgB1 | ospC3 |
| ospD3 | ipaH_5 | ospB | ospD1 |
| ospE1 | ipaH_7 | ipgB2 | |
| ospE2 | |||
| ospF | |||
| ospG | |||
| virA | |||
| ospE1 ospE2d | |||
| ospC1 ospB ospFe | |||
| ipaH1.4 | |||
| ipaH2.5 | |||
| ipaH4.5 | |||
| ipaH7.8 | |||
| ipaH9.8 | |||
| ipaH_4 |
Strains were analyzed for the ability to inhibit STS-induced apoptosis by DAPI staining of the nuclei of infected cells to determine if the nuclei had the characteristic apoptotic phenotype—namely DNA fragmentation and/or chromatin condensation (7).
The E. coli strain expresses the Shigella entry region and invades epithelial cells (see text).
Gene product also associates with Spa15.
Double mutant.
Triple mutant.
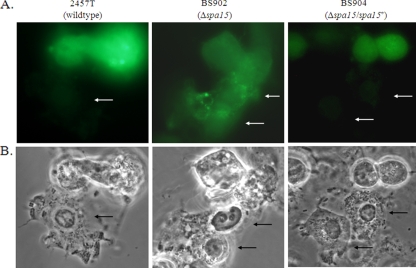
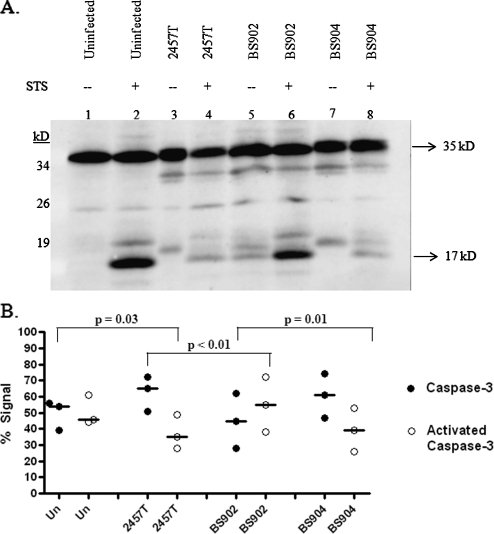
BS902 was next analyzed in the apoptosis assay and was found to be unable to prevent STS-induced apoptosis. We first analyzed infected cells in the presence of STS by DAPI staining. The nuclei of infected cells had characteristic apoptotic nuclei due to DNA fragmentation and chromatin condensation (data not shown). To confirm the DAPI staining, immunofluorescence analysis for caspase-3 activation was performed. Unlike HeLa cells infected with wild-type bacteria, activated caspase-3 was detected in HeLa cells infected with BS902 (Fig. 1). Interestingly, the mutant also appeared to induce apoptosis in infected cells and closely neighboring uninfected cells, even in the absence of STS. DAPI staining of the nuclei of these cells showed classic signs of apoptosis with shape distortion, DNA fragmentation, and/or chromatin condensation (Fig. 2). Complementation of the Δspa15 mutation with the wild-type gene (BS904) restored protection in the apoptosis assay (Fig. 1). The inability of BS902 to prevent STS-induced apoptosis was verified in a Western blot analysis for caspase-3 activation (Fig. 3A). Densitometry analysis revealed there was a significant increase in caspase-3 activation in BS902-infected monolayers treated with STS compared to both 2457T-infected and BS904-infected monolayers treated with STS (Fig. 3B). In addition, the Western blot demonstrated that BS902-infected cells were apoptotic even in the absence of STS, as evidenced by the presence of the 17-kDa band (lane 5). Lanes of uninfected cells, wild-type-infected cells (2457T), or cells infected with the Δspa15/spa15+-complemented strain (BS904) did not have bands at 17 kDa when STS was absent (lanes 1, 3, and 7). In lanes 4 and 8, infected cells treated with STS have some caspase-3 activation, since 90% of the monolayer is infected and the remaining 10% of cells in the monolayer are uninfected and sensitive to STS. This level of activation is expected and similar to what was seen in Fig. 1, in which the uninfected cells in the population have a positive signal for activated caspase-3.
FIG. 1.
A Δspa15 mutant cannot prevent caspase-3 activation in HeLa monolayers treated with STS. (A) Activated caspase-3 (green) immunofluorescence of monolayers infected with 2457T (left), BS902 (center), and BS904 (right). Activated caspase-3 is not present in cells infected with 2457T or BS904 but is present in uninfected cells and cells infected with BS902. Arrows point to infected cells. (B) Phase-contrast view of panel A to visualize the bacteria inside the cells. Images are representative of three repeated experiments. All images were taken with a 100× objective.
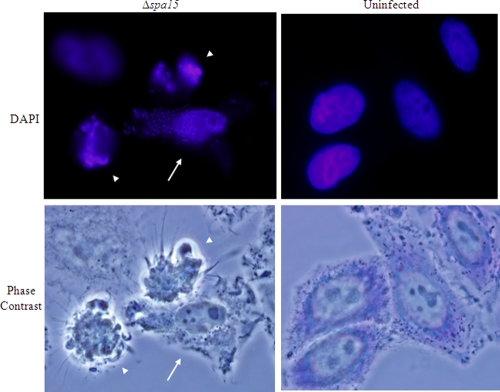
FIG. 2.
BS902-infected cells are apoptotic upon normal infection. HeLa monolayers were infected with the Δspa15 mutant (BS902) without STS for 6 h. (Top) DAPI staining reveals altered nuclear shape, chromatin condensation, and/or DNA fragmentation of the nuclei in infected cells (arrow) and in closely neighboring uninfected cells (arrowheads). The bacteria are visible in the infected cell due to the DAPI staining. Nuclei of uninfected cells have a normal, round shape and visible nucleoli. Bottom images are the phase-contrast views with Giemsa staining of monolayers. Images are representative of three repeated experiments. All images were taken with a 100× objective.
FIG. 3.
Western blot analysis for caspase-3 activation. (A) Whole-cell lysates were separated by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting with antibodies that recognize the full-length, inactive form of caspase-3 (35 kDa) and the 17-kDa fragment resulting from cleavage during activation. There are some extra bands at ∼33 and 18 kDa in lanes containing bacterial strains. These bands are Shigella proteins that cross-react with the anti-caspase-3 antibodies in the Western blot. Adsorption of the antibody significantly reduced the intensity of the bands, but was not able to remove them completely (data not shown). Equal loading was verified by Coomassie staining for total protein (data not shown). (B) Densitometry analysis of the caspase-3 Western blot, with the percent total caspase-3 signal represented on the y axis. The graph is the scatter plot of the amount of inactive caspase at 35 kDa (closed circles) compared to the amount of activated caspase-3 (open circles) at 17 kDa for each treatment of three repeated experiments. A horizontal line represents the median value for each protein in each treatment. P values were determined using Tukey's ANOVA post hoc test with normalization of the data to the mean of the gels. “Un” represents uninfected HeLa cells, while infected monolayers are represented by the strain number used in the treatment groups. All groups were treated with STS.
Finally, we constructed mutations in ipaA and ipgB1, since their products are known to associate with Spa15 and are secreted into epithelial cells (33, 34, 42). The mutants had reduced invasion efficiencies, as previously described (33, 42). We found that neither ipaA nor ipgB1 was required for apoptosis inhibition since both of the mutants protected cells in the apoptosis assay (data not shown). The four other proteins that associate with Spa15 were ruled out as candidate protective proteins since the genes encoding these proteins are not present in the E. coli strain expressing the SER (Table 3). Therefore, the Δspa15 mutant was the only strain besides the ΔmxiE mutant that was unable to protect HeLa cells in the apoptosis assay.
Spa15 is secreted into the cytoplasm of the host cell.
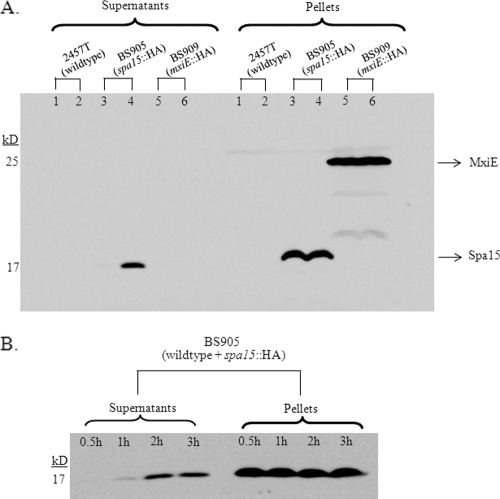
We reasoned that in order to inhibit apoptosis, the bacterial proteins need to be secreted into the cytoplasm of the epithelial cell. Therefore, we constructed C-terminal HA tags of spa15 and mxiE and looked for secretion of these proteins in the CR secretion assay. After construction, the sequences were verified, and the plasmids carrying the tagged spa15 and mxiE were transformed into 2457T-producing strains BS905 and BS909, respectively. The CR assay was performed at time points of 30 min and 2.5 h to determine if protein secretion could be seen within the first 30 min like the Ipa proteins or if secretion required longer incubation times. Using an antibody against the HA tag, we found that MxiE was not secreted at either 30 min or 2.5 h (Fig. 4A). However, Spa15 was secreted by 2.5 h in the CR assay, despite the absence of detectable secretion in the first 30 min (Fig. 4A).
FIG. 4.
Spa15 is secreted in the CR secretion assay. (A) Western blot analysis was performed for the HA tag on Spa15 and MxiE in supernatant and pellet samples from the CR secretion assay. 2457T alone at 30 min (lane 1) and 2.5 h (lane 2) served as a negative control for the anti-HA antibody. 2457T/pSpa15-2HA plasmid (strain BS905) did not secrete the tagged version of Spa15 at 30 min (lane 3) but did secrete at 2.5 h (lane 4). 2457T/pMxiE-2HA plasmid (strain BS909) did not secrete the tagged version of MxiE at either 30 min (lane 5) or 2.5 h (lane 6). The data are representative of three independent experiments. Equal loading was verified by Coomassie staining of total protein (data not shown). (B) Time course of Spa15 secretion in the CR assay. Secretion of Spa15 in 2457T harboring the pSpa15-2HA plasmid (strain BS905) in the CR assay was measured at 0.5, 1, 2, and 3 h. Equal loading was verified by Coomassie staining of total protein (data not shown).
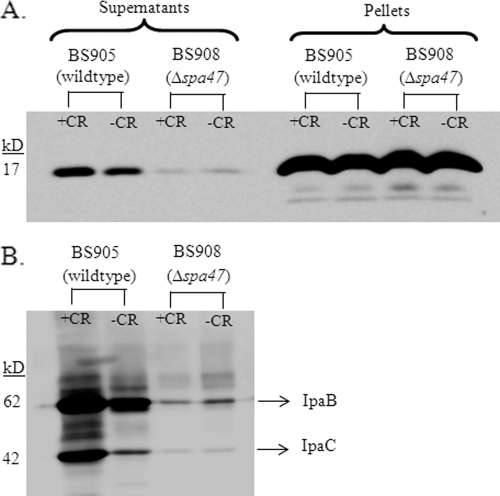
To further define Spa15 secretion, we performed a time course experiment in wild-type bacteria (BS905) in the CR assay and collected samples at 30 min, 1 h, 2 h, and 3 h. We found that Spa15 was only slightly secreted by 1 h and significantly secreted by 2 h in the CR assay (Fig. 4B). To determine if Spa15 secretion is dependent on the T3SS and is CR inducible like other effectors, we transformed the pSpa15-2HA plasmid into a Δspa47 mutant (BS908) and performed the CR assay. Spa47 is the ATPase that provides energy for the T3SS, and a Δspa47 mutant is unable to secrete type III effectors (40, 43). Western blot analysis demonstrated that there was no Spa15 secretion in the Δspa47 mutant at 2.5 h (Fig. 5A). In addition, the secretion of Spa15 like other T3 effectors was inducible with CR since there was a reduction in Spa15 secretion in BS905 when CR was absent (Fig. 5A). This pattern of CR-inducible secretion is the same as those for known Shigella T3SS effectors. To demonstrate that the secretion of Spa15 is similar to those of other T3SS effector proteins, we blotted the same samples for secretion of IpaB and IpaC and found the same pattern of secretion as Spa15 (Fig. 5B). Therefore, Spa15 is secreted in a CR-inducible, T3SS-dependent fashion.
FIG. 5.
Spa15 secretion requires the T3SS. (A) Secretion of Spa15 in 2457T harboring the pSpa15-2HA plasmid (strain BS905) with and without CR was compared to the secretion of Spa15 in the Δspa47 mutant harboring the pSpa15-2HA plasmid (strain BS908) with and without CR. All samples were taken at the 2.5-h time point. These results are representative of three repeated experiments. (B) The supernatants from panel A were blotted and analyzed with antibodies to IpaB (62 kDa) and IpaC (42 kDa).
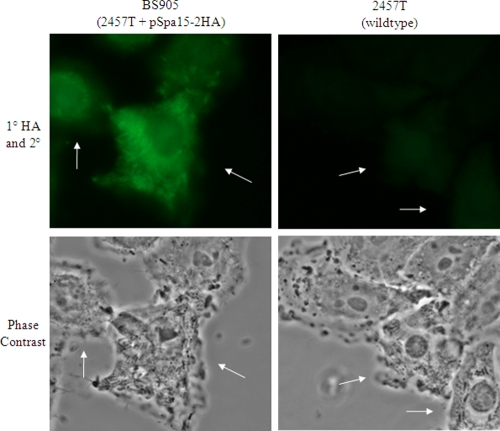
Finally, to determine if secretion of the HA-tagged Spa15 in the CR assay also occurred in infected HeLa cells, BS905 was used in an infection assay in which the intracellular bacterial population was allowed to grow for 3 h in the presence of gentamicin to kill any extracellular bacteria. We chose the 3-h time point since this is when we apply STS in the apoptosis assay. Therefore, the protective bacterial factor should be present in the cytoplasm of the host cell by 3 h for apoptosis inhibition to occur. The infected monolayers were fixed and stained with the primary anti-HA antibody and counterstained with a secondary antibody conjugated to Alexa Fluor 488. There was a strong signal with the anti-HA antibody concentrated around the bacteria and distributed throughout the HeLa cell cytoplasm, indicating that Spa15 was secreted inside epithelial cells by 3 h postinfection (Fig. 6).
FIG. 6.
Spa15 is secreted into the cytoplasm of infected epithelial cells. Immunofluorescence (top) and phase-contrast views (bottom) of HeLa cells infected with BS905 at 3 h postinfection are shown. The HA signal for BS905 is concentrated around the bacteria and distributed throughout the cytoplasm of the host cell, indicating that Spa15 is secreted during infection. There was no significant signal above background when the 2457T-infected HeLa cells were stained with the primary and secondary antibodies (right). The immunofluorescence images were taken with the same exposure time. All images were taken with a 100× objective. Arrows point to infected cells.
Spa15 secretion is delayed in the ΔmxiE mutant.
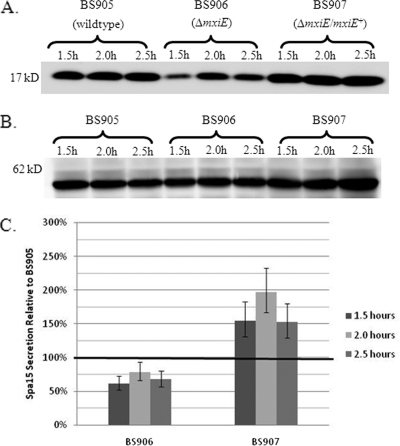
Since the ΔmxiE mutant is unable to prevent STS-induced apoptosis, we hypothesized that the secretion of Spa15 in this mutant was altered. We performed a time course experiment with the CR assay comparing Spa15 secretion in 2457T harboring the pSpa15-2HA plasmid (BS905) to secretion in the ΔmxiE mutant harboring the pSpa15-2HA plasmid (BS906) at 1.5, 2.0, and 2.5 h. We chose these time points since secretion occurs by 2 h in wild-type bacteria. As seen in Fig. 7A, secretion of Spa15 is significantly reduced in BS906 compared to that in BS905. After complementation of the ΔmxiE mutation in the presence of the pSpa15-2HA plasmid (BS907), secretion of Spa15 was restored to wild-type levels (Fig. 7A). We verified equal loading of protein in all samples by both Coomassie staining of total protein (data not shown) and analyzing the secretion of IpaB since IpaB secretion is not affected by the ΔmxiE mutation (Fig. 7B). As shown, the same amount of IpaB was secreted at all time points in all strains. Figure 7C is a graph of the densitometry analysis of Spa15 secretion in which the geometric mean ratios of secretion are plotted. The amount of Spa15 secretion was standardized to the amount of IpaB secretion for each sample. Relative to BS905, there was a significant difference in Spa15 secretion between BS906 and BS907 across all time points, with a P value of <0.001. In addition, Spa15 secretion was consistently decreased in BS906 relative to BS905 since the values fall below the black line representing 100% in Fig. 7C. The P value for these data, as determined by a repeated-measures ANOVA using Tukey's post hoc analysis, is 0.01. Spa15 secretion in BS907 was consistently increased relative to BS905 since the values are above the black line. The P value for these data is 0.002. The reduced secretion in the ΔmxiE mutant most likely explains why this mutant is unable to inhibit STS-induced apoptosis, and the data suggest that the levels of MxiE determine the amount of Spa15 secretion.
FIG. 7.
Spa15 secretion in the ΔmxiE mutant. (A) Time course of Spa15 (17 kDa) secretion in BS905 (wild type) compared to secretion of Spa15 in BS906 (ΔmxiE) and BS907 (ΔmxiE/mxiE+). The CR assay was performed at 1.5, 2.0, and 2.5 h, and Western blot analysis was performed against the HA tag. This Western blot represents three independent experiments. (B) The samples from panel A were blotted for IpaB to confirm equal loading of total protein. (C) Densitometry analysis of Spa15 secretion in BS905, BS906, and BS907. The geometric mean ratios of Spa15 secretion from the Western blot in panel A in BS906 and BS907 are plotted relative to the secretion of Spa15 in BS905 (set at 100% and represented by the black line). The amount of Spa15 secretion was standardized to the amount of IpaB secretion in panel B for each sample. Error bars represent the standard error for the three repeated experiments. A repeated-measures ANOVA using Tukey's post hoc analysis demonstrated that the P value for Spa15 secretion in BS906 relative to BS905 is 0.01. The P value for Spa15 secretion in BS907 relative to BS905 is 0.002. The P value for the difference in Spa15 secretion between BS906 and BS907 is <0.001.
DISCUSSION
We have identified spa15 as the S. flexneri gene required for the inhibition of apoptosis in epithelial cells. Given that the ΔmxiE mutant is unable to prevent STS-induced apoptosis (7), we were surprised to discover that none of the MxiE-regulated genes was required for protection. The repertoire of MxiE-regulated genes has been extensively categorized (3, 22, 24, 28); these genes have a 17-bp sequence defined as the MxiE box that is upstream of the promoter (28). Therefore, we are confident that Table 3 represents all of the MxiE-regulated genes in S. flexneri. Complementation of the ΔmxiE mutation in strain BS613 restored apoptosis protection (data not shown), which led us to investigate proteins known to associate with MxiE to explain the ΔmxiE mutant phenotype. Spa15 associates with MxiE and has been described as a coantiactivator for MxiE to prevent early transcriptional activation of MxiE-regulated genes (35). The Δspa15 mutant was unable to inhibit apoptosis, while complementation with spa15+ restored protection. The small plaque size (data not shown) also suggests that the bacteria are unable to prevent apoptosis in the absence of STS. As the cells become apoptotic, the bacteria are most likely exposed to the extracellular environment where gentamicin is present during the plaque assay (32). Subsequently, the bacteria are killed by the antibiotic and the plaques fail to develop fully.
Even in the absence of STS, Δspa15 mutant-infected cells and closely neighboring uninfected cells were apoptotic. We hypothesize that the bacterial lipopolysaccharide (LPS) is activating the extrinsic pathway of apoptosis since LPS induces apoptosis via caspase-8 activation (45). Furthermore, LPS stimulates tumor necrosis factor alpha (TNF-α) production (8) and epithelial cells secrete TNF-α, interleukin-1α (IL-1α), and IL-6 during infection with Shigella and other pathogens (1, 9, 12, 20, 23). TNF-α induces apoptosis via the extrinsic pathway, and a combination of IL-1α and TNF-α induces apoptosis in epithelial cells (44). IL-6 protects the colonic epithelium during Citrobacter rodentium infection by inducing the expression of antiapoptotic genes BCL-xL, MCL-1, cIAP-2, and BCL-3 (9). We have never observed apoptosis in neighboring uninfected cells in the absence of STS when wild-type bacteria are used to infect the epithelial cells. We hypothesize that the Δspa15 mutant-infected cells are releasing more TNF-α and/or IL-1α (or less IL-6) to induce apoptosis in neighboring uninfected cells. Therefore, it is possible that Shigella not only protects cells that it infects, but also protects closely neighboring uninfected cells. This prosurvival effect on neighboring cells is ideal for the bacteria since Shigella eventually spreads to adjacent cells during infection.
Spa15 has been described as a T3SS chaperone based on the requirement of Spa15 for secretion of several proteins (35). These proteins include IpaA, IpgB1, IpgB2, OspB, OspC2, OspC3, OspD1, and MxiE (16, 34, 35). We found that none of the secreted target proteins was required to inhibit apoptosis (Table 3). Therefore, the only mutants that were unable to inhibit STS-induced apoptosis were the ΔmxiE mutant and the Δspa15 mutant. This result led us to investigate whether MxiE, Spa15, or both proteins were secreted through the T3SS. We suspected that MxiE is not secreted given that it is a transcriptional activator and that a previous report showed that MxiE is not secreted by 30 min in the CR assay (35). However, for both proteins, we wanted to determine if secretion occurred at later time points, as is observed for some T3SS effectors (41). We chose the 2.5-h time point, since we apply STS at 3 h postinfection in the apoptosis assay (7). If the proteins are secreted and inhibit apoptosis, adequate secretion must therefore occur prior to 3 h postinfection. We constructed an HA-tagged version of Spa15 and determined that it was functional, as evidenced by complementation in BS914. BS914 invaded HeLa cells at wild-type levels and protected in the apoptosis assay (data not shown), verifying that the HA tag does not interfere with Spa15 function. Afterwards, we detected secretion of HA-tagged Spa15 in the CR assay.
This report is the first to demonstrate that Spa15 is secreted by the T3SS. We were able to detect Spa15 secretion because we assayed for the tagged protein, and we analyzed later time points in the CR secretion assay. A previous report only analyzed Coomassie-stained protein gels for secreted products of S. flexneri M90T and a Δspa15 mutant at the 30-min time point (35). It is therefore not surprising that secretion of Spa15 was not detected prior to our work. One could argue that sample processing after the CR assay resulted in the appearance of secretion, perhaps due to lysis of the bacteria. The absence of Spa15 secretion in the Δspa47 mutant argues against this possibility since Spa15, like IpaB and IpaC, remained in the pellet fraction and was not secreted due to the absence of Spa47. In addition, the HA tag does not cause secretion of an otherwise nonsecreted protein since the HA-tagged MxiE was not secreted. We are therefore confident that Spa15 is secreted into epithelial cells based on the CR assay and the immunofluorescence analysis of Spa15 secretion in infected epithelial cells (Fig. 6).
Since Spa15 is the antiapoptosis factor and is secreted, we hypothesized that the ΔmxiE mutant did not prevent STS-induced apoptosis because Spa15 secretion was altered. We detected a significant decrease in Spa15 secretion in the ΔmxiE mutant (Fig. 7). Secretion was restored above wild-type levels when the ΔmxiE mutation was complemented (BS907), which is verified by the fact that BS613 inhibits STS-induced apoptosis (data not shown). The comparison of Spa15 secretion between strains BS905, BS906, and BS907 is valid, since the HA tag does not interfere with function. Therefore, not enough Spa15 is secreted from the ΔmxiE mutant in order for this strain to inhibit apoptosis in the presence of STS. Interestingly, epithelial cells infected with the ΔmxiE mutant never appeared apoptotic in the absence of STS (data not shown). The low levels of Spa15 secreted by the ΔmxiE mutant are most likely sufficient to inhibit apoptosis normally during infection but insufficient to inhibit apoptosis in infected cells in the presence of a strong apoptosis inducer like STS.
Based on our data, it appears that the levels of MxiE affect the amount of Spa15 that is secreted. With MxiE absent in strain BS906, Spa15 is not needed to prevent early activation of MxiE-regulated genes. Spa15 secretion above wild-type levels in BS907 suggests that when more MxiE is present, more Spa15 is needed to prevent early transcriptional activation of MxiE-regulated genes. Once this larger amount of Spa15 is released from MxiE, Spa15 is available for secretion by the T3SS. It is also possible that MxiE contributes to the stability of Spa15. The absence of MxiE may cause Spa15 to remain attached to another protein, possibly OspD1, which would reduce the level of Spa15 secretion in the ΔmxiE mutant. Future studies analyzing how MxiE modulates the secretion of Spa15 are clearly needed.
Most T3SS chaperones bind to one or two targets and are encoded next to the genes for these targets. For example, ipgC is immediately upstream of ipaB and ipaC on the Shigella virulence plasmid, and IpgC binds to both IpaB and IpaC to prevent early association of these proteins (30). Spa15 is an atypical chaperone, since it is encoded in the middle of the mxi-spa operon and the genes that encode the Spa15 targets are scattered throughout the 220-kb virulence plasmid (6, 34). Homology searches with BLAST revealed several homologues to Spa15 in other pathogens, including InvB from Salmonella enterica subsp. enterica serovar Typhimurium, SpaK from S. enterica subsp. enterica serovar Paratyphi A, BsaR from several Burkholderia species, and YsaK from Yersinia enterocolitica. At least two of the homologues, InvB and YsaK, are also atypical T3SS chaperones in that they bind to many targets and are not encoded near the genes for those targets (26, 34). These homologues are similar in structure, and there are six conserved residues in the chaperones that define a binding motif (26). Given the sequence identities and similarities, it is possible that some of the homologues are also secreted into eukaryotic cells. The percent identity between Spa15 and the homologues is ∼30%. It remains to be seen if the homologues have additional functions. These T3SS chaperones may truly represent a new, unique class of chaperones.
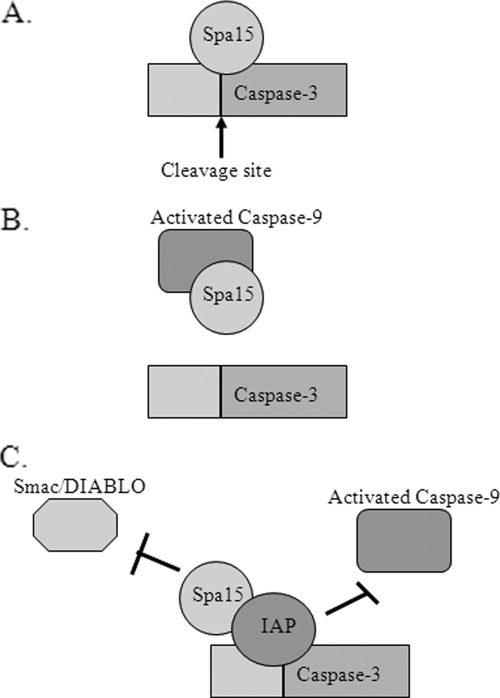
During apoptosis, cytochrome c release from the mitochondria results in caspase-9 activation and subsequent caspase-3 activation (2). Given that S. flexneri inhibits STS-induced apoptosis prior to caspase-3 activation despite the release of cytochrome c and caspase-9 activation, we believe there are a few likely eukaryotic targets for Spa15 to directly inhibit caspase-3 (Fig. 8). First, Spa15 could bind directly to caspase-3 and block the cleavage site. Alternatively, Spa15 could associate with caspase-9 to prevent it from activating caspase-3. This association would not prevent caspase-9 from being activated by the apoptosome (2) but would prevent the activated form of caspase-9 from cleaving caspase-3. Finally, Spa15 could enhance the binding of an inhibitor of apoptosis protein (IAP) to caspase-3. This enhanced binding would prevent the dissociation of the IAP from caspase-3 by proteins like Smac/Diablo that are released from the mitochondria (11). The binding of Spa15 to an IAP would also prevent activated caspase-9 from cleaving caspase-3 since the caspase-3 cleavage site would be blocked by the IAP. We hypothesize that the simple binding of Spa15 to a eukaryotic target would be sufficient for inhibiting caspase-3 activation. We are in the process of identifying potential eukaryotic targets through protein interaction screens.
FIG. 8.
Model of Spa15 function in the eukaryotic cell. Since caspase-3 activation is inhibited in infected cells in the presence of STS, there are three possible mechanisms by which Spa15 could inhibit caspase-3 activation. Spa15 could bind directly to caspase-3 to prevent cleavage and activation (A), inhibit the activated form of caspase-9 to prevent caspase-3 activation (B), or enhance the binding of an IAP to caspase-3 (C). This enhanced binding would prevent Smac/Diablo from dissociating the IAP from caspase-3 and prevent caspase-9 from activating caspase-3.
In conclusion, we have identified Spa15 as the antiapoptosis factor that inhibits apoptosis in epithelial cells during S. flexneri infection. Of equal importance, we were able to demonstrate for the first time that Spa15 is secreted through the T3SS and has effector function in addition to being a T3SS chaperone. Prevention of epithelial cell apoptosis is important for the bacteria to survive inside the host. During infection, many apoptosis stimuli are present. These stimuli include the cytokines TNF-α (21) and Fas ligand (37), leukocyte elastases (15), and the transmigration of PMNs across the colonic epithelium (25) that occurs during Shigella infection (29). If S. flexneri is unable to prevent epithelial cell apoptosis and the infected cells die, the bacteria will be exposed to the extracellular environment that is infiltrated with immune cells. In fact, in vivo studies have verified that only immune cells are apoptotic during Shigella infection (37, 38, 47). Therefore, the inhibition of epithelial cell apoptosis is vital for S. flexneri to establish a replicative niche inside the host in order to survive.
Acknowledgments
This work was supported by grant AI24656 from the National Institute of Allergy and Infectious Diseases. We thank the Henry M. Jackson Foundation and the Uniformed Services University for the Val Hemming Fellowship awarded to C.S.F., as well as the Graduate Education Office at USU for stipend support.
We also thank members of the Maurelli lab for technical assistance and Cara Olsen for help with statistical analysis.
The opinions or assertions contained herein are the private ones of the authors and are not to be construed as official or reflecting the views of the Department of Defense or the Uniformed Services University of the Health Sciences.
Editor: S. M. Payne
Footnotes
Published ahead of print on 5 October 2009.
REFERENCES
- 1.Arondel, J., M. Singer, A. Matsukawa, A. Zychlinsky, and P. J. Sansonetti. 1999. Increased interleukin-1 (IL-1) and imbalance between IL-1 and IL-1 receptor antagonist during acute inflammation in experimental shigellosis. Infect. Immun. 67:6056-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashe, P. C., and M. D. Berry. 2003. Apoptotic signaling cascades. Prog. Neuropsychopharmacol. Biol. Psychiatry 27:199-214. [DOI] [PubMed] [Google Scholar]
- 3.Ashida, H., T. Toyotome, T. Nagai, and C. Sasakawa. 2007. Shigella chromosomal IpaH proteins are secreted via the type III secretion system and act as effectors. Mol. Microbiol. 63:680-693. [DOI] [PubMed] [Google Scholar]
- 4.Bahrani, F. K., P. J. Sansonetti, and C. Parsot. 1997. Secretion of Ipa proteins by Shigella flexneri: inducer molecules and kinetics of activation. Infect. Immun. 65:4005-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardini, M. L., J. Mounier, H. d'Hauteville, M. Coquis-Rondon, and P. J. Sansonetti. 1989. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. USA 86:3867-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchrieser, C., P. Glaser, C. Rusniok, H. Nedjari, H. d'Hauteville, F. Kunst, P. Sansonetti, and C. Parsot. 2000. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol. Microbiol. 38:760-771. [DOI] [PubMed] [Google Scholar]
- 7.Clark, C. S., and A. T. Maurelli. 2007. Shigella flexneri inhibits staurosporine-induced apoptosis in epithelial cells. Infect. Immun. 75:2531-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comstock, K. L., K. A. Krown, M. T. Page, D. Martin, P. Ho, M. Pedraza, E. N. Castro, N. Nakajima, C. C. Glembotski, P. J. Quintana, and R. A. Sabbadini. 1998. LPS-induced TNF-alpha release from and apoptosis in rat cardiomyocytes: obligatory role for CD14 in mediating the LPS response. J. Mol. Cell. Cardiol. 30:2761-2775. [DOI] [PubMed] [Google Scholar]
- 9.Dann, S. M., M. E. Spehlmann, D. C. Hammond, M. Iimura, K. Hase, L. J. Choi, E. Hanson, and L. Eckmann. 2008. IL-6-dependent mucosal protection prevents establishment of a microbial niche for attaching/effacing lesion-forming enteric bacterial pathogens. J. Immunol. 180:6816-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekert, P. G., and D. L. Vaux. 2005. The mitochondrial death squad: hardened killers or innocent bystanders? Curr. Opin. Cell Biol. 17:626-630. [DOI] [PubMed] [Google Scholar]
- 12.Fahey, J. V., T. M. Schaefer, J. Y. Channon, and C. R. Wira. 2005. Secretion of cytokines and chemokines by polarized human epithelial cells from the female reproductive tract. Hum. Reprod. 20:1439-1446. [DOI] [PubMed] [Google Scholar]
- 13.Formal, S. B., G. J. Dammin, E. H. LaBrec, and H. Schneider. 1958. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J. Bacteriol. 75:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ginzberg, H. H., P. T. Shannon, T. Suzuki, O. Hong, E. Vachon, T. Moraes, M. T. Abreu, V. Cherepanov, X. Wang, C. W. Chow, and G. P. Downey. 2004. Leukocyte elastase induces epithelial apoptosis: role of mitochondrial permeability changes and Akt. Am. J. Physiol. Gastrointest. Liver Physiol. 287:G286-G298. [DOI] [PubMed] [Google Scholar]
- 16.Hachani, A., L. Biskri, G. Rossi, A. Marty, R. Menard, P. Sansonetti, C. Parsot, G. T. Van Nhieu, M. L. Bernardini, and A. Allaoui. 2008. IpgB1 and IpgB2, two homologous effectors secreted via the Mxi-Spa type III secretion apparatus, cooperate to mediate polarized cell invasion and inflammatory potential of Shigella flexenri. Microbes Infect. 10:260-268. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Hromockyj, A. E., and A. T. Maurelli. 1989. Identification of Shigella invasion genes by isolation of temperature-regulated inv::lacZ operon fusions. Infect. Immun. 57:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennison, A. V., and N. K. Verma. 2004. Shigella flexneri infection: pathogenesis and vaccine development. FEMS Microbiol. Rev. 28:43-58. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. M. 2004. Murine oviduct epithelial cell cytokine responses to Chlamydia muridarum infection include interleukin-12-p70 secretion. Infect. Immun. 72:3951-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung, H. C., L. Eckmann, S. K. Yang, A. Panja, J. Fierer, E. Morzycka-Wroblewska, and M. F. Kagnoff. 1995. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J. Clin. Investig. 95:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kane, C. D., R. Schuch, W. A. Day, Jr., and A. T. Maurelli. 2002. MxiE regulates intracellular expression of factors secreted by the Shigella flexneri 2a type III secretion system. J. Bacteriol. 184:4409-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le-Barillec, K., J. G. Magalhaes, E. Corcuff, A. Thuizat, P. J. Sansonetti, A. Phalipon, and J. P. Di Santo. 2005. Roles for T and NK cells in the innate immune response to Shigella flexneri. J. Immunol. 175:1735-1740. [DOI] [PubMed] [Google Scholar]
- 24.Le Gall, T., M. Mavris, M. C. Martino, M. L. Bernardini, E. Denamur, and C. Parsot. 2005. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology 151:951-962. [DOI] [PubMed] [Google Scholar]
- 25.Le'Negrate, G., E. Selva, P. Auberger, B. Rossi, and P. Hofman. 2000. Sustained polymorphonuclear leukocyte transmigration induces apoptosis in T84 intestinal epithelial cells. J. Cell Biol. 150:1479-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lilic, M., M. Vujanac, and C. E. Stebbins. 2006. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell 21:653-664. [DOI] [PubMed] [Google Scholar]
- 27.Maurelli, A. T., B. Baudry, H. d'Hauteville, T. L. Hale, and P. J. Sansonetti. 1985. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect. Immun. 49:164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavris, M., P. J. Sansonetti, and C. Parsot. 2002. Identification of the cis-acting site involved in activation of promoters regulated by activity of the type III secretion apparatus in Shigella flexneri. J. Bacteriol. 184:6751-6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormick, B. A., A. M. Siber, and A. T. Maurelli. 1998. Requirement of the Shigella flexneri virulence plasmid in the ability to induce trafficking of neutrophils across polarized monolayers of the intestinal epithelium. Infect. Immun. 66:4237-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menard, R., P. Sansonetti, C. Parsot, and T. Vasselon. 1994. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell 79:515-525. [DOI] [PubMed] [Google Scholar]
- 31.Mills, J. A., J. M. Buysse, and E. V. Oaks. 1988. Shigella flexneri invasion plasmid antigens B and C: epitope location and characterization with monoclonal antibodies. Infect. Immun. 56:2933-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oaks, E. V., M. E. Wingfield, and S. B. Formal. 1985. Plaque formation by virulent Shigella flexneri. Infect. Immun. 48:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohya, K., Y. Handa, M. Ogawa, M. Suzuki, and C. Sasakawa. 2005. IpgB1 is a novel Shigella effector protein involved in bacterial invasion of host cells. Its activity to promote membrane ruffling via Rac1 and Cdc42 activation. J. Biol. Chem. 280:24022-24034. [DOI] [PubMed] [Google Scholar]
- 34.Page, A. L., P. Sansonetti, and C. Parsot. 2002. Spa15 of Shigella flexneri, a third type of chaperone in the type III secretion pathway. Mol. Microbiol. 43:1533-1542. [DOI] [PubMed] [Google Scholar]
- 35.Parsot, C., E. Ageron, C. Penno, M. Mavris, K. Jamoussi, H. d'Hauteville, P. Sansonetti, and B. Demers. 2005. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol. Microbiol. 56:1627-1635. [DOI] [PubMed] [Google Scholar]
- 36.Pilonieta, M. C., and G. P. Munson. 2008. The chaperone IpgC copurifies with the virulence regulator MxiE. J. Bacteriol. 190:2249-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raqib, R., C. Ekberg, P. Sharkar, P. K. Bardhan, A. Zychlinsky, P. J. Sansonetti, and J. Andersson. 2002. Apoptosis in acute shigellosis is associated with increased production of Fas/Fas ligand, perforin, caspase-1, and caspase-3 but reduced production of Bcl-2 and interleukin-2. Infect. Immun. 70:3199-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sansonetti, P. J., J. Arondel, J. R. Cantey, M.-C. Prévost, and M. Huerre. 1996. Infection of rabbit Peyer's patches by Shigella flexneri: effect of adhesive or invasive bacterial phenotypes on follicle-associated epithelium. Infect. Immun. 64:2752-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroeder, G. N., and H. Hilbi. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin. Microbiol. Rev. 21:134-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamano, K., S. Aizawa, E. Katayama, T. Nonaka, S. Imajoh-Ohmi, A. Kuwae, S. Nagai, and C. Sasakawa. 2000. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyotome, T., T. Suzuki, A. Kuwae, T. Nonaka, H. Fukuda, S. Imajoh-Ohmi, T. Toyofuku, M. Hori, and C. Sasakawa. 2001. Shigella protein IpaH9.8 is secreted from bacteria within mammalian cells and transported to the nucleus. J. Biol. Chem. 276:32071-32079. [DOI] [PubMed] [Google Scholar]
- 42.Tran Van Nhieu, G., A. Ben-Ze'ev, and P. J. Sansonetti. 1997. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 16:2717-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venkatesan, M. M., J. M. Buysse, and E. V. Oaks. 1992. Surface presentation of Shigella flexneri invasion plasmid antigens requires the products of the spa locus. J. Bacteriol. 174:1990-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright, K., G. Kolios, J. Westwick, and S. G. Ward. 1999. Cytokine-induced apoptosis in epithelial HT-29 cells is independent of nitric oxide formation. Evidence for an interleukin-13-driven phosphatidylinositol 3-kinase-dependent survival mechanism. J. Biol. Chem. 274:17193-17201. [DOI] [PubMed] [Google Scholar]
- 45.Yu, L. C., A. N. Flynn, J. R. Turner, and A. G. Buret. 2005. SGLT-1-mediated glucose uptake protects intestinal epithelial cells against LPS-induced apoptosis and barrier defects: a novel cellular rescue mechanism? FASEB J. 19:1822-1835. [DOI] [PubMed] [Google Scholar]
- 46.Zurawski, D. V., C. Mitsuhata, K. L. Mumy, B. A. McCormick, and A. T. Maurelli. 2006. OspF and OspC1 are Shigella flexneri type III secretion system effectors that are required for postinvasion aspects of virulence. Infect. Immun. 74:5964-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zychlinsky, A., K. Thirumalai, J. Arondel, J. R. Cantey, A. O. Aliprantis, and P. J. Sansonetti. 1996. In vivo apoptosis in Shigella flexneri infections. Infect. Immun. 64:5357-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]