Abstract
Prophage typically are induced to a lytic cycle under stressful environmental conditions or when the host's survival is threatened. However, stress-independent, spontaneous induction also occurs in nature and may be cell density dependent, but the in vivo signal(s) that can trigger induction is unknown. In the present study, we report that acyl-homoserine lactones (AHL), the essential signaling molecules of quorum sensing in many gram-negative bacteria, can trigger phage production in soil and groundwater bacteria. This phenomenon also was operative in a λ lysogen of Escherichia coli. In model coculture systems, we monitored the real-time AHL production from Pseudomonas aeruginosa PAO1 using an AHL bioluminescent sensor and demonstrated that λ-prophage induction in E. coli was correlated with AHL production. As a working model in E. coli, we show that the induction responses of λ with AHL remained unaffected when recA was deleted, suggesting that this mechanism does not involve an SOS response. In the same λ lysogen we also demonstrated that sdiA, the AHL receptor, and rcsA, a positive transcriptional regulator of exopolysaccharide synthesis, are involved in the AHL-mediated induction process. These findings relate viral reproduction to chemical signals associated with high host cell abundance, suggesting an alternative paradigm for prophage induction.
The reproductive cycle of bacteriophage may be lytic, resulting in the rapid destruction of host cells, or lysogenic, in which the viral genome instead is stably maintained as a prophage in the host genome and replicated as the host cell grows and divides (1). The lysogenic state can be converted to lytic either by various inducing agents, such as mitomycin C (mitC), UV light, antibiotics, and other chemicals (22, 27), or by subjecting the host to physiological stresses such as amino acid deprivation (33). Most inducing agents as well as physiological stresses evaluated to date result in damage to host cell DNA, and the RecA-mediated molecular mechanism of this induction process is well characterized (27, 28). However, RecA-independent inductions also occur in E. coli that do not involve an SOS response (41, 45), but the signal(s) that trigger RecA-independent induction is not known.
In environmental samples, it is not possible to directly measure the portion of bacteria that are lysogenic, nor is it possible to determine the number of lysogenic (temperate) phages relative to that of lytic (virulent) phages. Instead, studies of lysogeny in environmental samples involve an estimation of the lysogenic fraction of bacterial populations based on a comparison of viral production and host cell lysis in induced and control samples (60). Estimates of the lysogenic fraction vary widely in aquatic environments (0.7 to 82%), and temporal variations in the prevalence of lysogeny in heterotrophic bacterial populations have been reported in marine environments (13, 62). Seasonal studies of marine cyanobacteria have shown the inducible fraction to be inversely proportional to host cell abundance (31, 32). In other words, lysogeny seemed to be more prevalent in late winter during periods of low host cell abundance. While studies of soils are rather limited, they suggest that the environmental conditions within the soil ecosystem select for lysogeny as a more prevalent reproductive strategy among soil phages (19).
It is believed that lysogeny is an adaptive reproductive strategy that allows viruses to survive in a quiescent state within the cell during suboptimal physiological conditions of the host, especially during situations when host cell abundance is very low (52). This seems particularly relevant in the harsh environment of the soil ecosystem, where extracellular viruses may be rapidly inactivated before infecting a new host. Therefore, there also may exist chemical signals that initiate the lytic cycle under favorable conditions, especially when host cell abundance is high. Such a signal of high host cell abundance should be an SOS-independent response but may exploit the same molecular switch that determines the SOS-dependent lytic/lysogenic decision in well-characterized lambda-like phages (36, 58). In near-surface aquatic environments, UV irradiation is likely an important factor leading to prophage induction. However, this probably is not the case in soil, since virtually all soil prokaryotes reside in intra- and interaggregate pores and thus are protected from UV exposure even near the soil surface. Therefore, alternative prophage induction mechanisms may exist in terrestrial ecosystems.
Quorum sensing in bacteria is a cell density-dependent phenomenon that regulates the coordinated expression of diverse biological phenotypes, such as motility, biofilm formation, chemiluminescence, and the production of toxins, exopolysaccharides, biosurfactants, and other virulence factors (55). Thus, quorum sensing is a critical component of the adaptive survival and activity of many bacteria as well as an essential factor in the virulence of bacterial pathogens. Interactions of host bacteria with temperate bacteriophage also may influence microbial processes. Most notable are virulence factors of many pathogenic bacteria, such as exotoxins, that are phage encoded (7, 8). Indirect evidence for a link between quorum sensing and the regulation of the lytic/lysogenic switch appeared recently when quorum sensing was shown to increase Shiga toxin (Stx toxin) production along with the transcription of λ-like phage genes in E. coli O157:H7 (48). In this particular study it was shown indirectly that Stx expression was induced by an SOS response, and genes involved in the SOS response were regulated by quorum sensing (48). Further evidence for this linkage includes the spontaneous induction of prophage during biofilm development (15, 23, 57), the upregulation of phage-related genes in Desulfovibro vulgaris during stationary phase, and the induction of prophage Mu in stationary phase (11, 40). In other instances, the spontaneous induction of prophage has been observed as cultures enter stationary phase and conditions associated with high cell density (10, 12, 40, 52, 57), situations in which quorum-sensing compounds for some bacteria might reach threshold concentrations that are necessary for the induction of cell-density-dependent processes.
These results formed the basis for the present study involving the effect of exogenously added AHL on prophage induction in bacteria extracted directly from soil samples, ground-water samples, and bacterial communities colonizing field-deployed porous beads designed to simulate the highly porous nature of the soil environment (19). An induction response also was observed for bacteria grown under pure culture conditions. In coculture systems, we monitored real-time AHL production from Pseudomonas aeruginosa PAO1 (i.e., no exogenously added AHL) using an AHL bioluminescent sensor and demonstrated that λ-prophage induction in E. coli was correlated with AHL production. By using single-gene knockout mutations in an E. coli-λ system, we establish the molecular basis of this induction mechanism, which suggests that AHL-mediated prophage induction is an SOS-independent process and involves an AHL receptor, SdiA (34, 54), and a transcriptional regulator of exopolysachharide synthesis, RcsA (29).
MATERIALS AND METHODS
Bacteria, plasmids, and phages.
E. coli strain BW25113 (lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78) was used as the host for prophage induction experiments, and plasmid pKD46 was used for deletion mutagenesis by the method of Datsenko and Wanner (16). These resources were obtained from the E. coli Genetic Stock Center (Yale University). P. aeruginosa strains PAO1 (21) and PAO214 (ΔlasI) (20) were kindly provided by Herbert Schweizer (Colorado State University, Ft. Collins, CO). Agrobacterium tumefaciens A136(pCF218)(pMV26) (an AHL biosensor) (9, 47) was a gift from Pamela Sokol (University of Calgary, Calgary, Canada). E. coli strain MG1655 (F− l− ilvG rfb-50 rph-1) was used as an indicator strain during plaque assays, and bacteriophage λimm434, a λ derivative that contains the immunity region of bacteriophage 434, was a gift from Max Gottesman (Columbia University, New York, NY). This λ mutant is functionally equivalent to the wild type with respect to prophage induction, and both are immune to superinfection. The sdiA-overexpressing plasmid pDEW140 (59) was provided by Robert A. LaRossa (DuPont Company, Wilmington, DE).
Media, chemicals, and other reagents.
Unless otherwise indicated, all bacterial strains were grown routinely in TSB medium containing 1% Bacto tryptone (Difco) and 0.5% NaCl with or without ampicillin (50 μg/ml) and kanamycin (30 μg/ml). Glucose, l-arabinose, and other chemicals were from Acros Organics, NJ. The restriction endonuclease DpnI was purchased from New England Biolabs, and PCRs were conducted using the Taq polymerase TaKaRa Ex Taq (TaKaRa Bio Inc., Otsu, Japan).
In vitro effect of exogenously added AHL on induction.
An AHL mixture was prepared in ethyl acetate acidified with 0.1% (vol/vol) acetic acid that contained N-(butyl, heptanoyl, hexanoyl, β-ketocaproyl, octanoyl, and tetradecanoyl)-dl-homoserine lactones (Sigma-Aldrich, Inc.), and each was used at a final concentration of 1 μM in the media. First, the necessary volume of the AHL solution was placed in glass tubes or flasks, and then the ethyl acetate was evaporated in a constant shaking condition to leave AHL compounds as a film at the bottom of the tubes or flasks. In a separate control experiment, we determined that flasks or tubes with evaporated ethyl acetate but lacking AHL did not affect prophage induction (data not shown).
For the induction of whole-community samples from soils, bacteria were extracted and concentrated by centrifugation on Nycodenz as described elsewhere (19), suspended in their respective soil extracts, and transferred into the AHL-coated flasks. Control samples were transferred into the flasks without AHL. Soil suspensions with or without AHL were incubated for 18 h in the dark at room temperature. The enumeration of all viruses and bacteria was done using epifluorescence microscopy as described elsewhere (19). Bacteria and viruses from the soil samples were saved for phylogenetic analysis with a 16S rRNA gene Phylochip microarray as described below.
For Bio-Sep beads, samples were washed with sterile water and crushed in phosphate-buffered, 1% potassium citrate with a sterile glass rod (19). Crushed beads then were continuously shaken horizontally for 30 min and centrifuged for 10 min at 12,000 × g. Supernatants containing free viruses were discarded, and the bead pellet containing bacteria was suspended in the corresponding soil extracts with AHL or mitC. The enumeration of bacteria extracted and purified from Bio-Sep beads using Nycodenz was performed as described above.
For groundwater samples collected from a uranium-contaminated aquifer near Rifle, CO, the induction assay was conducted on site immediately following collection. The site characteristics have been described elsewhere (3). The bacteria and extracellular viruses were extracted and concentrated by a tangential flow filtration method as described elsewhere (63). An aliquot of the virus-free bacterial concentrate (100 ml) was added to glass bottles containing AHL and control bottles without AHL. Both bottles were incubated for 18 h on site, and subsamples then were snap-frozen in liquid N2 and brought to the laboratory for the enumeration of bacteria and viruses.
For experiments with E. coli, after the complete evaporation of AHL, 2 ml TSB (containing 1% Bacto tryptone [Difco] and 0.5% NaCl) was added to experimental tubes. For experiments with lysogenized E. coli BW25113 or its sdiA, rcsA, and recA knockout mutants (described in the next section), a single colony was picked with a sterile loop and placed in 0.5 ml TSB and vortexed for 30 s, and 5 μl from this cell suspension was added to the tubes with or without AHL. After 18 h of incubation at 29°C, cells were centrifuged at 8,000 × g and the phage-containing supernatant was removed and preserved by the addition of 1 drop of chloroform. The amount of phage released into the supernatant (i.e., PFU) was determined by a standard agar overlay technique with the susceptible nonlysogenic indicator strain MG1655 as described previously (4). The cell pellet in the original assay tube was suspended, and an aliquot was plated to determine the number of CFU.
We isolated a bacterium from Tennessee soil that also was analyzed for AHL-dependent phage production. This isolate was characterized phylogenetically by sequencing the 16S rRNA gene (1,484 bp) by following a standard protocol of TA cloning and sequencing.
Generation of PCR fragments for constructing knockout mutants of the λ lysogen.
The primer design and the PCR-based single-locus deletion method, as described by Datsenko and Wanner (16), were adopted to construct knockout mutants. Primers used for constructing gene deletions consisted of 50 nucleotides (nt) homologous to the adjacent upstream or downstream flanking region of the target gene, followed by the 20-nt sequence upstream or downstream of the kanamycin resistance gene (kan). The N-terminal primer consisted of the 50-nt upstream region of the target gene including the initiation codon (H1) and the 20 nt upstream of the kan gene, 5′-ATTCCGGGGATCCGTCGACC-3′ (P1), whereas the C-terminal primer consisted of 29 nt of the adjacent downstream region plus the C-terminal 21 nt of the target gene, including the termination codon (H2), followed by the 20 nt of kan downstream sequence, 5′-TGTAGGCTGGAGCTGCTTCG-3′ (P2). All extensions for individual genes (sdiA, rcsA, and recA) knocked out in this study are given in Table 1. PCRs were carried out in 50-μl reaction mixtures containing 2.5 U of TaKaRa Ex Taq polymerase, 1 ng pKD13 plasmid DNA, 1.0 μM of each primer, and 200 μM deoxynucleoside triphosphates. Reactions were run for 30 cycles consisting of 94°C for 30 s, 58°C for 30 s, 72°C for 2 min, plus an additional 2 min extension at 72°C after the final cycle. PCR products were digested with DpnI, ethanol precipitated, suspended in 6 μl H2O, and analyzed by 1.5% agarose gel electrophoresis using 0.5× Tris-acetate buffer.
TABLE 1.
Extension sequences for P1 and H2 primers used to prepare knockout mutants for sdiA, recA, and rcsA
| Gene | N-terminal primer sequence | C-terminal primer sequence |
|---|---|---|
| sdiA | 5′-ATTATCATTATAAATGATACTCACTCTCAGGGGCGTTGCGGTTTACTATG-3′ | 3′-TGCATCTGGCACGCAGGACAGAAAAGAGATCAAATTAAGCCAGTAGCGGC-5′ |
| recA | 5′-CAGAACATATTGACTATCCGGTATTACCCGGCATGACAGGAGTAAAAATG-3′ | 3′-ATGCGACCCTTGTGTATCAAACAAGACGATTAAAAATCTTCGTTAGTTTC-5′ |
| rcsA | 5′-TATTCAGGTAAGGGGAATTATCGTTACGCATTGAGTGAGGGTATGCCATG-3′ | 3′-ACTGGTGGGAAACCACCAGTCAGAATGTGTTAGCGCATGTTGACAAAAAT-5′ |
Lysogenization and construction of sdiA, rcsA, and recA knockout mutants.
E. coli K-12 BW25113 was lysogenized with λimm434 as described previously (4). Single-gene knockout mutants of the BW25113 lysogen were constructed using the PCR-based direct deletion method described by Datsenko and Wanner (16), with a slight modification of recommended temperatures in various steps. To suppress the heat induction of λ, all incubations were done at 30°C. Deleted genes were verified by PCR with kanamycin-specific primers k1 and k2 and locus-specific primers U and D, as described previously (16) and summarized above. PCR products were analyzed by agarose gel electrophoresis (1.5%).
In situ demonstration of AHL-dependent prophage induction in cocultures of E. coli and P. aeruginosa PAO1.
Single colonies from P. aeruginosa PAO1, PAO214 (ΔlasI), E. coli BW25113 (λimm434 lysogen), and sdiA, rcsA, and recA mutants of the same E. coli strain were picked with a sterile loop and placed in 1 ml TSB medium and vortexed for 30 s. Different combinations of E. coli and Pseudomonas cocultures were prepared by mixing 200 μl of E. coli suspension with 100 μl of Pseudomonas suspension in 20 ml TSB. All of the cocultures also were inoculated with 20 μl of an overnight-grown (24 h) culture of A. tumefaciens A136 biosensor and incubated for up to 18 h at 29°C. Samples were taken every 3 h, including the t0 sample for bacterial (CFU per milliliter) and viral (PFU per milliliter) enumeration and bioluminescence analysis. Luminescence was determined by taking 100-μl samples in glass tubes and measuring them with a portable luminometer (Femtomast FB14; Zylux Corporation, Huntsville, AL). Viruses (PFU per milliliter) were counted as described in the previous section. After viruses were taken, aliquots of the remaining coculture were plated on Levine eosin methylene blue (EMB) (Becton, Dickinson and Co., Sparks, MD) agar plates to determine the number of CFU (CFU per milliliter) of E. coli and P. aeruginosa (26). E. coli BW25113 produced dark green colonies that were methyl red-positive lactose fermenters. P. aeruginosa, a non-lactose-fermenting bacterium, produced no acid from fermentation, therefore the colonies were lighter colored and translucent, and they could be differentiated from E. coli colonies. The colony color and morphology of pure E. coli and P. aeruginosa also were determined separately after 18 h for comparisons to the cocultures. In a separate control experiment, it was determined that A. tumefaciens could not produce any visible colonies on EMB agar plates in 18 h.
16S rRNA gene-Phylochip analysis.
In experiments involving agricultural soil, phylogenetic analyses using 16S rRNA gene microarrays (Phylochip) were performed (6). Three types of samples were prepared and used as a template for the amplification of 16S rRNA genes: (i) DNA extracted directly from the soil, representing the entire prokaryotic community; (ii) viral DNA extracted and purified from mitC-inducible prophage; and (iii) viral DNA from AHL-inducible prophage. For samples ii and iii, bacteria were extracted and purified from soil by centrifugation on a cushion of Nycodenz, and subsamples were induced with either mitC or AHL as described above. Special care was taken for purifying DNA from viral fractions to avoid bacterial DNA contamination (19). Briefly, after extraction and treatment with DNase, phage extracts were concentrated and washed in phosphate-buffered saline using Amicon spin filters (95-kDa molecular mass cutoff). The recovered phage concentrates (250 μl) then were heated to 90°C for 15 min and cooled, and DNA was precipitated with a 0.1 volume of 3 M sodium acetate (pH 7.3) and 2.5 volumes of absolute ethanol. With control experiments we have shown previously that this procedure is sufficient to remove free bacterial DNA (19). The 16S rRNA genes were amplified by PCR from both bacterial and viral fractions as described previously (19). After PCR, the samples were further processed at University of Tennessee Affymetrix Core Laboratory for hybridization to the Phylochip microarrays. Briefly, PCR products from three individual replicates from each sample were pooled and concentrated through Microcon filters (Millipore, MA) and fragmented to 50 to 200 bp using DNase I (0.02 U μg−1 DNA; Invitrogen, Carlsbad, CA) and One-Phor-All buffer (GE Healthcare, Piscataway, NJ) by following the manufacturers' protocols. The mixture then was incubated at 25°C for 20 min and 98°C for 10 min before being biotin labeled with a GeneChip DNA labeling reagent kit (Affymetrix, Santa Clara, CA) by following the manufacturer's instructions. The labeled DNA then was denatured at 99°C for 5 min and hybridized to custom-made Affymetrix GeneChips identical to those used by Brodie et al. (6) at 48°C and 60 rpm for 16 h. Phylochip washing and staining then were performed according to the standard Affymetrix protocols described elsewhere (30). The distribution of all phyla with a normalized intensity value exceeding 2% of the total chip intensity then was plotted in pie graphs for visual comparison.
Overexpression of SdiA and immunoblot analysis of RcsA expression.
SdiA protein was overexpressed in E. coli hosts by electrotransforming pDEW140 plasmid (59). Protein for immunoblot analyses was isolated and purified from bacterial samples as described elsewhere (17) and quantified using a Pierce bicinchoninic acid protein estimation kit (Thermo Scientific). Briefly, bacterial pellets were washed twice with 10 mM MgSO4, suspended in 50 mM Tris (pH 8.0)-2 mM EDTA, and briefly sonicated. As RcsA protein generally is found in inclusion bodies, the sonicated suspension was spun down at 12,000 × g for 2 min and the pellet was suspended in sodium dodecyl sulfate (SDS) lysis buffer. The suspensions were heated in a boiling water bath for 10 min prior to electrophoretic separation by SDS-12% polyacrylamide gel electrophoresis. Proteins were transferred electrophoretically to polyvinyl difluoride (Bio-Rad) membranes, and RcsA was detected chromogenically (Bio-Rad) using a polyclonal antiserum of RcsA. Positive controls of crude RcsA protein were obtained from overexpressing E. coli JM109 host cells containing pATC119 plasmid (53).
RESULTS AND DISCUSSION
AHL-mediated prophage induction in soil samples and phylogenetic diversity of inducible host bacteria.
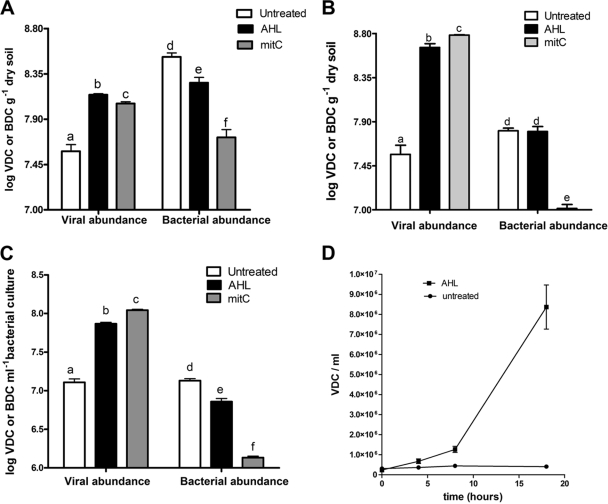
Surface soils from a National Science Foundation long-term ecological research site located at the Kellogg Biological Station in Hickory Corners, MI (http://lter.kbs.msu.edu/), and from the East Tennessee Agricultural Research Station (Knoxville, TN) were collected, and the bacterial community was extracted and purified using density gradient centrifugation as described above. The resulting washed cells were suspended in a buffered aqueous soil extract and induced in the presence of the AHL mixture. The viral abundance in samples exposed to AHL significantly increased (P = 0.0011 and P = 0.002, respectively) relative to that of the control samples, while the bacterial abundance decreased (P = 0.0176 and 0.0189) (Fig. 1A and B). This observation suggested that at least some fraction of the soil microbial community was inducible by exposure to AHL. The corresponding induction response in the samples exposed to mitC was comparable to the response elicited by AHL, but a more significant decrease in bacterial abundance (P = 0.0009) was observed (Fig. 1A and B). We isolated a bacterium closely related to Sinorhizobium meliloti (as determined by sequencing 1,484 bp of the 16S rRNA gene) from the same Tennessee soil discussed previously that showed a significant prophage induction response (P = 0.0001) when grown in the presence of AHL (Fig. 1C). The induction response elicited by mitC resulted in a much sharper decline in bacterial abundance relative to those of AHL-induced and control samples (P = 0.0043) (Fig. 1C). We also monitored a time-dependent induction response in this bacterium. The cultures were sampled at 4-h intervals, and viral abundance increased over time from 6 to 18 h of incubation (Fig. 1D).
FIG. 1.
Induction response upon exposure to AHL and mitC in two microbial communities Tennessee soil and in Sinorhizobium. Shown are viral and bacterial abundances as viral direct counts (VDC) or bacterial direct counts (BDC) in samples incubated with or without AHL and mitC for 18 h in bacteria extracted from KBS soil (A), Tennessee soil (B), and Sinorhizobium (C). Bars are means from triplicate experiments, and vertical bars represent the standard errors. Data sets with different letter designations were significantly different from one another (P < 0.05). (D) Viral production in AHL-induced cultures of Sinorhizobium taken at 0, 4, 8, and 18 h. The data points represent means from triplicate experiments, and the vertical bars represent one standard error. Statistical analyses of viral and bacterial abundance in all samples were performed separately.
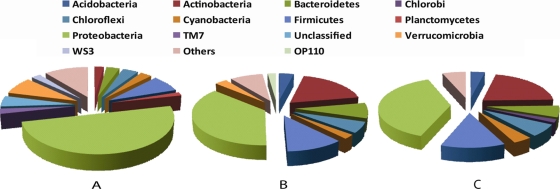
The phylogenetic diversity of the bacteria extracted from the Tennessee soil used in these induction assays also was analyzed with a 16S rRNA gene Phylochip microarray. The analysis revealed a highly diverse community dominated by proteobacterial groups (Fig. 2A). In three previous studies, 16S rRNA genes could be detected by PCR in the viral DNA fraction collected from wastewater (42) and soil (19) communities, and a broad-host-range phage also was detected (5), suggesting that at least generalized transducing phage have the capacity to carry the 16S rRNA gene. Thus, we used this approach as a proxy measure of the phylogenic diversity of potential host bacteria carrying either mitC- or AHL-inducible prophage, and the viral DNA fractions purified from inducible lysogenic bacteria also were analyzed in a similar fashion. A total of 26 phyla were detected in the virus-free bacterial fraction (Fig. 2A). In the induced prophage fractions fewer phyla were detected, but the AHL- and mitC-induced samples had similar distributions of the major phyla common to both samples (Fig. 2B and C). Interestingly, inducible Proteobacteria constituted a smaller percentage of the total chip intensity than the bacterial community (38 and 50%, respectively), suggesting either that not all proteobacterial lysogens were inducible by AHL or mitC or that not all Proteobacteria in the sample contained prophage. Conversely, the relative abundance of Actinobacteria and Firmicutes was overrepresented in the induced samples compared to the overall bacterial community, possibly indicating that more species within these phyla are inducible by AHL or mitC (Fig. 2B and C). The apparent AHL induction response in the Actinobacteria and Firmicutes is surprising, since gene expression in gram-positive bacteria is not known to be regulated by AHL. Thus, with the data presented here, it is not possible to determine if these bacteria responded directly to the presence of AHL or if there was an indirect effect brought about by the AHL-mediated release of other inducing agents from AHL-responsive bacteria.
FIG. 2.
16S rRNA gene Phylochip analysis of extracted bacteria from Tennessee soil (A) and the DNA fractions of inducible temperate phage concentrated and purified from mitC-induced (B) or AHL-induced (C) bacteria.
AHL-dependent prophage induction in groundwater community.
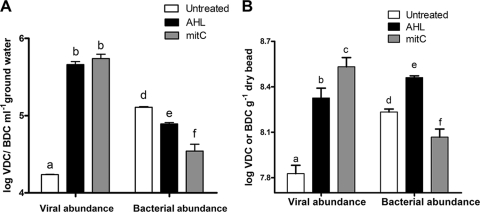
To further evaluate the AHL-mediated prophage induction in environmental samples, we tested the induction response in a bacterial community concentrated from a groundwater aquifer undergoing biostimulation with acetate to enhance uranium reduction (Rifle, CO) (3). The viral abundance increased significantly upon the exposure of the groundwater microbial community to either the AHL mixture (P = 0.0023) or mitC (P = 0.0001) (Fig. 3A). A corresponding decrease in bacterial abundance also was observed, suggesting that the community included a large population of lysogenic bacteria inducible by AHL (P = 0.0037) and mitC (P = 0.0002) (Fig. 3A).
FIG. 3.
Viral and bacterial abundances as viral direct counts (VDC) and bacterial direct counts (BDC) in samples incubated with or without AHL and mitC for 18 h in bacteria extracted from groundwater (A) or from soil-incubated Bio-Sep beads (B). Bars are means from triplicate experiments, and vertical bars represent the standard errors. Data sets with different letter designations were significantly different from one another (P < 0.05).
AHL-mediated induction in Bio-Sep beads.
To more accurately assess induction responses under soil conditions, we employed Bio-Sep beads as a novel in situ enrichment matrix. Given their high surface area and porosity, Bio-Sep beads mimic the soil environment and previously have been used in many environmental applications as a cell immobilization matrix and to sample and characterize microbial communities from various environments (14, 37). We recently used them to assess the prevalence of lysogeny within soil bacterial communities (19). Beads that previously had been equilibrated in a yeast extract solution were incubated in the field at the same sites where soil samples were taken for the experiment described above. Recovered beads were rinsed in sterile water and used for induction experiments. Induction assays were performed in a buffered soil extract solution that was prepared by extracting the same soils in which the beads were buried. Triplicate bead samples were immersed in their respective soil extracts containing either mitC or AHL. All samples were incubated statically at room temperature in the dark for 18 h. Viral production from bacterial communities associated with beads exposed to AHL was similar to that from mitC-treated beads, and the abundance of viruses was considerably greater in the treated samples compared to that of the controls (P = 0.0001) (Fig. 3B). The bacterial count was significantly lower in the mitC-induced samples (P = 0.0015) relative to that of the controls but was increased in the AHL treatments (P = 007). The increase in bacterial abundance in AHL-induced samples most likely was due to a stimulatory effect of AHL on the growth of at least a portion of the bacteria in the samples that exceeded the loss of cells due to viral production (49), whereas mitC had an inhibitory effect on bacterial growth in addition to cell loss through viral lysis (Fig. 3B). This result also indicates that only a portion, but not all, of the bacterial community in the beads was inducible by AHL. Recent work indicates that mitC-inducible lysogens comprise ca. 5 to 40% of natural soil bacterial communities (61) or ca. 80% of the community enriched in Bio-Sep beads (19).
AHL-mediated induction response in the E. coli-λ system and its mutants.
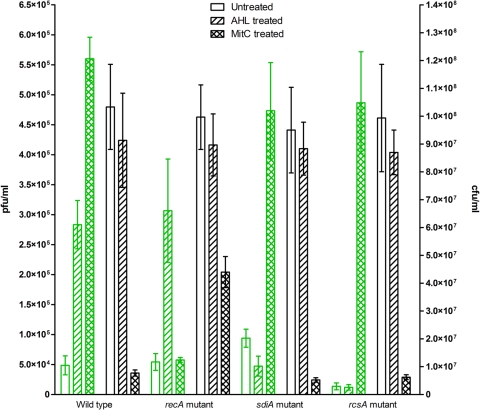
Motivated by the results from environmental samples, we attempted to demonstrate this quorum-sensing mediated prophage induction in the well-characterized λ system of E. coli as a model. We lysogenized wild-type E. coli BW25113 with λimm434 and examined the inducibility of the lysogen by adding a mixture of six AHL compounds of various chain lengths to the culture medium. The results showed a significant increase (P = 0.0007) in the abundance of phage in the culture supernatant after 18 h of incubation compared to that of the uninduced control culture without AHL (Fig. 4). The initial and final cell counts (CFU) did not decrease significantly (P = 0.41) in the presence of AHL.
FIG. 4.
Prophage induction response of wild-type and mutant recA, sdiA, and rcsA λ lysogens of E. coli BW25113 exposed to exogenously added AHL and mitC. After 18 h of incubation at 29°C with or without AHL or mitC, supernatants of each culture were diluted, and phage production was counted as PFU (PFU per milliliter) using the susceptible indicator strain E. coli MG1655 (in green). After serial dilution, bacterial pellets of each culture were counted as CFU (CFU per milliliter). Values plotted are means from triplicate determinations, and the error bars are one standard deviation.
Although E. coli cannot produce AHL, it has an AHL receptor encoded by sdiA that responds to AHL produced by other microbial species (34, 54). To address more directly the involvement of the quorum-sensing receptor, sdiA was deleted by single-gene knockout from the same lysogen and the experiment was repeated. The sdiA knockout mutant of E. coli lysogen showed no increase in phage production after incubation with AHL for 18 h with respect to the uninduced control lacking AHL (Fig. 4). This result directly demonstrates that the quorum-sensing receptor SdiA is required for the AHL-dependent induction of λimm434. mitC-dependent prophage induction was unaffected by the deletion of sdiA. A similar induction response with mitC was observed for both the sdiA mutant and the wild-type lysogen (Fig. 4), indicating that RecA-mediated SOS induction does not require SdiA. To confirm this conclusion, we knocked out the key component of the SOS cascade, recA (28), from wild-type E. coli. A positive induction response was observed when the sample was incubated with AHL, but mitC failed to induce the lysogen (Fig. 4). This indicates that AHL-mediated induction does not involve any DNA damage-dependent mechanism controlled by recA. Unlike mitC-induced samples, however, the bacterial count in all AHL-dependent inductions did not decrease significantly (P = 0.3 to 0.4) (Fig. 4).
We acknowledge that mitC-induced E. coli should result in at least 100-fold more phage particles than we observed. The probable reason for a low titer is the readsorption of phage by the surviving and growing E. coli population during the 18-h incubation. In a separate time course induction experiment using mitC, we observed that the phage titer increased significantly and peaked during the first 3 h of exposure and subsequently decreased over the next few hours (data not shown). With AHL as the inducing agent provided via coculture with P. aeruginosa PAO1, significant viral production was not observed prior to 6 h of incubation, at which time the phage titer began to increase (Fig. 5). Exogenously added AHL also yields results similar to those of coculture experiments (data not shown). As these experimental conditions were not optimal for mitC induction, we conducted a control experiment under optimal induction conditions in which the cells of the same λ lysogen were harvested at mid-log phase (optical density at 600 nm = 0.6), washed twice with medium, and incubated with mitC (1 μg ml−1) for 4 h. The supernatants from these incubations produced at least 103-fold more phage than the uninduced control, whereas the recA mutant of this lysogen did not exhibit an increase in phage production after 4 h of incubation with mitC (data not shown). These results indicate that the model E. coli-λ system was functionally equivalent to other canonical model systems that have been used to investigate RecA-mediated prophage induction.
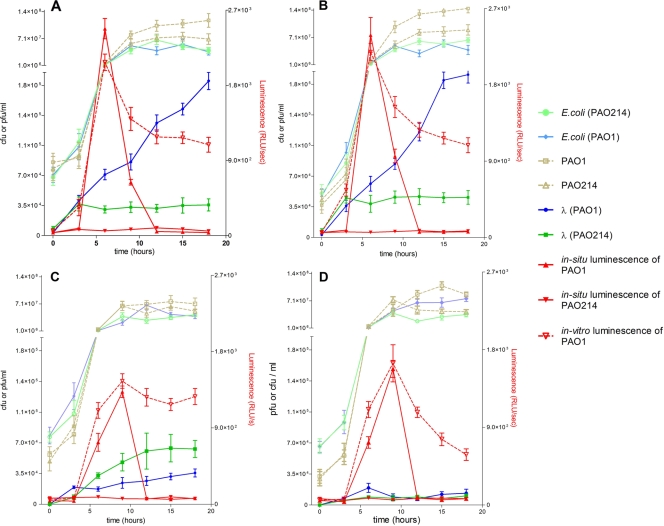
FIG. 5.
Time-dependent prophage induction in E. coli BW25113 λ lysogen (A) or its knockout mutants of recA (B), sdiA (C), and rcsA (D) when cocultured with either Pseudomonas aeruginosa PAO1 or its lasI knockout mutant, P. aeruginosa PAO214. The production of N-butanoyl-l-homoserine lactone and N-3-oxo-dodecanoyl-homoserine lactone (AHL) was monitored in situ using a bioluminescent reporter strain, A. tumefaciens A136, carrying the traI-luxCDABE plasmid grown in all cocultures with other bacteria (solid red lines). Supernatant of each coculture experiment also was analyzed in vitro for AHL concentration by incubation with the biosensor separately (broken red lines). Strain designations in parentheses in the key indicate the strain of P. aeruginosa cocultured with E. coli. Values plotted are means from triplicate experimental cultures, and the error bars are one standard deviation.
In situ demonstration of AHL-dependent prophage induction in cocultures of E. coli and P. aeruginosa PAO.
To determine if AHL-mediated prophage induction could occur in the absence of exogenously added AHL but from AHL generated in coculture (i.e., cell-to-cell communication), we conducted batch experiments containing bacterial cocultures of P. aeruginosa PAO1, which produces N-butanoyl-l-homoserine lactone (C4-AHL) and N-3-oxo-dodecanoyl-homoserine lactone (3-oxo-C12-AHL), E. coli λimm434 lysogen inducible by AHL, and A. tumefaciens A136 (9, 47) containing the traI-luxCDABE construct that produces light (luminescence) at a level that is directly proportional to the AHL concentration. The production of phage and luminescence was monitored every 3 h for 18 h in two coculture systems, one with wild-type P. aeruginosa PAO1 that actively produces AHL, E. coli λ lysogen, and A. tumefaciens A136, and the other with P. aeruginosa PAO1 replaced by a lasI knockout mutant of PAO1 (PAO214) that could not produce AHL (25, 38). In both systems, the initial cell density of P. aeruginosa was standardized (Fig. 5A). In both cocultures the E. coli λ lysogen was provided from the same stock suspension made by vortexing a single colony of E. coli in TSB, and A. tumefaciens was inoculated to the coculture as a 1:500 dilution from a single overnight culture. The level of the production of λ lysogen in the first system was five- to sixfold greater than that in the second system containing the lasI mutant and was similar to the induction response observed in the presence of exogenously added AHL (Fig. 4 and 5A). In the first system, AHL synthesis monitored by bioluminescence in situ reached a maximum at 6 h, while the second system containing a lasI mutant of PAO1 did not produce any light, which is consistent with the inability of this mutant to produce AHL (Fig. 5A). In a control experiment that included only the AHL bioreporter strain and the lasI mutant of strain PAO1, no luminescence was detected, confirming that neither the biosensor (A. tumefaciens) nor P. aeruginosa PAO214 was able to produce any AHL compounds. The onset of an increase in phage production in the first system coincided with the maximal AHL production (Fig. 5A). In a pair of similar coculture experiments, a recA knockout mutant of the same λ lysogen also showed an increase in phage production similar to the increase in luminescence when incubated with P. aeruginosa PAO1, but phage abundance did not increase when incubated with the lasI mutant of PAO1, and no luminescence was detected (Fig. 5B). In all cases, the increase in viral production occurred between 6 and 15 h (Fig. 5A and 5B). However, in a similar system the sdiA knockout mutant of the λ lysogen did not show any increase in phage production with either wild-type P. aeruginosa PAO1 or the lasI mutant of P. aeruginosa PAO1 (Fig. 5C). In all coculture experiments, the initial cell numbers of P. aeruginosa, E. coli, and A. tumefaciens were standardized, and the initial cell counts (CFU per milliliter) were similar to those of the control cultures (Fig. 5A, B, and C). In all cases, the luminescence decreased significantly after 9 h, probably because A. tumefaciens grew more slowly or was inhibited in coculture with P. aeruginosa and E. coli. Thus, the entire set of bioluminescence measurements was repeated by taking archived cell-free aliquots of culture supernatants and incubating them with A. tumefaciens A136 separately. As in the in situ measurements, luminescence (i.e., AHL concentration) peaked at 6 h and declined thereafter but was sustained at a level above 1,000 relative luminescence units throughout the experiment (Fig. 5A, B, and C). In this relatively simple model system, the results confirm that AHL produced by a different species can influence prophage induction in E. coli and directly demonstrate the involvement of sdiA, the AHL receptor of Enterobacteriaceae (56).
P. aeruginosa PAO1 is known to produce other potential SOS-dependent inducing agents, such as quorum-sensing-controlled toxins, rhamnolipid, cyanide, and pyocyanin (18, 24, 39, 43). However, these cannot be responsible for the induction response we observed in coculture experiments, because the AHL-dependent viral production was unaffected in the recA mutant (Fig. 5B). As previously demonstrated with A. tumefaciens (2), we observed no effect on the growth of E. coli in TSB amended with filtered culture supernatants from P. aeruginosa PAO1 relative to that of unamended control cultures (data not shown). This result confirms that the induction of the λ lysogen was not due to a potential SOS response created by toxins but from AHL produced by P. aeruginosa PAO1. Furthermore, were this the case, we should have observed a comparable induction response in coculture experiments with the mutant P. aeruginosa PAO214 that is compromised only in its ability to produce AHL. No such induction response was observed.
Probable molecular induction mechanism in λ involves SdiA and RcsA.
A positive transcriptional regulator of exopolysaccharide synthesis, rcsA, is the only regulator known to be directly involved in SOS-independent spontaneous λ induction in E. coli (41). In Pantoea stewartii, the expression of rcsA was shown to be directly dependent on the AHL concentration (35). These previous findings led to the hypothesis that rcsA also is involved in AHL-mediated prophage induction in E. coli. To test this, we constructed an rcsA knockout mutant of the λ lysogen used in the previous experiments. When this lysogen was exposed to exogenous AHL (Fig. 4) or cocultured with P. aeruginosa PAO1 (Fig. 5D), phage production did not increase compared to that of their respective controls that had either no AHL (Fig. 4) or were cocultured with a lasI mutant of P. aeruginosa PAO1 (Fig. 5D). The AHL production in the coculture of rcsA mutant lysogen and PAO1 was monitored based on luminescence with A. tumefaciens A136 biosensor and showed a trend similar to that of the cocultures of wild-type or recA mutant lysogens (Fig. 5A, B, and D) but decreased more than that in the cocultures of sdiA mutant lysogen (Fig. 5C and D). This result may indicate that when the functional SdiA receptor is present, it binds AHL, thereby reducing the extracellular AHL concentration and leading to the lower luminescence values. Taken together, these findings directly demonstrate that rcsA is an essential component of the quorum-sensing circuit that induces prophage.
Notably, the rcsA mutant showed significantly less spontaneous induction (no AHL added) than the wild type (Fig. 4), which is consistent with results obtained by Rozanov et al. (41). We did not complement the rcsA knockout mutant with an RcsA-overexpressing plasmid, since this was performed already by Rozanov et al. (41) and was shown to increase spontaneous prophage induction in a complemented λ lysogen of E. coli.
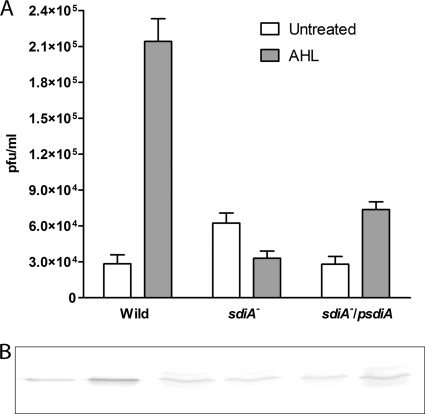
In contrast, we noticed a higher spontaneous induction (at least twofold) in the sdiA knockout lysogen (Fig. 6A). To restore its original phenotype, we complemented the mutation by transforming it with overexpressing sdiA-containing plasmid and monitored induction with AHL. A small but significant decrease in spontaneous induction relative to that of the sdiA mutant was observed (Fig. 6A). However, in our results, the complementation of this mutation did not fully restore the wild phenotype when it was incubated with AHL (Fig. 6A). To determine how AHL-bound SdiA affects RcsA, we monitored RcsA expression during induction by AHL via immunoblotting with polyclonal anti-RcsA. In the wild-type λ lysogen, RcsA expression increased in the presence of AHL (Fig. 6B). From this result we hypothesized that SdiA is a negative regulator of the rcsA promoter (35), as it is in Pantoea stewartii, that becomes derepressed when AHL binds to a SdiA homolog (EsaR) (35). If this hypothesis is correct, an increase in RcsA expression should be expected in the sdiA mutant relative to that in the wild type. However, our analysis showed no difference in RcsA expression (Fig. 6B). Likewise, in the complemented sdiA mutant no difference in RcsA expression was observed relative to that of the sdiA mutant or the wild type. These two results suggest that sdiA does not have any direct negative or positive regulatory role in RcsA expression. However, RcsA expression did not increase in the sdiA knockout mutant when AHL was applied (Fig. 6B), suggesting that SdiA is required for AHL-mediated RcsA expression. Thus, there must be intermediate transducers involved in carrying AHL-SdiA-mediated signal to activate rcsA for prophage induction. From these results a working model could be deduced for E. coli where sdiA and rcsA play the terminal roles in the AHL-mediated signal transduction of RecA-independent prophage induction. It has been demonstrated that SdiA is insoluble unless it is bound to AHL, but it appears to have a regulatory role in many other physiological processes in its insoluble state (64). It is unclear at this point how the insoluble fraction of SdiA, as would be the case in the absence of AHL which cannot be produced by E. coli, is involved in spontaneous prophage induction.
FIG. 6.
(A) Spontaneous prophage induction of wild-type and mutant sdiA and sdiA/psdiA (a sdiA mutant complemented with a SdiA-overexpressing plasmid) lysogens with or without AHL added in vitro. (B) Immunoblot assay for RcsA expression in the corresponding subsamples. The lanes were loaded alternately as controls and AHL-induced samples and correspond to the bars in the graph of panel A. In all lanes, 150 μg total protein was loaded. The position of the RcsA band was determined by the total protein extracted from an RcsA-overexpressing E. coli control in all cases (not shown).
Conclusion.
Our findings are the first report of an in vivo chemical signal that can trigger the lytic/lysogenic switch under conditions indicative of high host cell density, thereby maximizing the probability of subsequent infection by progeny viruses. Our hypothesis regarding cell density-dependent prophage induction at first glance appears to be in conflict with the generalized model of homoimmunity in pure bacterial cultures. However, in highly diverse microbial ecosystems such as soil or complex biofilms, a broad host range may be the rule rather than the exception. Nevertheless, our results directly demonstrate the induction of λ lysogen in the presence of an AHL-producing P. aeruginosa strain in the absence of any exogenously added AHL. This finding, combined with the demonstration of AHL-mediated prophage induction in microbial communities from two very different natural ecosystems (i.e., soil and groundwater), suggests that this is a widespread chemical signaling phenomenon governing an important interaction between host bacteria and temperate phage. Our observation illustrates the clever nature with which phage may exploit the chemical communication of their host bacteria by using a system that regulates many vital bacterial processes. These processes are essential for the successful establishment of symbiotic or pathogenic relationships with their respective eukaryotic hosts. For example, it raises questions regarding a possible role of AHL-producing bacteria in the phage-mediated horizontal transfer of genes such as that encoding Stx toxin in E. coli (48), which already was shown to be transferred spontaneously through prophage induction by the simple coculture of lysogenic and nonlysogenic strains nearly 40 years ago (46). With the recent discovery of AHL production by cyanobacteria (44), our results also may have implications for understanding broad-host-range lysogenic cyanophages (51) involved in the horizontal transfer of photosystem II genes (50), which has global importance in the open ocean. We acknowledge that our work is limited to either an in vivo model of E. coli and Pseudomonas or to field-collected soil and groundwater samples assayed in vitro. Additional studies with other phage-host systems and direct in situ measurements will be required to better understand any possible ecological or pathological implications of these findings. For example, understanding how and when prophage are activated to lytic reproduction may lead to better treatment strategies for containing disease agents such as Shigella dysenteriae type 1 and enterohemorrhagic E. coli O157:H7, which possess prophage-encoded toxins.
Acknowledgments
This project was supported by National Research Initiative Competitive Grant nos. 2004-35107-14884 to K.E.W. and 2007-35319-18432 to M.R. from the USDA Cooperative State Research, Education, and Extension Service.
We also thank Julia Gouffon of the Affymetrix Core Laboratory, University of Tennessee, for the processing and analysis of the Phylochip samples. Bio-Sep beads were generously provided by K. L. Sublette, University of Tulsa. We are grateful to those who supplied bacterial strains as indicated in Materials and Methods and for the insightful comments provided by three anonymous reviewers that greatly aided the revision process.
Footnotes
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Ackermann, H. W., and M. S. Dubow. 1987. General properties of bacteriophages, vol. 1. CRC Press, Boca Raton, Fla.
- 2.An, D., T. Danhorn, C. Fuqua, and M. R. Parsek. 2006. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. USA 103:3828-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R. T., H. A. Vrionis, I. Ortiz-Bernad, C. T. Resch, P. E. Long, R. Dayvault, K. Karp, S. Marutzky, D. R. Metzler, A. Peacock, D. C. White, M. Lowe, and D. R. Lovley. 2003. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 69:5884-5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arber, W., L. Enquist, B. Hohn, N. E. Murray, and K. Murray. 1983. Experimental methods for use with lambda, p. 433-466. In J. W. R. R. W. Hendrix, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Beumer, A., and J. B. Robinson. 2005. A broad-host-range, generalized transducing phage (SN-T) acquires 16S rRNA genes from different genera of bacteria. Appl. Environ. Microbiol. 71:8301-8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodie, E. L., T. Z. Desantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 72:6288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brüssow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casas, V., J. Miyake, H. Balsley, J. Roark, S. Telles, S. Leeds, I. Zurita, M. Breitbart, D. Bartlett, F. Azam, and F. Rohwer. 2006. Widespread occurrence of phage-encoded exotoxin genes in terrestrial and aquatic environments in Southern California. FEMS Microbiol. Lett. 261:141-149. [DOI] [PubMed] [Google Scholar]
- 9.Chambers, C. E., M. B. Visser, U. Schwab, and P. A. Sokol. 2005. Identification of N-acylhomoserine lactones in mucopurulent respiratory secretions from cystic fibrosis patients. FEMS Microbiol. Lett. 244:297-304. [DOI] [PubMed] [Google Scholar]
- 10.Chen, B. Y., C. S. Lin, and H. C. Lim. 1995. Temperature induction of bacteriophage lambda mutants in Escherichia coli. J. Biotechnol. 40:87-97. [DOI] [PubMed] [Google Scholar]
- 11.Chen, F., J. R. Lu, B. J. Binder, Y. C. Liu, and R. E. Hodson. 2001. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR gold. Appl. Environ. Microbiol. 67:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, M. E., Q. He, Z. He, K. H. Huang, E. J. Alm, X. F. Wan, T. C. Hazen, A. P. Arkin, J. D. Wall, J. Z. Zhou, and M. W. Fields. 2006. Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl. Environ. Microbiol. 72:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cochran, P. K., and J. H. Paul. 1998. Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl. Environ. Microbiol. 64:2308-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner, J. A., R. R. Beitle, K. Duncan, R. Kolhatkar, and K. L. Sublette. 2000. Biotreatment of refinery spent-sulfidic caustic using an enrichment culture immobilized in a novel support matrix. Appl. Biochem. Biotechnol. 84-86:707-719. [DOI] [PubMed] [Google Scholar]
- 15.D'Argenio, D. A., M. W. Calfee, P. B. Rainey, and E. C. Pesci. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebel, W., and J. E. Trempy. 1999. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions To activate its own expression. J. Bacteriol. 181:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh, D., K. Roy, K. E. Williamson, D. C. White, K. E. Wommack, K. L. Sublette, and M. Radosevich. 2008. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl. Environ. Microbiol. 74:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 21.Holloway, B. W., U. Romling, and B. Tummler. 1994. Genomic mapping of Pseudomonas aeruginosa PAO. Microbiology 140:2907-2929. [DOI] [PubMed] [Google Scholar]
- 22.Kirby, E. P., W. L. Ruff, and D. A. Goldthwait. 1972. Cell division and prophage induction in Escherichia coli: effects of pantoyl lactone and various furan derivatives. J. Bacteriol. 111:447-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirov, S. M., J. S. Webb, C. Y. O'May, D. W. Reid, J. K. K. Woo, S. A. Rice, and S. Kjelleberg. 2007. Biofilm differentiation and dispersal in mucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 153:3264-3274. [DOI] [PubMed] [Google Scholar]
- 24.Kownatzki, R., B. Tummler, and G. Doring. 1987. Rhamnolipid of Pseudomonas aeruginosa in sputum of cystic fibrosis patients. Lancet i:1026-1027. [DOI] [PubMed] [Google Scholar]
- 25.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 26.Levine, M. 1918. Differentiation of B. coli and B. aerogenes on a simplified eosin-methylene blue agar. J. Infect Dis. 23:43-47. [Google Scholar]
- 27.Little, J. W. 2005. Lysogeny, prophage induction, and lysogenic conversion, p. 37-54. In M. K. Waldor, D. I. Friedman, and S. Adhya (ed.), Phages. ASM Press, Washington, DC.
- 28.Little, J. W. 1991. Mechanism of specific LexA cleavage-autodigestion and the role of recA coprotease. Biochimie 73:411-422. [DOI] [PubMed] [Google Scholar]
- 29.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 30.Masuda, N., and G. M. Church. 2002. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDaniel, L., L. A. Houchin, S. J. Williamson, and J. H. Paul. 2002. Plankton blooms—Lysogeny in marine Synechococcus. Nature 415:496. [DOI] [PubMed] [Google Scholar]
- 32.McDaniel, L., and J. H. Paul. 2005. Effect of nutrient addition and environmental factors on prophage induction in natural populations of marine Synechococcus species. Appl. Environ. Microbiol. 71:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melechen, N. E., and G. Go. 1980. Induction of lambdoid prophages by amino-acid deprivation-differential inducibility-role of RecA. Mol. Gen. Genet. 180:147-155. [DOI] [PubMed] [Google Scholar]
- 34.Michael, B., J. N. Smith, S. Swift, F. Heffron, and B. M. Ahmer. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minogue, T. D., A. L. Carlier, M. D. Koutsoudis, and S. B. von Bodman. 2005. The cell density-dependent expression of stewartan exopolysaccharide in Pantoea stewartii ssp. stewartii is a function of EsaR-mediated repression of the rcsA gene. Mol. Microbiol. 56:189-203. [DOI] [PubMed] [Google Scholar]
- 36.Oppenheim, A. B., O. Kobiler, J. Stavans, D. L. Court, and S. Adhya. 2005. Switches in bacteriophage lambda development. Annu. Rev. Genet. 39:409-429. [DOI] [PubMed] [Google Scholar]
- 37.Peacock, A. D., Y. J. Chang, J. D. Istok, L. Krumholz, R. Geyer, B. Kinsall, D. Watson, K. L. Sublette, and D. C. White. 2004. Utilization of microbial biofilms as monitors of bioremediation. Microb. Ecol. 47:284-292. [DOI] [PubMed] [Google Scholar]
- 38.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ran, H., D. J. Hassett, and G. W. Lau. 2003. Human targets of Pseudomonas aeruginosa pyocyanin. Proc. Natl. Acad. Sci. USA 100:14315-14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranquet, C., A. Toussaint, H. de Jong, G. Maenhaut-Michel, and J. Geiselmann. 2005. Control of bacteriophage Mu lysogenic repression. J. Mol. Biol. 353:186-195. [DOI] [PubMed] [Google Scholar]
- 41.Rozanov, D. V., R. D'Ari, and S. P. Sineoky. 1998. RecA-independent pathways of lambdoid prophage induction in Escherichia coli. J. Bacteriol. 180:6306-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander, M., and H. Schmieger. 2001. Method for host-independent detection of generalized transducing bacteriophages in natural habitats. Appl. Environ. Microbiol. 67:1490-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharif, D. I., J. Gallon, C. J. Smith, and E. Dudley. 2008. Quorum sensing in Cyanobacteria: N-octanoyl-homoserine lactone release and response, by the epilithic colonial cyanobacterium Gloeothece PCC6909. ISME J. 2:1171-1182. [DOI] [PubMed] [Google Scholar]
- 45.Shkilnyj, P., and G. B. Koudelka. 2007. Effect of salt shock on stability of λimm434 lysogens. J. Bacteriol. 189:3115-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith, H. W., and M. A. Linggood. 1971. The transmissible nature of enterotoxin production in a human enteropathogenic strain of Escherichia coli. J. Med. Microbiol. 4:301-305. [DOI] [PubMed] [Google Scholar]
- 47.Sokol, P. A., U. Sajjan, M. B. Visser, S. Gingues, J. Forstner, and C. Kooi. 2003. The CepIR quorum-sensing system contributes to the virulence of Burkholderia cenocepacia respiratory infections. Microbiology 149:3649-3658. [DOI] [PubMed] [Google Scholar]
- 48.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson, B. S., S. A. Eichorst, J. T. Wertz, T. M. Schmidt, and J. A. Breznak. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan, M. B., D. Lindell, J. A. Lee, L. R. Thompson, J. P. Bielawski, and S. W. Chisholm. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 52.Thingstad, T. F. 2000. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 45:1320-1328. [Google Scholar]
- 53.Torres-Cabassa, A. S., and S. Gottesman. 1987. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Houdt, R., A. Aertsen, P. Moons, K. Vanoirbeek, and C. W. Michiels. 2006. N-acyl-L-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 256:83-89. [DOI] [PubMed] [Google Scholar]
- 55.Venturi, V. 2006. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 30:274-291. [DOI] [PubMed] [Google Scholar]
- 56.Walters, M., and V. Sperandio. 2006. Quorum sensing in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 296:125-131. [DOI] [PubMed] [Google Scholar]
- 57.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wegrzyn, G., and A. Wegrzyn. 2005. Genetic switches during bacteriophage lambda development. Prog. Nucleic Acid Res. Mol. Biol. 79:1-48. [DOI] [PubMed] [Google Scholar]
- 59.Wei, Y., A. C. Vollmer, and R. A. LaRossa. 2001. In vivo titration of mitomycin C action by four Escherichia coli genomic regions on multicopy plasmids. J. Bacteriol. 183:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 61.Williamson, K. E., M. Radosevich, D. W. Smith, and K. E. Wommack. 2007. Incidence of lysogeny within temperate and extreme soil environments. Environ. Microbiol. 9:2563-2574. [DOI] [PubMed] [Google Scholar]
- 62.Williamson, S. J., L. A. Houchin, L. McDaniel, and J. H. Paul. 2002. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microbiol. 68:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winget, D. M., K. E. Williamson, R. R. Helton, and K. E. Wommack. 2005. Tangential flow diafiltration: an improved technique for estimation of virioplankton production. Aquatic Microb. Ecol. 41:221-232. [Google Scholar]
- 64.Yao, Y., M. A. Martinez-Yamout, T. J. Dickerson, A. P. Brogan, P. E. Wright, and H. J. Dyson. 2006. Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J. Mol. Biol. 355:262-273. [DOI] [PubMed] [Google Scholar]