Abstract
RfaH is a transcriptional antiterminator that reduces the polarity of long operons encoding secreted and surface-associated cell components of Salmonella enterica serovar Typhimurium, including O antigen and lipopolysaccharide core sugars. A ΔrfaH mutant strain is attenuated in mice (50% lethal dose [LD50], >108 CFU). To examine the potential for using rfaH in conjunction with other attenuating mutations, we designed a series of strains in which we replaced the native rfaH promoter with the tightly regulated arabinose-dependent araC PBAD promoter so that rfaH expression was dependent on exogenously supplied arabinose provided during in vitro growth. Following colonization of host lymphoid tissues, where arabinose was not available, the PBAD promoter was no longer active and rfaH was not expressed. In the absence of RfaH, O antigen and core sugars were not synthesized. We constructed three mutant strains that expressed different levels of RfaH by altering the ribosome-binding sequence and start codon. One mutation, ΔPrfaH178, was introduced into the attenuated vaccine strain χ9241 (ΔpabA ΔpabB ΔasdA) expressing the pneumococcal surface protein PspA from an Asd+ balanced-lethal plasmid. Mice immunized with this strain and boosted 4 weeks later induced higher levels of serum immunoglobulin G specific for PspA and for outer membrane proteins from other enteric bacteria than either an isogenic ΔrfaH derivative or the isogenic RfaH+ parent. Eight weeks after primary oral immunization, mice were challenged with 200 LD50 of virulent Streptococcus pneumoniae WU2. Immunization with ΔPrfaH178 mutant strains led to increased levels of protection compared to that of the parent χ9241 and of a ΔrfaH derivative of χ9241.
When recombinant attenuated Salmonella enterica serovar Typhimurium vaccines (RASV) are used to deliver heterologous antigens, it may be advantageous to reduce the host immune response against the RASV carrier, thereby enhancing the immune response against the heterologous antigen. The dominant immunogen on the Salmonella cell surface is lipopolysaccharide (LPS) O antigen (41). However, strains with mutations that eliminate LPS O antigen may be less immunogenic due to their failure to colonize the intestinal tract and to invade intestinal mucosal cells (43, 44). We hypothesized that in vivo-programmed downregulation of O-antigen expression, occurring after colonization of host lymphoid tissues, would serve to reduce the immune response against the RASV carrier while triggering a strong immune response against heterologous antigens (10) and outer membrane proteins cross-reactive with other enteric bacteria (28).
The genes for LPS core and O-antigen biosynthesis are clustered into long operons (37, 49) that cannot be fully transcribed if the native promoter is replaced by a heterologous promoter. RfaH, a transcriptional antiterminator, reduces the polarity of long operons by binding to the ops sequence, located in an untranslated 5′ region of the transcript, and interacting with the transcription complex (1). RfaH is required for the expression of secreted and surface-associated cell components of S. enterica serovar Typhimurium, including O antigen and core sugar components of LPS (3, 39). rfaH mutant strains produce truncated LPS and reduced amounts of O antigen and core (24), rendering them sensitive to human serum (27), hypersensitive to bile, and attenuated in mice (26, 45). ΔrfaH mutants are immunogenic in mice, inducing a protective immune response against Salmonella challenge (27).
The major immunogenic surface molecules of S. enterica are the O antigen and flagella. Complete LPS is of considerable importance, as rough mutants of Salmonella lacking LPS O-antigen side chains or portions of the core are avirulent, fail to colonize the intestinal tract, and are deficient in invading cells of the intestinal mucosa (43). To circumvent this problem, we and others have explored different ways to achieve regulated O-antigen synthesis so that O antigen is synthesized in vitro but not in vivo, creating vaccine strains that are phenotypically wild type at the time of immunization and become attenuated after colonization of host tissues. We have termed this strategy regulated delayed attenuation (11, 12, 22). One means to achieve regulated delayed attenuation is the deletion of certain genes essential for O-antigen synthesis, such as pmi (manA) (9, 12, 22) or galE (15, 19, 42). Strains with pmi or galE deletions have a reversibly rough phenotype because they are able to synthesize complete O antigen or O antigen and entire core only when grown in the presence of mannose or galactose, respectively. When grown in the presence of their respective sugars, these mutants are fully fit to carry out host colonization and invasion of host tissues during the early stages of infection (12, 18). Upon reaching deeper tissues where free mannose and galactose are not available, O antigen is no longer synthesized and the strains become phenotypically rough.
Another strategy for achieving regulated delayed attenuation relies on replacement of the promoter of a gene of interest with the arabinose-regulated araC PBAD promoter (11, 12). The araC PBAD promoter has been used to develop regulated delayed attenuation strains in which the expression of a number of Salmonella virulence genes, such as fur, crp, and rpoS, is dependent upon arabinose availability (11). In this work, the rfaH promoter, including sequences for activator or repressor protein binding, was deleted and replaced with an araC PBAD cassette to yield Salmonella strains in which rfaH transcription was arabinose dependent. By manipulation of translation signals, we constructed a series of strains, each synthesizing different amounts of RfaH. Growth of these strains in the presence of arabinose permitted transcription of rfaH and synthesis of full-length O antigen. We evaluated these strains for virulence, immunogenicity, and the ability to deliver a test antigen, the pneumococcal protein PspA. Immunized mice were challenged with virulent Streptococcus pneumoniae to determine protective efficacy.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. Typhimurium cultures were grown at 37°C in LB broth (4) or nutrient broth (Difco) or on LB agar with or without 0.1% arabinose. Selenite broth, with or without supplements, was used for enrichment of Salmonella from mouse tissues. Diaminopimelic acid was added (50 μg/ml) for the growth of Δasd strains (30). LB agar containing 5% sucrose was used for sacB gene-based counterselection in allelic-exchange experiments. S. pneumoniae WU2 was cultured on brain heart infusion agar containing 5% sheep blood or in Todd-Hewitt broth plus 0.5% yeast extract. MOPS (morpholinepropanesulfonic acid) minimal medium (32) with or without 10 μg/ml p-aminobenzoic acid was used to confirm the phenotype of ΔpabA ΔpabB mutants.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. Typhimurium | ||
| χ3761 | Wild type; UK-1 | 16 |
| χ9430 | χ3761 ΔpagL7::pYA4284 | Laboratory stock |
| χ9241 | ΔpabA1516 ΔpabB232 ΔasdA16 ΔaraBAD23 ΔrelA198::araC PBADlacI TT | 51 |
| χ9660 | ΔPrfaH176TT araC PBADrfaH TT | χ3761 |
| χ9734 | ΔPrfaH177TT araC PBADrfaH TT | χ3761 |
| χ9735 | ΔPrfaH178TT araC PBADrfaH TT | χ3761 |
| χ9945 | ΔrfaH49 | χ3761 |
| χ9884 | ΔrfaH49 | χ9241 |
| χ9852 | ΔPrfaH178TT araC PBADrfaH TT | χ9241 |
| χ9761 | Δ(galE-uvrB)-1005 ΔmsbB48 ΔfliC2426 ΔpefA1225 ΔfimA2119 ΔfimH1019 ΔagfBAC811 | Laboratory stock |
| E. coli | ||
| χ7232 | endA1 hsdR17 (rK− mK+) glnV44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 λ pirdeoR (φ80dlac Δ(lacZ)M15) | 34 |
| χ7213 | thi-1 thr-1 leuB6 glnV44 fhuA21 lacY1 recA1 RP4-2-Tc::Mu[λ pir] ΔasdA4 Δ(zhf-2::Tn10) | 34 |
| S. pneumoniae WU2 | Wild-type virulent, encapsulated type 3 | 5 |
| pYA3700 | TT araC PBAD cassette plasmid; Apr | 46 |
| pRE112 | sacBmobRP4 R6K ori Cmr | 14 |
| pYA4284 | pRE112; ΔpagL7 | Laboratory stock |
| pYA4718 | ΔrfaH49 | pRE112 |
| pYA4721 | ΔPrfaH176 ctcgagAGGAgtcattATGa | pRE112 |
| pYA4301 | ΔPrfaH177 ctcgagAAGAgtcattATG | pRE112 |
| pYA4304 | ΔPrfaH178 ctcgagAAGAgtcattGTG | pRE112 |
| pYA3493 | Plasmid Asd+; pBRori β-lactamase signal sequence-based periplasmic secretion plasmid | 20 |
| pYA4088 | 852-bp DNA encoding the α-helical region of PspA from aa 3-285 in pYA3493 | 51 |
The Shine-Dalgarno sequence and start codon for each rfaH allele are indicated in uppercase, bold letters.
Plasmids and mutant strain construction.
DNA manipulations were carried out as described previously (36). Transformation of Escherichia coli and S. enterica was done by electroporation. Transformants were selected on LB agar plates containing appropriate antibiotics. Selection for Asd+ plasmids was done on LB agar plates. The primers used in this work are listed in Table 2. A 500-bp DNA fragment containing the region upstream of the rfaH promoter was PCR amplified using the S. Typhimurium χ3761 genome as a template with primers RfaH1-FXmaI-pstI and RfaH1-RPstI (Table 2). The PCR-amplified fragment was digested with PstI and cloned into the PstI site of vector pYA3700 (Table 1), which lies just upstream of araC. Primer T4TT-R, which binds to the T4 transcriptional terminator antisense sequence present downstream of the PstI site in pYA3700, and primer RfaH1-RPstI were used to screen plasmid isolates for inserts in the correct orientation. This intermediate plasmid was digested with XhoI and KpnI, restriction sites that lie downstream of the araC PBAD cassette. Three different 0.5-kb PCR fragments, including the entire 489-bp rfaH gene and an additional 21 bp downstream, were amplified from the S. Typhimurium χ3761 genome, using three different upstream primers, RfaH2-FXhoI, RfaH2-1, and RfaH2-2, and the same downstream primer, RfaH2-RkpnI. Each PCR fragment was engineered to encode a different Shine-Dalgarno (SD) sequence and/or start codon (Table 1) due to differences in the upstream primers. The PCR fragments were digested with XhoI and KpnI and inserted into the intermediate plasmid described above. The three resulting constructs were confirmed by DNA sequence analysis. Then, 2.5-kb DNA fragments including araC PBAD, rfaH, and rfaH 5′ and 3′ flanking regions were excised from each of the plasmids by digestion with KpnI and XmaI and inserted into pRE112, resulting in plasmids pYA4721, pYA4301, and pYA4304. To construct the ΔrfaH49 deletion, two pairs of primers, RfaH-1F/RfaH-1R and RfaH-2F/RfaH-2R, were used to amplify approximately 300-bp fragments upstream and downstream of rfaH, respectively, from the χ3761 genome. The two PCR fragments were purified in agarose gels and used as templates at a 1:1 molar ratio for joining by PCR using primers RfaH-1F and RfaH-2R. The resulting PCR product was digested with KpnI and XmaI and ligated with plasmid pRE112, digested with the same two enzymes, resulting in plasmid pYA4718, which carried a deletion of the entire rfaH gene from ATG to TAA. The mutations were introduced into S. Typhimurium χ3761 by allelic exchange using the four suicide vectors pYA4718, pYA4721, pYA4301, and pYA4304 to generate χ9945, χ9660, χ9734, and χ9735, respectively. The ΔrfaH49 and PrfaH178 mutations were also introduced into S. Typhimurium strain χ9241 to yield strains χ9884 and χ9852, respectively. The presence of both pabA1516 and pabB232 mutations in Salmonella strains χ9241, χ9884, and χ9852 was verified by the inability of the strains to grow in MOPS minimal medium without p-aminobenzoate. The presence of the ΔasdA16 mutation was confirmed by inability to grow in media without diaminopimelic acid and by PCR. The ΔaraBAD23 mutation was verified by PCR and by its white colony phenotype on MacConkey agar supplemented with 1% arabinose. LPS profiles of Salmonella strains were examined by the methods of Hitchcock and Brown (17) using cultures standardized based on the optical density at 600 nm (OD600).
TABLE 2.
Primers used in this work
| Primer name | Sequence (5′-3′) |
|---|---|
| RfaH1-FXmaI-pstI | ACTGCCTGCAGCCCGGGCTTAAACTGCGGCAGCTTGTC |
| RfaH1-RPstI | ACTGCCTGCAGCTTCATCCTTTAAGCCGTATC |
| RfaH2-FXhoI | ATGCACTCGAGAGGAGTCATTATGCAATCCTGGTATT |
| RfaH2-RkpnI | CGACGGTACCGGTTGTATTCTGAACGATCGC |
| RfaH2-1 | ATGCACTCGAGAAGAGTCATTATGCAATCCTGGTATT |
| RfaH2-2 | ATGCACTCGAGAAGAGTCATTGTGCAATCCTGGTATT |
| RfaH-1F | ACTGGGTACCCCATCGCCACGCGTTTTGGC |
| RfaH-1R | TGAACGATCGCCTGCAGGAATGACTCTTATCCGCTTG |
| RfaH-2F | AGTCATTCCTGCAGGCGATCGTTCAGAATACAACCT |
| RfaH-2R | ATCT CCCGGGTTATGCACTGCCGGGACGCG |
| T4TT-R | ATCACAATTCTAGGATAGAAT |
P22 transduction studies.
P22HT int (40) was propagated on S. Typhimurium strain χ9430 carrying the integrated suicide plasmid pYA4284, which confers chloramphenicol resistance. The strains to be tested were grown overnight in nutrient broth at 37°C. The cultures were diluted 1:100 in fresh, prewarmed nutrient broth with or without 0.1% arabinose and grown at 37°C to an OD600 of 0.9. Then, 10 μl of phage (1 × 108 PFU) was added to 1 ml of cells (5 × 108 CFU), and the mixture was incubated at room temperature for 30 min, centrifuged, and resuspended in 200 μl of buffered saline with gelatin (BSG). A 100-μl aliquot was spread onto LB agar plates containing 15 μg/ml chloramphenicol and incubated overnight at 37°C. The colonies were counted the following day. This experiment was performed twice.
MIC test.
The MICs of different antimicrobial substances were determined by using 96-well tissue culture plates (50). Twofold serial dilutions of the bile salt, deoxycholate (0.1 to 50 mg/ml), and polymyxin B (0.1 to 10 μg/ml) were made across the plates. Bacteria were grown at an OD600 of 0.8 to 0.9 in nutrient broth with or without 0.1% arabinose and washed in phosphate-buffered saline (PBS). The cells were diluted to 1 × 105 to 1 × 106 CFU in nutrient broth with or without arabinose, and 0.1 ml of the diluted cell suspension was added to each well. The microtiter plates were incubated overnight at 37°C. The OD of each culture was determined using a SpectraMax M2e (Molecular Devices, CA) plate reader. The threshold of inhibition was 0.1 at OD600. The actual titers were determined by spreading culture dilutions onto LB plates, followed by overnight incubation at 37°C. Assays were repeated at least three times.
Swarming.
Swarming motility was assessed on LB plates solidified with 0.3% agar and supplemented with 0.5% glucose with or without 0.1% arabinose. The plates were allowed to dry at room temperature for 2 h. Freshly grown bacteria were collected from LB agar plates without arabinose, washed, and diluted in saline. Six microliters of bacterial suspension (approximately 1 × 106 CFU) was spotted onto the middle of the plates, which were subsequently incubated at 37°C for 6 h. The diameters of the swarming colonies were measured. The experiments were repeated three times.
Determination of virulence in mice.
Seven-week-old female BALB/c mice were obtained from the Charles River Laboratories (Wilmington, MA). All animal procedures were approved by the Arizona State University Animal Care and Use Committees. The mice were acclimated for 7 days after arrival before the experiments were started.
For determination of the 50% lethal dose (LD50), bacteria were grown statically overnight at 37°C in LB broth, diluted 1:50 in fresh medium containing 0.1% arabinose, and grown with aeration (180 rpm) at 37°C. When the cultures reached an OD600 of 0.8 to 0.9, they were harvested by room temperature centrifugation at 4,000 rpm, washed once, and normalized to the required inoculum density in BSG by adjusting the suspension to the appropriate OD600 value. Groups of five mice each were infected orally with 20 μl containing various doses of S. Typhimurium χ3761 or its derivatives, ranging from 1 × 103 CFU to 1 × 108 CFU. The animals were observed for 4 weeks postinfection, and deaths were recorded daily.
To evaluate colonization, mice were orally inoculated with 20 μl of BSG containing 1 × 109 CFU of each strain. At days 4 and 8 after inoculation, three animals per group were euthanized and spleen and liver samples were collected. Each sample was homogenized in a total volume of 1 ml BSG, and dilutions of 10−1 to 10−6 (depending on the tissue) were plated onto MacConkey agar and LB agar, each containing 0.1% arabinose, to determine the number of viable bacteria. A 0.1-ml aliquot of each tissue sample was enriched for Salmonella by introduction into selenite cysteine broth and incubated for 14 h at 37°C. Samples that were negative by direct plating and positive by enrichment in selenite cysteine broth were recorded as <10 CFU/g.
Immunogenicity of vaccine strains in mice.
RASV strains were grown statically overnight in LB broth with 0.1% arabinose at 37°C. The following day, 2 ml of the overnight culture was inoculated into 100 ml of LB broth with 0.1% arabinose and grown with aeration at 37°C to an OD600 of 0.8 to 0.9. The cells were harvested by room temperature centrifugation at 4,000 rpm for 15 min, and the pellet was resuspended in 1 ml of BSG. Mice were orally inoculated with 20 μl of BSG containing 1 × 109 CFU of each strain on day 0 and boosted on day 28 with the same dose of the same strain. Blood was obtained by mandibular vein puncture at biweekly intervals. The blood was allowed to coagulate at 37°C for 2 hours. Following centrifugation, the serum was removed from the whole blood and stored at −20°C.
Antigen preparation.
Recombinant PspA (rPspA) protein was purified as described previously (20). The rPspA clone that encodes the α-helical region of PspA (amino acids 1 to 302) in pET20b was a kind gift from Susan Hollingshead at the University of Alabama at Birmingham. S. Typhimurium LPS was purchased from Sigma. Outer membrane proteins were prepared as described previously (20). To prepare whole-cell antigens, various enteric bacteria were grown statically overnight at 37°C in LB broth, diluted 1:50 in fresh medium, and grown with aeration (180 rpm) at 37°C to an OD600 of 0.8 to 0.9. The cells were harvested by centrifugation at 4,000 rpm, washed once in PBS, and suspended in coating buffer (50 mM Na2CO3, 50 mM NaHCO3, 0.1% sodium azide, pH 9.6) to an OD600 of 0.8. Then, 100 μl/well of the resulting cell suspension was used to coat the enzyme-linked immunosorbent assay (ELISA) plate overnight at 4°C.
SDS-PAGE and Western blot analyses.
Protein samples were boiled for 5 min and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For Western blotting, proteins separated by SDS-PAGE were transferred electrophoretically to nitrocellulose membranes. The membranes were blocked with 3% skim milk in 10 mM Tris-0.9% NaCl (pH 7.4) and incubated with rabbit polyclonal antibodies specific for PspA (51), LacI, or GroEL (Sigma, St. Louis, MO). Rabbits were immunized with purified His-tagged LacI protein to obtain the anti-LacI antibodies. The secondary antibody was an alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Sigma). Immunoreactive bands were detected by the addition of BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium solution (Sigma). The reaction was stopped after 2 min by washing the blots with large volumes of deionized water.
ELISA.
ELISA was used to assay serum antibodies against S. Typhimurium LPS; rPspA; bacterial outer membrane proteins, including those from Salmonella (SOMPs); and whole-cell bacterial suspensions (1 × 109 CFU/ml), as previously described (23). Color development (absorbance) was recorded at 405 nm using a SpectraMax M2e automated ELISA plate reader (Molecular Devices, Menlo Park, CA). Absorbance readings 0.1 higher than PBS control values were considered positive.
Pneumococcal challenge.
We assessed the protective efficacy of immunization with the attenuated Salmonella strains expressing pspA at week 8 by intraperitoneal (i.p.) challenge with 4 × 104 CFU of S. pneumoniae WU2 in 200 μl of BSG (31). The LD50 of S. pneumoniae WU2 in BALB/c mice was 2 × 102 CFU by i.p. administration (data not shown). The challenged mice were monitored daily for 30 days.
Statistical analysis.
Antibody titer data were expressed as geometric means, and the relative immunoreactivity was expressed as an arithmetic mean. The means were evaluated by two-way analysis of variance and a chi-square test for multiple comparisons among groups. A P value of <0.05 was considered statistically significant.
RESULTS
Mutant construction and LPS phenotypes.
We constructed S. Typhimurium χ3761 derivatives, each designed to synthesize different amounts of RfaH, with rfaH transcribed from the arabinose-regulated araC PBAD promoter. Figure 1A illustrates the chromosomal structures of the araC PBAD rfaH mutant strains. Ninety-eight base pairs upstream of the start codon were replaced with araC PBAD to create the ΔPrfaH177 mutation. A different SD sequence or a GTG start codon was introduced to create ΔPrfaH176 and ΔPrfaH178, respectively. The SD and start codon sequences for each mutant strain are given in Table 1.
FIG. 1.
Arabinose regulation of rfaH. (A) Map of deletion-insertion mutations resulting in arabinose-regulated rfaH expression. (B) LPS phenotypes of wild-type S. Typhimurium χ3761 and the indicated isogenic derivatives. LPSs from different mutant strains grown in nutrient broth with (+) or without (−) 0.1% arabinose were silver stained after separation by 12% SDS-PAGE. (C) Western blots of LPS preparations from panel B. The blots were probed with anti-Salmonella group B antibodies.
The levels of O-antigen synthesis in the mutants were determined by silver staining (Fig. 1B) and by Western blotting using anti-Salmonella group B O-antigen serum (Fig. 1C). Lack of rfaH expression, because of either deletion (ΔrfaH49) or growth of PBAD rfaH strains in the absence of arabinose, resulted in reduced O-antigen synthesis in all four mutant strains. In the three araC PBAD rfaH mutants, nearly wild-type O-antigen levels were restored by the addition of arabinose to the growth medium. None of the three arabinose-regulated rfaH mutants were completely rough when grown in the absence of arabinose, although the ΔPrfaH178 strain produced the smallest amount of high-molecular-weight O antigen (Fig. 1B and C). The ΔPrfaH176 mutant, with a canonical SD sequence and an ATG start codon, would be expected to synthesize the most RfaH, and the ΔPrfaH178 mutant, with a nonideal SD sequence and a GTG start codon, would be expected to synthesize the least RfaH among the three strains. Although we did not directly measure the amount of RfaH synthesized by each strain, the differing amounts of O antigen produced were consistent with our expectations, with the PrfaH176 mutant producing the most O antigen and the PrfaH178 mutant producing the least in the absence of arabinose. The ΔrfaH49 mutant did not produce any detectable high-molecular-weight O antigen with or without arabinose (Fig. 1B and C).
To further evaluate arabinose-regulated O-antigen synthesis, we performed infection studies with the O-antigen-specific phage P22. The strains were grown in nutrient broth with or without arabinose and used as recipients for transduction assays. When strains χ9660 (ΔPrfaH176), χ9734 (ΔPrfaH177), and χ9735 (ΔPrfaH178) were grown with arabinose, the number of transductants obtained was similar to that obtained with the wild-type parent strain, χ3761 (Table 3). In the absence of arabinose, the number of transductants was reduced about 10-fold, which was still about 3- to 4-fold more than that obtained with the ΔrfaH49 strain, χ9445. These results are consistent with our observations that in the absence of arabinose, the strains with arabinose-regulated rfaH make less full-length O antigen than χ3761 and more than χ9445 (Fig. 1).
TABLE 3.
MICs of antibiotic substances, swarming motilities, transduction efficiencies, and virulences of S. Typhimurium strain χ3761 and its rfaH mutant derivatives
| Strain | Presence of 0.1% arabinosea | No. of P22 transductantsb | MIC |
Swarming motility (mm) | Oral LD50 (CFU) | |
|---|---|---|---|---|---|---|
| DOCc (mg/ml) | Polymyxin B (μg/ml) | |||||
| χ9660 (ΔPrfaH176) | − | 305 | 3.2 | 0.25 | 14.5 | NDd |
| + | 3,270 | 6.25 | 0.60 | 21.3 | ND | |
| χ9734 (ΔPrfaH177) | − | 266 | <1.6 | <0.12 | 12.2 | ND |
| + | 3,980 | 3.2 | 1.18 | 20.2 | 1.0 × 106 | |
| χ9735 (ΔPrfaH178) | − | 339 | 1.6 | <0.12 | 8.0 | ND |
| + | 4,340 | 6.25 | 0.60 | 18.1 | 1.0 × 106 | |
| χ9445 (ΔrfaH49) | − | 88 | <1.6 | <0.12 | 3.0 | ND |
| + | ND | <1.6 | <0.12 | 3.0 | >1.0 × 108 | |
| χ3761 (RfaH+) | − | 3,180 | 6.25 | 0.25 | 18.3 | 1.0 × 104 |
| + | ND | 6.25 | 0.60 | 21.0 | ND | |
+, present; −, absent.
The phage lysate used for transduction was grown on a chloramphenicol-resistant strain. Transduction was performed as described in Materials and Methods. The results reflect the numbers of chloramphenicol-resistant colonies obtained after transduction.
DOC, deoxycholate.
Not determined.
Phenotypic evaluation of rfaH mutant strains.
A previous report indicated that an rfaH deletion resulted in increased sensitivity to antimicrobial peptides (26). Therefore, we evaluated our mutant strains for sensitivity to the bile salt deoxycholate and to the antimicrobial peptide polymyxin B. The deoxycholate and polymyxin B MICs for the wild-type strain, χ3761, were >5-fold higher than for the ΔrfaH49 mutant, χ9945 (Table 3). These results are in agreement with previous reports that a Salmonella sp. strain SL1344 ΔrfaH mutant strain was more sensitive to polymyxin B than the wild type (26). Strain χ9660, the arabinose-regulated PrfaH176 mutant, had MICs for both test substances similar to those of the wild type, exhibiting only a twofold reduction in the MIC in the absence of arabinose, consistent with the leaky phenotype observed for O-antigen synthesis (Fig. 1). The other two arabinose-regulated rfaH strains, χ9734 and χ9735, had nearly wild-type MICs for deoxycholate and polymyxin B in the presence of arabinose. The MICs in the absence of arabinose were identical to those of the ΔrfaH49 strain, χ9945.
Rough mutants are unable to swarm because of insufficient surface wetness (47). Therefore, we evaluated the capacities of our mutant strains to exhibit swarming motility on soft agar (Table 3). Strain χ9945 (ΔrfaH49) did not swarm, as expected. Swarming was dramatically reduced (>2-fold reduction in diameter) in the absence of arabinose for the arabinose-regulated rfaH strain χ9735 (ΔPrfaH178). The remaining two strains, χ9660 (ΔPrfaH176) and χ9734 (ΔPrfaH177), showed slight reductions in swarming (<2-fold reduction in diameter) in the absence of arabinose. Swarming was restored to wild-type levels in the presence of arabinose in all three arabinose-regulated rfaH strains.
Virulence of ΔPrfaH and ΔrfaH49 mutant strains in mice.
To assess the virulence of the rfaH mutants, we determined their LD50s in BALB/c mice (Table 3). The ΔrfaH49 mutant was highly attenuated, with no death occurring at the maximum dose tested, 1 × 108 CFU, in agreement with previous results (27). The parent strain, χ3761, was highly virulent, with an LD50 of 1 × 104 CFU. The arabinose-regulated mutant strains χ9734 and χ9735 were both attenuated, with LD50s of 1 × 106 CFU, despite the fact that they were grown in the presence of arabinose prior to inoculation. Strain χ9660, the ΔPrfaH176 mutant, was not tested, since our earlier phenotypic analyses indicated that rfaH expression was not tightly regulated (Fig. 1 and Table 3).
Expression of the pneumococcal gene pspA in RASV strain χ9241 derivatives carrying different rfaH mutations.
S. Typhimurium strain χ9241 is an attenuated vaccine strain that has been successfully used to deliver the pneumococcal surface protein PspA and to induce protective immunity against S. pneumoniae challenge (23, 51). To evaluate the effects of rfaH mutations on the efficacy of this vaccine strain, the ΔrfaH49 and ΔPrfaH178 mutations were introduced into χ9241 to yield strains χ9884 and χ9852, respectively. Subsequently, the Asd+ recombinant plasmid pYA4088, which carries a recombinant pspA gene fused to DNA encoding the β-lactamase signal sequence (51), was introduced into the new strains. Expression of pspA is driven by the Ptrc promoter, and the bla signal sequence directs periplasmic secretion of PspA. Note that strain χ9241 carries the ΔrelA198::araC PBAD lacI TT deletion/insertion (TT stands for transcriptional terminator). When this strain is grown in the presence of arabinose, lacI is expressed. The LacI protein binds to the Ptrc promoter on pYA4088, preventing pspA expression. Once the strain invades and colonizes host tissues, where arabinose is not available, LacI is no longer synthesized and pspA is expressed. This feature has been termed regulated delayed antigen synthesis (51).
To evaluate PspA synthesis, whole-cell lysates from strain χ9241(pYA4088) and the rfaH mutant strains χ9884(pYA4088) and χ9852(pYA4088) grown in LB broth with or without 0.1% arabinose were prepared and examined by Western blot analysis (Fig. 2). All of the strains expressed a protein with an approximate molecular mass of 38.5 kDa, the expected size of the LacI protein, that reacted specifically with an anti-LacI polyclonal antibody. No LacI was detected when the strains were grown in LB medium without 0.1% arabinose. Conversely, in LB medium with 0.1% arabinose, where all strains expressed LacI, no PspA protein was detected. Strains carrying pYA4088 grown in the absence of arabinose produced PspA, but not LacI. There was no obvious difference in the amounts of PspA synthesized by any of the strains carrying pYA4088.
FIG. 2.
PspA and LacI synthesis is regulated by 0.1% arabinose. The Western blots show the synthesis of PspA in S. Typhimurium strains χ9884(pYA4088) (ΔrfaH49), χ9852(pYA4088) (ΔPrfaH178), χ9421(pYA4088) (RfaH+), and χ9241(pYA3493). The bacteria were grown in LB broth with (+) or without (−) 0.1% arabinose overnight at 37°C. Equal numbers of cells from each culture were pelleted, suspended in loading buffer, and boiled. After centrifugation, equal volumes were subjected to SDS-PAGE in triplicate gels. Each gel was transferred to nitrocellulose and probed with a different polyclonal antibody specific for either PspA, LacI, or GroEL. GroEL was used as a standardization marker. Relevant portions of each blot are shown.
Colonization of mouse tissues and immune responses in mice after oral immunization with RASV expressing PspA.
To evaluate the effect of arabinose-regulated rfaH expression on colonization of mouse tissues, strains χ9241(pYA4088), χ9884(pYA4088), and χ9852(pYA4088) were grown in the presence of arabinose and used to inoculate groups of BALB/c mice. On days 4 and 8, three mice from each group were euthanized, and spleen and liver samples were harvested, homogenized, and plated on MacConkey and LB plates, each containing 0.1% arabinose. We found significant differences between the strains in their abilities to colonize the liver and spleen (Fig. 3). Strain χ9852(pYA4088) (ΔPrfaH178) and its parent strain, χ9241(pYA4088), colonized the spleen and liver in significantly higher numbers than χ9884(pYA4088) (ΔrfaH49) (P < 0.0001). There was a slight reduction in tissue colonization by χ9852(pYA4088) compared to its parent strain, χ9241(pYA4088), but the difference was not statistically significant (P > 0.05).
FIG. 3.
Colonization of mouse spleens and livers by attenuated S. Typhimurium harboring plasmid pYA4088 grown in LB broth containing 0.1% arabinose. Shown is spleen (A) and liver (B) colonization by the indicated strains in BALB/c mice at 4 and 8 days postinoculation. *, no bacteria were detected by direct plating, but results were positive by selenite cysteine enrichment. These samples were scored as <10 CFU. The horizontal lines represent the means, and the error bars represent standard errors of the means.
In a separate experiment, we evaluated the in vivo stability of the ΔPrfaH178 mutation in χ9852(pYA4088). A group of three mice were inoculated with 1 × 109 CFU of χ9852(pYA4088). Five days later, the mice were euthanized, and vaccine was recovered from the spleens and livers. Seven colonies were chosen at random and evaluated for arabinose-regulated LPS synthesis by growing each isolate in LB with or without arabinose. Each culture was analyzed on LPS gels. All seven isolates exhibited the same arabinose-regulated LPS phenotype as the parent strain (data not shown).
Effects of rfaH mutations on the immunogenicity and protective efficacy of RASV strains.
We orally inoculated groups of mice with 1 × 109 to 2 × 109 CFU of either χ9241(pYA4088), χ9884(pYA4088) (ΔrfaH49), χ9852(pYA4088) (ΔPrfaH178), or the control strain, χ9241(pYA3493), which does not express pspA. The mice were boosted with a similar dose of the same strain 4 weeks later. The antibody responses to rPspA and Salmonella LPS in the sera of immunized mice were measured (Fig. 4). This experiment was performed twice; 5 mice per group were used in the first experiment, and 8 to 11 mice per group were used in the second experiment. The results from the two experiments were similar and were pooled for analysis. High serum IgG titers against PspA were observed by 2 weeks after the primary immunization in mice inoculated with χ9241(pYA4088) and χ9852(pYA4088) (Fig. 4A). Anti-PspA titers in mice immunized with χ9884(pYA4088) (ΔrfaH) were slower to develop and by 6 weeks had reached high titers, although they never achieved titers comparable to those in mice immunized with χ9241(pYA4088). However, mice immunized with χ9852(pYA4088) (ΔPrfaH178) achieved significantly higher titers than the other two groups. No anti-PspA IgG was detected in sera from mice inoculated with the control strain, χ9241(pYA3493).
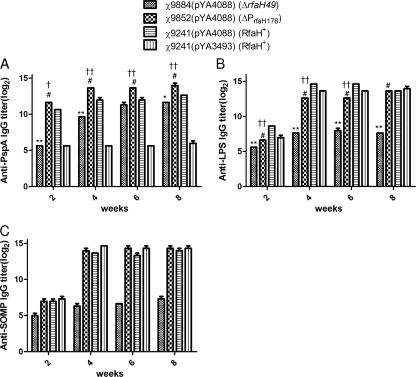
FIG. 4.
Serum IgG responses in immunized and control mice. Total serum IgGs specific for rPspA (A), S. Typhimurium LPS (B), and SOMP (C) were measured by ELISA. The data represent reciprocal anti-IgG antibody levels in pooled sera from mice orally immunized with attenuated Salmonella carrying either pYA4088 (pspA) or pYA3493 (control) the indicated number of weeks after immunization. The error bars represent variations between triplicate wells. The mice were boosted at week 4. **, χ9884(pYA4088) versus χ9241(pYA4088), P < 0.001; *, χ9884(pYA4088) versus χ9241(pYA4088), P < 0.01; #, χ9884(pYA4088) versus χ9852(pYA4088), P < 0.001; ††, χ9852(pYA4088) versus χ9241(pYA4088), P < 0.001; †, χ9852(pYA4088) versus χ9241(pYA4088), P < 0.01. For panel C, χ9884(pYA4088) was different from all other groups (P < 0.001). There was no statistical difference between the other strains shown in panel C.
Anti-LPS titers were low but detectable at 2 weeks (Fig. 4B). The titers remained low in mice immunized with the ΔrfaH strain, χ9884(pYA4088), throughout the course of the experiment, while anti-LPS serum titers increased in mice immunized with the other strains. Before week 8, the anti-LPS titers were significantly lower in mice immunized with the ΔPrfaH178 strain, χ9852(pYA4088), than in either group immunized with a χ9241 derivative (P < 0.001), and they were significantly higher than those of mice immunized with the ΔrfaH strain (P < 0.001). By week 8, mice immunized with either of the χ9241 derivatives or with χ9852(pYA4088) had similar anti-LPS IgG titers. Anti-SOMP titers were similar for all strains except χ9884(pYA4088), which was statistically different (P < 0.001) (Fig. 4C).
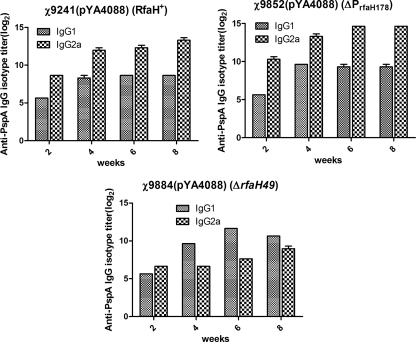
IgG isotype analyses.
We evaluated the responses of the IgG isotype subclasses IgG1 and IgG2a (Fig. 5) to rPspA. Th1 helper cells direct cell-mediated immunity and promote class switching to IgG2a, and Th2 cells provide potent help for B-cell antibody production and promote class switching to IgG1. Immunization with strain χ9884(pYA4088) induced a strong Th2 response, since the levels of anti-PspA IgG1 were higher than those of IgG2a. Conversely, immunization with strains χ9852(pYA4088) (ΔPrfaH178) and χ9241(pYA4088) (RfaH+) induced primarily a Th1-type response against PspA.
FIG. 5.
Serum IgG1 and IgG2a responses to rPspA. The data represent ELISA results determining the levels of IgG1 and IgG2a subclass antibody to rPspA in the sera of BALB/c mice orally immunized with χ9241(pYA4088), χ9884(pYA4088), or χ9852(pYA4088) the indicated number of weeks after immunization. The error bars represent the standard deviations.
Evaluation of protective immunity.
To evaluate the effects of the ΔrfaH49 and ΔPrfaH178 mutations on the ability of the RASV expressing PspA to induce protective immunity, mice were challenged by the i.p. route with 4 × 104 CFU (200 LD50) of S. pneumoniae WU2 4 weeks after the boost. Immunization with any of the pspA-expressing strains provided significant protection against challenge compared to immunization with the control strain, χ9241(pYA3493) (Fig. 6; P < 0.001). Immunization with χ9852(pYA4088) (ΔPrfaH178) induced significantly greater protection than did immunization with χ9884(pYA4088) (ΔrfaH49) (P < 0.05). There was no significant difference between the protective efficacy of χ9241(pYA4088) and that of the other strains expressing pspA (P > 0.05). All of the mice that died in these experiments succumbed within 4 days after challenge.
FIG. 6.
Oral immunization with PspA-expressing Salmonella strains protects BALB/c mice against i.p. challenge with S. pneumoniae WU2. Nine (control) or 11 to 13 (vaccine) mice per group were orally immunized twice at 4-week intervals with the indicated vaccine strains. The mice were challenged with 4 × 104 CFU of S. pneumoniae WU2 (200 LD50) 4 weeks after the second oral immunization. Mortality was monitored for 3 weeks after pneumococcal challenge. The numbers in parentheses refer to the number of surviving mice and the total number of mice per group. All vaccine groups were significantly different from the χ9241(pYA3493) (RfaH+) control (P < 0.01). χ9884(pYA4088) (ΔrfaH49) versus χ9852(pYA4088) (ΔPrfaH178), P = 0.0326.
Increased immunogenicity of conserved antigens against different enteric bacteria.
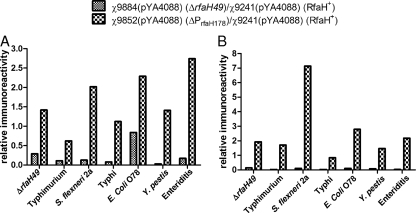
Previous work had shown that a ΔrfaH mutation increased the immunogenic reactivities of conserved minor epitopes from other enteric bacteria (29). Therefore, we examined the cross-reactive antibodies elicited by the ΔrfaH49 vaccine strain, χ9884(pYA4088), with those elicited by the arabinose-regulated ΔPrfaH178 strain, χ9852(pYA4088), and the RfaH+ strain, χ9241(pYA4088). The reactivities of pooled immune sera (11 to 16 mice per group) from groups of mice inoculated in the previous experiment taken 2 weeks after boosting was evaluated by ELISA against a panel of homologous and heterologous wild-type strains, as well as their outer membrane proteins (Fig. 7). In each case, immunization with the arabinose-regulated rfaH strain, χ9852(pYA4088), generated higher titers against both whole cells and OMPs isolated from a diverse group of gram-negative organisms, including S. Typhimurium, S. enterica serovars Typhi and Enteritidis, Shigella flexneri, E. coli, and Yersinia pestis, than the ΔrfaH49 strain, χ9884(pYA4088).
FIG. 7.
Relative reactivities of immune sera, measured 6 weeks after the primary immunization, to homologous and heterologous bacteria from mice immunized with χ9241(pYA4088) and its mutant derivatives. Pooled immune sera from mice immunized with χ9884(pYA4088) (ΔrfaH49), χ9852(pYA4088) (ΔPrfaH178), and χ9241(pYA4088) (parent vaccine strain) were tested against whole cells (A) or purified outer membrane proteins (B) from different enteric bacteria by indirect ELISA. The reactivity is expressed relative to that of χ9241(pYA4088) immune serum at the same dilution. Means of values from three independent experiments are shown. All χ9852(pYA4088) groups were significantly different from the χ9884(pYA4088) groups (P < 0.001). The strains are (left to right) ΔrfaH49 χ9945, S. Typhimurium χ3761, S. flexneri 2a wild type 2457T, S. Typhi ISP1820 χ7122, E. coli O78, Y. pestis KIM6+, and S. Enteritidis χ3550 (group D1).
DISCUSSION
Recombinant attenuated S. Typhimurium strains have been used extensively as multivalent vectors expressing heterologous antigens. An ideal RASV should be able to invade and transiently persist in lymphoid tissues (Peyer's patch, spleen, and liver) to stimulate both strong primary and lasting memory immune responses, cause no disease symptoms, and be susceptible to all clinically useful antibiotics (2, 7, 8). Achieving this balance between adequate attenuation/safety and ability to elicit long-lasting protective immunity is not always easy or straightforward. We have developed several strategies to address this issue, including regulated delayed attenuation (11, 12, 22), whereby the live-vaccine strain displays abilities similar to those of a wild-type virulent parental pathogen to successfully colonize effector lymphoid tissues before display and imposition of the fully attenuated phenotype, and regulated delayed antigen synthesis, in which the expression of a heterologous antigen gene is delayed until the vaccine strain has colonized host tissues (51).
In this work, we applied the regulated delayed attenuation approach to produce strains with arabinose-regulated synthesis of RfaH, a transcriptional antiterminator required for the production of complete LPS (26, 38) and for transcription of other virulence-associated genes, including the sii operon, which contributes to the intestinal phase of infection (26). We compared arabinose-regulated rfaH expression strains to a ΔrfaH strain. Based on P22 transduction assays, the ΔrfaH mutant produced some full-length O antigen (Table 3) undetectable on silver-stained gels (Fig. 1), consistent with a previous report (29). When grown without arabinose, the regulated delayed rfaH strains produced O antigen detectable by silver staining, although the amount was reduced compared to that produced by the same strains grown with arabinose or by strains expressing wild-type rfaH (Fig. 1 and Table 3).
A ΔrfaH mutant of S. Typhimurium has been shown to induce protection against challenge with wild-type S. Typhimurium (27). However, a potential drawback of this strain is its innate sensitivity to host antimicrobial agents, such as bile and antimicrobial peptides (26) (Table 3). This, in addition to the lack of complete O antigen, could lead to reduced colonization of lymphoid tissues (Fig. 3), an important criterion for stimulation of a strong, lasting immune response. When the ΔPrfaH178 mutant was grown in the presence of arabinose, its phenotype was similar to that of its wild-type parent, χ3761 (Fig. 1 and 3 and Table 3). In the absence of arabinose, this mutant was nearly as susceptible as the ΔrfaH mutant to deoxycholate and polymyxin B. In addition, even when grown in the presence of arabinose, the ΔPrfaH178 mutant was attenuated, with an LD50 100 times greater than that of χ3761, although not as attenuated as the ΔrfaH mutant (Table 3).
To ensure the safety of live Salmonella vaccines, it is critical that the strain carry at least two genetically unlinked attenuating mutations (13, 35, 48). Therefore, we evaluated the ΔrfaH mutation and the arabinose-regulated rfaH mutation ΔPrfaH178 in combination with the ΔpabA ΔpabB mutations in strain χ9241 and introduced plasmid pYA4088, which directs the synthesis of a heterologous antigen, the pneumococcal protective antigen PspA (5, 25). The ΔrfaH derivative χ9884(pYA4088) colonized mouse tissues poorly compared to its parent, χ9241(pYA4088), typical of strains lacking full-length O antigen (33, 38). However, the strain with the ΔPrfaH178 mutation, χ9852(pYA4088), colonized the host spleen and liver nearly as well as χ9241(pYA4088). Further, mice immunized with two doses of χ9852(pYA4088) elicited an anti-PspA IgG response equal to or greater than that induced by χ9241(pYA4088) (Fig. 4). Mice immunized with the ΔrfaH mutant, χ9884(pYA4088), also developed anti-PspA serum IgG antibodies, although the titers were significantly lower than those in mice immunized with χ9241(pYA4088) or χ9852(pYA4088). In addition, mice immunized with either χ9241(pYA4088) or χ9852(pYA4088) developed much higher titers of IgG2a than those immunized with the ΔrfaH strain, χ9884(pYA4088) (Fig. 5). IgG2a antibody is the most potent isoform for directing complement deposition, an important host mechanism for clearing S. pneumoniae (6).
When challenged i.p. with virulent S. pneumoniae, all groups immunized with strains expressing pspA were protected (Fig. 6). Consistent with the colonization and serum antibody data, immunization with the regulated rfaH expression strain χ9852(pYA4088) provided significantly greater protection than immunization with the ΔrfaH strain, χ9884(pYA4088) (P < 0.05). These results support the notion that delaying expression of an attenuation phenotype increases protective efficacy (11, 12, 22).
In our laboratory, we are interested in developing vaccines against enteric pathogens, including pathogenic E. coli, enteric Yersinia species, and Shigella. This is an area in which S. Typhimurium rfaH mutants may be valuable by inducing cross-protective antibodies against other enteric bacteria (29). Downregulating the production of O antigen, the major immunodominant antigen in Salmonella, has been shown to enhance the immunogenicity of conserved antigens (12). A recent study showed that mice immunized with a ΔrfaH mutant produced higher titers of serum antibodies against conserved antigens from different S. enterica serovars and from other enteric organisms than mice immunized with an attenuated smooth strain (27, 29). The ΔrfaH ΔpabA ΔpabB strain, χ9884(pYA4088), induced an enhancement of cross-reactive antibodies to only itself and the avian-pathogenic E. coli strain χ7122, but not to smooth, wild-type S. Typhimurium, S. enterica serovar Typhi, S. enterica serovar Enteritidis, S. flexneri, or Y. pestis (Fig. 7A) or to outer membrane proteins derived from these strains (Fig. 7B). The differences between our results and those previously reported could possibly be due to differences in the time of serum collection, the number of boosts, or strain background and/or the presence in our strains of additional attenuating mutations and plasmid pYA4088, directing the synthesis of a heterologous antigen. In addition, we used χ9241(pYA4088) as the base strain to develop our ratios, while a ΔaroA strain was used in the previous study. Importantly, immunization with the arabinose-regulated rfaH strain χ9852(pYA4088) enhanced production of cross-reactive antibodies to all strains tested, including strong responses against S. Enteritidis, Shigella, avian-pathogenic E. coli, and Yersinia. These results are most likely due to the overall higher immunogenicity of χ9852(pYA4088) than χ9884(pYA4088) (Fig. 4), which in turn is probably related to the poor colonization of host tissues observed for χ9884(pYA4088) (Fig. 3). These results indicate that inclusion of the ΔPrfaH178 mutation in an RASV expressing pathogen-specific antigens designed to protect against Shigella, E. coli, and/or Yersinia could enhance the protective efficacy against these pathogens.
We stated previously that it would be advantageous to reduce the host immune response against the RASV carrier, thereby enhancing the immune response against the heterologous antigen. One of our goals for this study was to determine whether in vivo downregulation of rfaH would suppress the immune response against carrier-specific O antigen. For this application, the ΔPrfaH178 mutation was not as useful as we had hoped. Although the immune response against LPS O antigen was delayed in mice immunized with χ9852(pYA4088) compared to those immunized with χ9241(pYA4088) (Fig. 4), by week 8, mice immunized with either strain had developed nearly identical anti-LPS titers. There are several possible explanations for this result. One is that a mutation arose in vivo that allowed arabinose-independent rfaH expression in χ9852(pYA4088). This is unlikely, as we did not observe any such mutants in the spleen isolates we tested. While the araC PBAD promoter cassette used in this study is tightly regulated in vivo, as we established in a previous study (21), it is possible that low levels of nonphosphorylated arabinose in the mouse diet may have been sufficient to allow enough rfaH expression to permit full-length O-antigen synthesis. In addition, χ9852(pYA4088) was grown in the presence of arabinose, conditions permissive for rfaH expression, and given as a boost on day 28, which may also have played a role in generating the observed high anti-LPS antibody titers. A final possible explanation is that our ELISA coating antigen was complete LPS, which includes lipid A, core, and O antigen. One might expect better presentation of LPS core to the mouse immune system by the rfaH mutant strains than by χ9241(pYA4088), leading to an increase in anti-core antibody titers. Our ELISA was not designed to distinguish between anti-core and anti-O-antigen antibodies. This will be investigated in a future study to answer this question. It is an interesting question, since these anti-core antibodies may be cross-reactive with and potentially cross-protective against other enteric bacteria that share the same core structure.
In summary, we have shown that rfaH mutations can be combined with other attenuating mutations to produce an RASV capable of delivering a protective antigen to induce protective immunity. A strain with the delayed regulated mutation ΔPrfaH178 in combination with ΔpabA ΔpabB mutations was superior to a ΔpabA ΔpabB ΔrfaH strain in colonizing lymphoid tissues, eliciting serum antibodies to a heterologous antigen, and inducing protective immunity against S. pneumoniae challenge. Notably, the ΔPrfaH178 vaccine was more effective than the isogenic ΔrfaH strain at inducing antibodies cross-reactive with a number of other enteric pathogens, making it suitable for inclusion in vaccines to protect against enteric diseases.
Acknowledgments
This work was supported by grant 37863 from the Bill and Melinda Gates Foundation.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 5 October 2009.
REFERENCES
- 1.Artsimovitch, I., and R. Landick. 2002. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109:193-203. (Erratum, 110:801.) [DOI] [PubMed] [Google Scholar]
- 2.Atkins, H. S., M. Morton, K. F. Griffin, M. G. M. Stokes, J. P. Nataro, and R. W. Titball. 2006. Recombinant Salmonella vaccines for biodefence. Vaccine 24:2710-2717. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, M. J. A., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 4.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briles, D. E., J. D. King, M. A. Gray, L. S. McDaniel, E. Swiatlo, and K. A. Benton. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858-867. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. S., T. Hussell, S. M. Gilliland, D. W. Holden, J. C. Paton, M. R. Ehrenstein, M. J. Walport, and M. Botto. 2002. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc. Natl. Acad. Sci. USA 99:16969-16974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatfield, S. N., M. Roberts, G. Dougan, C. Hormaeche, and C. M. A. Khan. 1995. The development of oral vaccines against parasitic diseases utilizing live attenuated Salmonella. Parasitology 110:S17-S24. [DOI] [PubMed] [Google Scholar]
- 8.Cheminay, C., and M. Hensel. 2008. Rational design of Salmonella recombinant vaccines. Int. J. Med. Microbiol. 298:87-98. [DOI] [PubMed] [Google Scholar]
- 9.Collins, L. V., S. Attridge, and J. Hackett. 1991. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect. Immun. 59:1079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtiss, R., III. 2002. Bacterial infectious disease control by vaccine development. J. Clin. Investig. 110:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtiss, R., III, S.-Y. Wanda, B. M. Gunn, X. Zhang, S. A. Tinge, V. Ananthnarayan, H. Mo, S. Wang, and W. Kong. 2009. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect. Immun. 77:1071-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtiss, R., III, X. Zhang, S. Y. Wanda, H. Y. Kang, V. Konjufca, Y. Li., B. Gunn, S. Wang, G. Scarpellini, and L. I. Soo. 2007. Induction of host immune responses using Salmonella-vectored vaccines, p. 297-313. In Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC.
- 13.Curtiss, R., and S. M. Kelly. 1987. Salmonella typhimurium deletion mutants lacking adenylate-cyclase and cyclic-AMP receptor protein are avirulent and immunogenic. Infect. Immun. 55:3035-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 15.Germanier, R., and E. Fürer. 1971. Immunity in experimental salmonellosis. 2. Basis for avirulence and protective capacity of galE mutants of Salmonella typhimurium. Infect. Immun. 4:663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassan, J. O., and R. Curtiss. 1990. Control of colonization by virulent Salmonella Typhimurium by oral immunization of chickens with avirulent delta-Cya delta-Crp Salmonella Typhimurium. Res. Microbiol. 141:839-850. [DOI] [PubMed] [Google Scholar]
- 17.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hone, D., R. Morona, S. Attridge, and J. Hackett. 1987. Construction of defined galE mutants of Salmonella for use as vaccines. J. Infect. Dis. 156:167-174. [DOI] [PubMed] [Google Scholar]
- 19.Hone, D. M., S. R. Attridge, B. Forrest, R. Morona, D. Daniels, J. T. Labrooy, R. C. A. Bartholomeusz, D. J. C. Shearman, and J. Hackett. 1988. A galE-via (Vi-antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect. Immun. 56:1326-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang, H. Y., J. Srinivasan, and R. Curtiss III. 2002. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect. Immun. 70:1739-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong, W., S. Y. Wanda, X. Zhang, W. Bollen, S. A. Tinge, K. L. Roland, and R. Curtiss. 2008. Regulated programmed lysis of recombinant Salmonella in host tissues to release protective antigens and confer biological containment. Proc. Natl. Acad. Sci. USA 105:9361-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Y., S. Wang, G. Scarpellini, B. Gunn, W. Xin, S. Y. Wanda, K. L. Roland, and R. Curtiss III. 2009. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc. Natl. Acad. Sci. USA 106:593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Y., S. Wang, W. Xin, G. Scarpellini, Z. Shi, B. Gunn, K. L. Roland, and R. Curtiss III. 2008. A sopB deletion mutation enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Infect. Immun. 76:5238-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindberg, A. A., and C. G. Hellerqvist. 1980. Rough mutants of Salmonella typhimurium—immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J. Gen. Microbiol. 116:25-32. [DOI] [PubMed] [Google Scholar]
- 25.McDaniel, L. S., J. S. Sheffield, P. Delucchi, and D. E. Briles. 1991. PspA, a surface protein of Streptococcus pneumoniae, is capable of eliciting protection against pneumococci of more than one capsular type. Infect. Immun. 59:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy, G., V. Danino, U. Dobrindt, M. Pallen, R. Chaudhuri, L. Emody, J. C. Hinton, and J. Hacker. 2006. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect. Immun. 74:5914-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagy, G., U. Dobrindt, J. Hacker, and L. Emody. 2004. Oral immunization with an rfaH mutant elicits protection against salmonellosis in mice. Infect. Immun. 72:4297-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy, G., L. Emody, and T. Pal. 2008. Strategies for the development of vaccines conferring broad-spectrum protection. Int. J. Med. Microbiol. 298:379-395. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, G., T. Palkovics, A. Otto, H. Kusch, B. Kocsis, U. Dobrindt, S. Engelmann, M. Hecker, L. Emody, T. Pal, and J. Hacker. 2008. “Gently rough”: The vaccine potential of a Salmonella enterica regulatory lipopolysaccharide mutant. J. Infect. Dis. 198:1699-1706. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama, K., S. M. Kelly, and R. Curtiss III. 1988. Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat. Biotechnol. 6:693-697. [Google Scholar]
- 31.Nayak, A. R., S. A. Tinge, R. C. Tart, L. S. McDaniel, D. E. Briles, and R. Curtiss III. 1998. A live recombinant avirulent oral Salmonella vaccine expressing pneumococcal surface protein A induces protective responses against Streptococcus pneumoniae. Infect. Immun. 66:3744-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojas, G., S. Saldias, M. Bittner, M. Zaldivar, and I. Contreras. 2001. The rfaH gene, which affects lipopolysaccharide synthesis in Salmonella enterica serovar Typhi, is differentially expressed during the bacterial growth phase. FEMS Microbiol. Lett. 204:123-128. [DOI] [PubMed] [Google Scholar]
- 34.Roland, K., R. Curtiss III, and D. Sizemore. 1999. Construction and evaluation of a delta cya delta crp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 43:429-441. [PubMed] [Google Scholar]
- 35.Sambandamurthy, V. K., and W. R. Jacobs. 2005. Live attenuated mutants of Mycobacterium tuberculosis as candidate vaccines against tuberculosis. Microbes Infect. 7:955-961. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Samuel, G., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503-2519. [DOI] [PubMed] [Google Scholar]
- 38.Sanderson, K. E., and B. A. D. Stocker. 1981. Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J. Bacteriol. 146:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santangelo, T. J., and J. W. Roberts. 2002. RfaH, a bacterial transcription antiterminator. Mol. Cell 9:698-700. [DOI] [PubMed] [Google Scholar]
- 40.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 41.Singh, S. P., Y. U. Williams, P. E. Klebba, P. Macchia, and S. Miller. 2000. Immune recognition of porin and lipopolysaccharide epitopes of Salmonella typhimurium in mice. Microb. Pathog. 28:157-167. [DOI] [PubMed] [Google Scholar]
- 42.Stevenson, G., and P. A. Manning. 1985. Galactose epimeraseless (galE) mutant G30 of Salmonella typhimurium is a good potential live oral vaccine carrier for fimbrial antigens. FEMS Microbiol. Lett. 28:317-321. [Google Scholar]
- 43.Stocker, B. A. D., and P. H. Makela. 1986. Genetic determination of bacterial virulence, with special reference to Salmonella. Curr. Top. Microbiol. Immunol. 124:149-172. [DOI] [PubMed] [Google Scholar]
- 44.Stocker, B. A. D., and P. H. Makela. 1978. Genetics of (Gram-negative) bacterial surface. Philos. Trans. R. Soc. Lond. B 202:5-30. [DOI] [PubMed] [Google Scholar]
- 45.Stocker, B. A. D., B. M. Males, and W. Takano. 1980. Salmonella typhimurium mutants of rfaH—phenotype, genetics and antibiotic sensitivities. J. Gen. Microbiol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 46.Sun, W., S. Wang, and R. Curtiss III. 2008. Highly efficient method for introducing successive multiple scarless gene deletions and markerless gene insertions into the Yersinia pestis chromosome. Appl. Environ. Microbiol. 74:4241-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valentine, P. J., B. P. Devore, and F. Heffron. 1998. Identification of three highly attenuated Salmonella typhimurium mutants that are more immunogenic and protective in mice than a prototypical aroA mutant. Infect. Immun. 66:3378-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitfield, C., N. Kaniuk, and E. Frirdich. 2003. Molecular insights into the assembly and diversity of the outer core oligosaccharide in lipopolysaccharides from Escherichia coli and Salmonella. J. Endoxin Res. 9:244-249. [DOI] [PubMed] [Google Scholar]
- 50.Wiegand, I., K. Hilpert, and R. E. W. Hancock. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protocols 3:163-175. [DOI] [PubMed] [Google Scholar]
- 51.Xin, W., S. Y. Wanda, Y. Li, S. Wang, H. Mo, and R. Curtiss III. 2008. Analysis of type II secretion of recombinant pneumococcal PspA and PspC in a Salmonella enterica serovar Typhimurium vaccine with regulated delayed antigen synthesis. Infect. Immun. 76:3241-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]