Abstract
The Bacillus cereus group includes three closely related species, B. anthracis, B. cereus, and B. thuringiensis, which form a highly homogeneous subdivision of the genus Bacillus. One of these species, B. anthracis, has been identified as one of the most probable bacterial biowarfare agents. Here, we evaluate the sequence and length polymorphisms of the Bacillus collagen-like protein bcl genes as a basis for B. anthracis detection and fingerprinting. Five genes, designated bclA to bclE, are present in B. anthracis strains. Examination of bclABCDE sequences identified polymorphisms in bclB alleles of the B. cereus group organisms. These sequence polymorphisms allowed specific detection of B. anthracis strains by PCR using both genomic DNA and purified Bacillus spores in reactions. By exploiting the length variation of the bcl alleles it was demonstrated that the combined bclABCDE PCR products generate markedly different fingerprints for the B. anthracis Ames and Sterne strains. Moreover, we predict that bclABCDE length polymorphism creates unique signatures for B. anthracis strains, which facilitates identification of strains with specificity and confidence. Thus, we present a new diagnostic concept for B. anthracis detection and fingerprinting, which can be used alone or in combination with previously established typing platforms.
The Bacillus cereus group includes three closely related species, B. anthracis, B. cereus, and B. thuringiensis, as well as the more distantly related species B. mycoides and B. weihenstephanensis. These gram-positive, spore-forming bacteria form a highly homogeneous subdivision of the genus Bacillus, which also contains several other organisms belonging to the B. subtilis group. The importance and public awareness of B. cereus group organisms are associated with their distinct phenotypes and pathological effects. B. anthracis is the causative agent of anthrax, a disease that affects humans and animals worldwide and has also been developed as a biological warfare agent (17, 25). B. cereus is an opportunistic human pathogen which is responsible mainly for gastrointestinal illnesses resulting from food contamination (9), whereas B. thuringiensis is an insect pathogen whose toxin is a biological pesticide widely used in global agriculture (38). The systematics of the members of the B. cereus group poses significant challenges due to very high level of chromosomal synteny and protein identity (33). Intense efforts have focused on overcoming these challenges, and there has been a particular focus on developing methods for specific detection of B. anthracis and for differentiating among strains of these closely related organisms.
Biodefense and forensic needs prompted large-scale sequencing of multiple bacillus genomes in a search for polymorphic sites for use in typing procedures (33). One type of polymorphism involves variation in the number of repeating nucleotide units that are referred to as variable-number tandem repeats (VNTRs). The resulting variation in the length and mass of the PCR products of these units can be demonstrated by gel and capillary electrophoresis (20), mass spectrometry (29), or microchannel fluidics (30). To date, several different VNTRs have been identified and tested. For example, Keim et al. studied the genetic relationship among a large collection of B. anthracis isolates based on the VNTRs found in the vrr genes (19, 20). Using a similar approach, Valjevac et al. used VNTRs of Bcms loci as markers to assess the phylogeny of members of the B. cereus group (46). Finally, length variation of the collagen-like (CL) region of the bclA gene was employed to differentiate among B. anthracis strains (6, 42).
The CL sequences, which are composed of Gly-Xaa-Yaa (i.e., a glycine followed by two additional residues; GXY) repeats, have been identified in silico in more than 100 prokaryotic proteins (34). Recent studies demonstrated that some bacterial CL proteins (CLPs), such as streptococcal protein Scl and BclA, can form the collagen triple helix (4, 14, 48). Bacterial CLPs are typically surface exposed and are found in microorganisms pathogenic to humans and animals. BclA (Bacillus CLP of B. anthracis) is a major spore surface protein (41) and is found in all members of the B. cereus group (6; this study). A second CLP, designated BclB (47), was identified as a component of the B. anthracis exosporium; however, its distribution and structural properties have not been well characterized. Likewise, two closely related proteins, ExsH and ExsJ, contain GXY CL repeats and are presumably located in the exosporium of Bacillus strains (45).
In this work we investigated in silico the occurrence and distribution of the bcl genes, presumably encoding CLPs, in all members of the B. cereus group. A new classification of the resulting Bcl protein variants is proposed based on the domain composition and folding of these proteins. As many as 10 bcl genes were found in a single B. cereus strain. Five genes were consistently observed in B. anthracis strains and designated bclA to bclE. We further analyzed sequence polymorphisms among these bcl genes and assessed use of them for B. anthracis detection and strain fingerprinting. Representative members of the B. cereus group and less closely related control bacilli were used to demonstrate specific bclB gene-based detection of B. anthracis spores. Finally, a combination of experiments and mathematical modeling was used to demonstrate how combined use of the bclABCDE sequence polymorphisms can be a powerful tool for strain fingerprinting in biodefense and forensic applications.
MATERIALS AND METHODS
Bioinformatic analyses.
Sequence searches for collagen homologs were carried out using PSI-BLAST (1) and the NCBI nonredundant database. Several independent searches were performed using representative sequences of members of the collagen family (PF01391) in the PFAM database (2). The gapped BLAST algorithm (blastpgp) was used with default parameters (BLOSUM62 substitution matrix; gap open penalty, 11; gap extension penalty, 1; number of iterations, up to 5; expectation value threshold, 0.0001). CLANS (cluster analysis of sequences) (12) was used to identify (sub)families of closely related sequences and to visualize similarities within and between Bcl proteins. CLANS is a Java utility based on the Fruchterman-Reingold graph layout algorithm, which uses the P values of high-scoring segment pairs obtained from an N×N BLAST search to compute attractive and repulsive forces between each pair of sequences in a user-defined data set. A two-dimensional representation was obtained by seeding sequences randomly in the arbitrary distance space. The sequences were then moved within this environment according to the force vectors resulting from all pairwise interactions, and the process was repeated to convergence. Groups of sequences (i.e., clans) were extracted from the CLANS output. A multiple-sequence alignment of the retrieved sequences was constructed using MAFFT (18) and optimized manually. In addition, representatives of each clan were analyzed using GeneSlico MetaServer (22), a gateway to a variety of computational methods for protein structure prediction, including sequence comparisons and secondary structure prediction, as well as tertiary fold recognition. Finally, the fold recognition methods were compared, evaluated, and ranked by the PCONS server (28) to identify the preferred modeling templates and the consensus alignment.
Bacterial strains.
The strains used in this study are listed in Table 1. B. anthracis avirulent strain Sterne lacking the pXO2 plasmid was obtained from the Colorado Serum Company, Denver, CO. Genomic DNA of B. anthracis strain Ames was kindly provided by B. Lin of the Center for Bio/Molecular Science and Engineering at the Naval Research Laboratory, Washington, DC. Most Bacillus strains were obtained from the American Type Culture Collection, Manassas, VA.
TABLE 1.
Bacterial strains
| Species or serovar | Strain | Source |
|---|---|---|
| B. anthracis | Sterne 34F2 | Colorado Serum Companya |
| B. anthracis (DNA) | Ames | NRLb |
| B. cereus | ATCC 4342 | ATCCc |
| B. cereus | ATCC 14579 (type strain) | ATCC |
| B. cereus | ATCC 13061 | ATCC |
| B. thuringiensis serovar kurstaki | ATCC 33679 | ATCC |
| B. thuringiensis serovar kurstaki | ATCC 33680 | ATCC |
| B. subtilis | ATCC 6051 (type strain) | ATCC |
| B. subtilis | ATCC 21332 | ATCC |
| B. subtilis | ATCC 31028 | ATCC |
| B. megaterium | ATCC 14581 (type strain) | ATCC |
| B. mycoides | ATCC 6462 (type strain) | ATCC |
Colorado Serum Company, Denver, CO.
NRL, Naval Research Laboratory, Washington, DC.
ATCC, American Type Culture Collection, Manassas, VA.
Sporulation and spore preparation.
Spores were prepared as described previously (15). Briefly, Bacillus strains were grown overnight on Trypticase soy agar at 30°C. Colonies of each strain were suspended in phosphate-buffered saline (pH 7.0) and plated on the following sporulation agar media: Schaeffer medium (37) for B. mycoides, 2× SG (24) for B. anthracis and B. cereus, and NSM (32) for B. thuringiensis. The plates were incubated at 37°C, except for the B. mycoides plates, which was incubated at 30°C. The sporulation process was monitored using phase-contrast microscopy. Spores were collected when the cultures contained >95% phase-bright spores, typically after 4 days, and were suspended in 2 ml of sterile ice-cold MilliQ water. The suspensions were centrifuged at 4,000 × g for 5 min at 4°C, and the resulting spore pellets were resuspended in fresh water. Washing was repeated four more times to remove the remaining debris, and spore suspensions were stored at 4°C. The spore concentration was tested by plating spores on growth media. The purity of spore preparations was evaluated by phase-contrast microscopy with oil immersion using a Nikon Optiphot-2 optical microscope (Nikon Inc., Melville, NY) equipped with a Plan ×100 objective. For PCR amplification, aliquots of spore preparations were diluted in water to obtain the desired concentrations and used without any further processing.
DNA isolation and purification.
Bacteria were grown overnight in Trypticase soy broth at 30°C. To isolate genomic DNA, 0.5-ml cultures were used, and DNA was extracted and purified using an IT 1-2-3 R.A.P.I.D. DNA purification kit according to the manufacturer's recommendations (Idaho Technology, Inc., Salt Lake City, UT). A bead-beating step was conducted with a BIO101/FastPrep FP120 homogenizer (Thermo Fisher Scientific, Inc., Waltham, MA) for 45 s at speed 5.5.
PCR amplification.
PCR amplification was performed using a DNA Engine Tetrad 2 (MJ Research, Inc., Waltham, MA) and the following cycling protocol: initial denaturation at 94°C for 1 min, followed by 31 cycles of denaturation at 94°C for 45 s, annealing at 59°C for 45 s, and elongation at 72°C for 1 min 45 s and then a final extension step consisting of 5 min at 72°C. Bacillus total DNA templates were used at a final concentration of approximately 15 ng/μl of reaction buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl; pH 8.3). For PCR amplification with spores, approximately 104 spores per reaction mixture were used. For multiplex PCR, the same cycling protocol was employed with a temperature gradient from 50 to 65°C for primer annealing and an Mg2+ concentration range of 1.5 to 6.5 mM. Primers employed for PCR amplification are listed in Table 2. The PCR products were analyzed on 2% Invitrogen ultrapure agarose (Invitrogen Corp., Carlsbad, CA) containing 1 μg/ml ethidium bromide. Electrophoresis was carried out in 1× Tris-acetate-EDTA buffer at 95 V for 2 h. The DNA size standard used was the 2-log DNA ladder (New England BioLabs, Inc., Beverly, MA). Gel images were captured using the UVP Bio-Doc-It system (UVP LLC, Upland, CA) and were processed using iPhoto '08 v.7.1.4 (Apple Inc., Cupertino, CA) and Canvas 9 (ACD Systems of America, Miami, FL).
TABLE 2.
Primers used for PCR amplification of bclABCDE sequences
| PCR target | Primer | Sequence (5′-3′) |
|---|---|---|
| bclA genea | bclA F1 | GAATCTTTATCAGCTAGTGCATTTG |
| bclA R1 | AAGCAACTTTTTCAATAATAATGGATG | |
| bclB genea | bclB F1 | GGCCCAGAAAATATTGGACCTAC |
| bclB R1 | ATTAGACGATATTAAGACCTGCGC | |
| bclC genea | bclC F1 | CCATGCTTTCCAAGTAGCGCTGG |
| bclC R1 | ATTAAGCGATTCTAAATACAGTTAG | |
| bclD genea | bclD F1 | TGTAATAATCAAAATGGTGTACATC |
| bclD R1 | ATCAACTTAACCTTATTATCGTTAAC | |
| bclE genea | bclE F1 | AGTCCATTAAATTCTAATTTCAAGAT |
| bclE R1 | ATTAACTCAATCTAATAATCGTTAAAG | |
| bclA CL regionb | bclA F2 | ATGAATCTTTATCAGCTAGTGC |
| bclA R3 | AAATGCATATAGTCCTGCTGG | |
| bclB CL regionb | bclB F2 | AGGCCCAGAAAATATTGGAC |
| bclB R2 | AGGCTGACTAAATCCAAATCC | |
| bclC CL regionb | bclC F3 | AATAGCAGCATTACAAACCGC |
| bclC R3 | CTGTATCTGTGCCGTTGAAG | |
| bclD CL regionb | bclD F2 | GGTGTACATGTTGATTCATGC |
| bclD R2 | TGCATACGTCGCCGTAATAC | |
| bclE CL regionb | bclE F3 | CCTACTTTTCCTCCAGTTCC |
| bclE R4 | GTACGTTGCAGTTGTACTAAC | |
| bclB of B. anthracisc | bclB F2 | AGGCCCAGAAAATATTGGAC |
| bclB R4 | GAGTTCCTCCCACACCTGG | |
| Non-B. anthracis bclB | bclB F3 | CCAAGTRATGCAAACAGATTAG |
| bclB R5 | TACTWGTTCCACCHGTTAA |
The primers amplify bclABCDE genes.
The primers amplify mainly the fragment encoding the CL regions of bclABCDE.
The primers specifically amplify the bclB gene of B. anthracis.
Classification methods.
We proposed that the bclABCDE gene products can be used to classify B. anthracis samples as different strains with confidence. The accuracy of a PCR assay for classifying samples correctly based on the gel migration patterns of the bclABCDE gene products was estimated using bootstrap resampling (10). P values were calculated from the resulting probability density functions using a nonparametric approach (13), as described in Fig. S1 in the supplemental material. Bootstrap resampling was used to create synthetic replicates of the bclABCDE gene products for the eight different B. anthracis strains studied. For each pairwise comparison, the lowest level of confidence (i.e., highest P value) was used to label a dendrogram representation of the differences between strains. A P value corresponded to the likelihood that a sample drawn from one population was misclassified as a member of another population. A P value of 0.05 was considered significant.
RESULTS
Identification of Bacillus CLPs.
First, the Bacillus CLPs were identified using a bioinformatic approach. The PFAM family of collagen sequences (http://pfam.sanger.ac.uk/family?PF01391) contains 9,744 sequences of CL domains, defined as a region consisting of 60 amino acids, and often individual CL proteins harbor more than one collagenous domain. The BLASTCLUST program was used to identify 37 sequences with >55% sequence similarity that represent best the PF01391 family. These sequences were used as queries in independent PSI-BLAST searches of a nonredundant database, which led to identification of 4,214 full-length proteins with CL sequences. From this set of identified proteins, a subset of 236 sequences was extracted that were annotated as derived from the genus Bacillus, and a list of putative Bcl proteins was created. This high number of various Bcl proteins identified was not expected given the high level of chromosomal synteny and protein similarity among the members of the B. cereus group (33).
Classification of the Bcl proteins.
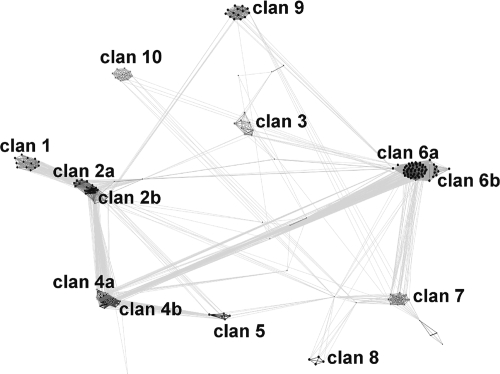
Next, we performed computational analyses to understand relationships between Bcl proteins. The noncollagen regions of Bcl sequences were clustered based on their pairwise BLAST similarity scores, using CLANS (12). We have experimentally found that for this group of sequences a P value threshold of 10−6 produces the best qualitative results. Lower P values resulted in disconnection of the most divergent sequences, while higher values resulted in overcompacting of the whole data set into a single clan with only a few outliers. CLANS identified 10 main subfamilies (clans) for the Bcl proteins (Fig. 1). A total of 171 Bcl proteins were clustered into one of these clans, while 65 of the Bcl proteins were not classified. In contrast, efforts to cluster the Bcl proteins based on comparison of their CL regions were inconclusive.
FIG. 1.
Two-dimensional projection of CLANS. Clustering results were obtained for the noncollagen regions of Bcl proteins. Connections between points represent the degree of pairwise sequence similarity, as quantified by BLAST P values (the darker the line, the higher the level of similarity).
Structural organization of the Bcl proteins.
Prediction of the detailed protein structure was performed using the GeneSilico MetaServer (22) for all Bcl proteins grouped into clans 1 to 10. We focused on (i) primary structure (e.g., domain prediction and identification), (ii) secondary structure (e.g., helices, strands, loops, transmembrane helices, and disordered regions), and (iii) fold recognition. In addition, groups of full-length sequences were extracted that formed clusters in the CLANS output, and multiple-sequence alignments were constructed for detection of the structural organization of Bcl sequences (Table 3). In summary, the domain architecture of Bcl proteins comprised (i) a short N-terminal region (N region) that occurs as 1 of 11 variants, (ii) a linker region (L region) that contains five conserved helices and is present in some Bcls, (iii) a highly variable CL region that is composed of 9 to 386 GXY triplets, and (iv) a C-terminal domain (CTD) that occurs in one of six folds that include the three known folds (a cupredoxin-like fold, a tumor necrosis factor/C1q-like fold, and a seven-blade beta-propeller fold).
TABLE 3.
Architecture of the Bcl proteins
| Clan | No. of amino acid residues in Bcl region or domain (predicted structure) |
No. of Bclsb | |||
|---|---|---|---|---|---|
| N region | L region | CL region | CTDa | ||
| 1 | 33 (disordered) | None | 145-228 | 87 or 88 (cupredoxin-like fold) | 13 |
| 2a | 33 (disordered) | None | 168-222 | 162 (unknown fold, 11 strands, and three helices) | 6 |
| 2b | 30 (disordered) | 122 (five helices and unknown fold) | 111-129 | 161 or 162 (unknown fold, 11 strands, and three helices) | 11 |
| 3 | 15 or 21 (variable, disordered) | None | 57-335 | 162 or 163 (unknown fold and 11 strands) | 7 |
| 4a | 25 (coiled coils) | 126 (five helices and unknown fold) | 33-447 | 131 or 132 (complement component C1q domain TNF-like fold; 26% sequence identity with PDB 1wck A) | 13 |
| 4b | 25 (coiled coils) | 132 (five helices and unknown fold) | 192-999 | 127-130 (complement component C1q domain TNF-like fold; 23% sequence identity with PDB 1wck A) | 20 |
| 5 | 25 (disordered) | 134 (five helices and unknown fold) | 66-225 | 301 (WD40 domain seven-blade beta-propeller fold; 45% sequence identity with PDB 1l0q A) | 5 |
| 6a | 36-41 (variable) | None | 78-1,158 | 135 (complement component C1q domain TNF-like fold; 22% sequence identity with PDB 1wck A) | 45 |
| 6b | 8 or 29 (variable) | None | 27-198 | 136-141 (complement component C1q domain TNF-like fold; 22% sequence identity with PDB 1wck A) | 9 |
| 7 | 40 (disordered) | None | 114-144 | 132 (complement component C1q domain TNF-like fold; 97% sequence identity with PDB 1wck A) | 15 |
| 8 | 32-38 (disordered) | None | 96-105 | 161-165 (unknown fold, two helices, and nine strands) | 4 |
| 9 | 19 or 21 (variable) | None | 417-675 | 133 (complement component C1q domain TNF-like fold; 17% sequence identity with PDB 1wck A) | 14 |
| 10 | 32 (disordered) | None | 45-129 | 134 (complement component C1q domain TNF-like fold; 17% sequence identity with PDB 1pk6 A) | 9 |
The PDB codes indicate the best predicted templates.
Numbers of Bcl proteins that cluster in the clans.
Distribution of Bcl proteins among Bacillus species.
There is no strict correlation between the distribution of a given clan's members and Bacillus species, which suggests that horizontal gene transfers occurred many times during the evolution of Bcl proteins (see Fig. S2 in the supplemental material). Nevertheless, clans 1, 2b, and 3 are the clans most characteristic of B. cereus. An exception is the BclB variant that forms clan 2a, which has been found only in B. anthracis. Based on the distribution pattern for members of the clans mentioned above, it is possible to distinguish B. anthracis from other Bacillus species. However, distinguishing between B. cereus and B. thuringiensis is difficult. To facilitate the classification of Bcl proteins, as well as strain identification, we developed a simple web server that is available at http://kudlaty.genesilico.pl/bcl/index.py. The query sequence submitted by a user is compared to hidden Markov model profiles of Bcl subfamilies with the HHsearch method (39). The ranking of “hits” against all of the clans characterized here is displayed, and the tentative classification of the query sequence is indicated graphically. This website also provides multiple-sequence alignments of members for each clan studied here. We believe that this resource will be useful for researchers interested in Bacillus organisms.
Detection and characterization of the bclABCDE genes in B. anthracis: proof of principle.
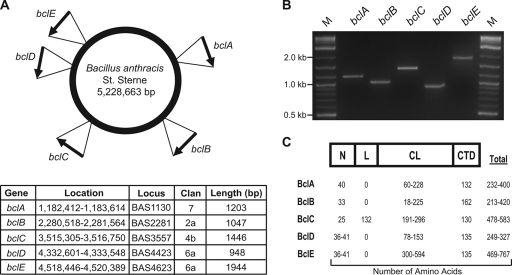
Five variable bcl genes have been found in the genome of B. anthracis strain Sterne (Fig. 2). In addition to the previously reported bclA (41) and bclB (47) genes, both of which encode exosporial proteins, three additional genes that encode presumed CLPs were identified. These genes were, by convention, designated bclC, bclD, and bclE consecutively in order of their clockwise localization around the chromosome (Fig. 2A). The coding sequences of the bclABC genes are located on a plus strand of the chromosome, while the coding sequences of bclD and bclE are located on a minus strand. Primers were designed to amplify each bcl gene using chromosomal DNA of the Sterne strain of B. anthracis as the template (Table 2). As expected, PCR amplifications yielded single-band products of the predicted lengths for each bcl gene (Fig. 2B). The largest amplified fragment from the Sterne strain of B. anthracis was that of the bclE gene, which was approximately 1.9 kb long. bclD, the smallest of the CLP-encoding genes, yielded an ∼0.9-kb amplified fragment. The remaining genes, bclA, bclB, and bclC were amplified as 1.2-, 1.0-, and 1.4-kb fragments, respectively.
FIG. 2.
Identification, detection, and characterization of the bcl genes and Bcl proteins of B. anthracis. Five genes designated bclA to bclE were identified in the genomes of B. anthracis strains. (A) Graphic representation of the B. anthracis strain Sterne chromosome showing the locations and orientations of the bclABCDE genes. Basic characteristics are shown in the table. (B) Detection of the bclABCDE genes in the genome of B. anthracis Sterne. The bclABCDE genes were PCR amplified, and resulting products were analyzed in a 2% agarose gel. Lanes M, 2-log DNA ladder. (C) Schematic representation (not to scale) and characterization of the BclABCDE proteins. The data are based on sequences obtained from the genomes of eight B. anthracis strains (Sterne, Ames, Australia 94, CNEVA-9066, A1055, Vollum, USA 6153, and Kruger). Unless indicated otherwise, Bcl protein regions were arbitrarily designated the N region (or amino-terminal region), L region, CL region, and CTD (or carboxyl-terminal region) (4). The table shows the ranges of the numbers of amino acids in the different regions of the BclABCDE proteins.
Two additional CLPs were identified in the B. anthracis genomes analyzed, and both of them contained short CL regions. The first protein, designated BclF (locus BAS3290), belongs to clan 10 and contains 12 GXY repeats interrupted by two 2-amino-acid insertions. The second protein, BclG (BA2449), belongs to clan 6b and contains nine GXY triplets. The apparent lack of length variation in the CL regions of these proteins among B. anthracis strains differed from the variation in BclA to BclE, and therefore, they were not included in the subsequent analyses. Nevertheless, there is significant length variation in the CL regions of both the BclF (up to 43 GXY repeats) and BclG (up to 66 GXY repeats) proteins in other members of the B. cereus group (Table 3), which could be used in typing of these organisms.
The bclABCDE genes were all found in the chromosomes in eight complete genomes of B. anthracis strains (Sterne, Ames, Australia 94, CNEVA-9066, A1055, Vollum, USA6153, and Kruger) and were characterized by significant length variation, especially in their CL regions (Fig. 2C). Each of the bcl genes potentially encodes a protein with an N region composed of 25 to 41 amino acids. The length of the central CL region in BclA, BclB, BclC, BclD, and BclE varies significantly, ranging from 18 to 594 amino acids (6 to 198 GXY repeats). The BclC protein is unique in that it contains the 132-amino-acid L region between the N and CL regions. The lengths of the CTDs in BclA, BclB, BclC, BclD, and BclE ranged from 130 to 162 amino acids. In summary, the genomes of B. anthracis strains contained five distinct bcl open reading frames encoding CLPs; however, significant size variation was observed for bcl alleles of different B. anthracis strains.
bclB gene-based detection of B. anthracis.
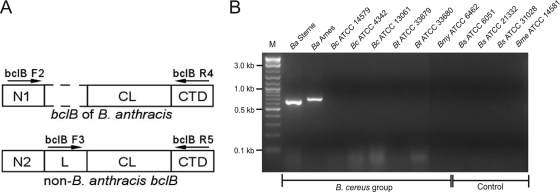
The bclABCDE genes were found in the genomes of other members of the B. cereus group, including B. cereus and B. thuringiensis, as well as in the genomes of B. mycoides and B. weihenstephanensis (see Fig. S2 in the supplemental material). However, although the CTD regions of all 25 available BclB sequences (clan 2) were highly conserved among the members of the B. cereus group, the BclB N regions in all 14 B. anthracis strains (clan 2a) varied significantly from 11 BclB sequences of B. cereus and B. thuringiensis strains (clan 2b). In addition, members of clan 2b also contain the L region composed of 122 amino acids, which is not present in BclB from clan 2a strains (Table 3). PCR primers were designed (Table 2) based on nucleotide sequence alignments of the 5′ ends of the bclB genes (see Fig. S3 in the supplemental material) to test the hypothesis that bclB-based amplification can be used to specifically detect DNA of B. anthracis (Fig. 3). As predicted, PCR amplification with primers bclB F2 and bclB R4 using DNA templates from B. anthracis strains Sterne and Ames yielded single products of the expected sizes, 645 bp and 699 bp, respectively, that were deduced from sequence data. Conversely, none of the PCRs that used as templates DNA from three B. cereus strains, two B. thuringiensis strains, and one B. mycoides strain resulted in amplification of the bclB genes of these strains. As expected, PCR was negative for DNA templates from the control strains of B. subtilis (n = 3) and B. megaterium (n = 1), which do not harbor the bclB gene. Altogether, our bioinformatic analyses of the bclB sequences that were obtained from 25 distinct members of the B. cereus group led to results indicating that bclB polymorphisms can provide quick and specific detection of anthrax etiology.
FIG. 3.
Specific detection of B. anthracis by PCR of the bclB gene. (A) Schematic representation (not to scale) of two main BclB variants of clan 2a and 2b proteins. Primers bclB F2 and bclB R4 (Table 2) were designed to differentiate between the bclB allele present in B. anthracis strains from the alleles present in other members of the B. cereus group, which are detected with primers bclB F3 and bclB R5. (B) PCR amplification using B. anthracis-specific primers. PCR products were analyzed by 2% agarose gel electrophoresis. The DNA templates used for PCR were genomic DNAs from B. anthracis (Ba) Sterne and Ames; B. cereus (Bc) strains ATCC 14579, ATCC 4342, and ATCC 13061; B. thuringiensis (Bt) strains ATCC 33679 and ATCC 33680; B. subtilis (Bs) strains ATCC 6051, ATCC 21332, and ATCC 31028; B. mycoides (Bmy) strain ATCC 6462; and B. megaterium (Bme) strain ATCC 14581. Lane M, 2-log DNA ladder.
Next, we performed PCR amplification using intact spores to assess the feasibility of a bclB-based method to detect B. anthracis (Fig. 4). B. anthracis spores, as well as control spores of B. cereus, B. thuringiensis, and B. mycoides, were prepared, and the purity of each preparation was evaluated with a light microscope (data not shown). Equal amounts of spores from the Bacillus strains were added to PCR mixtures, and amplification was carried out either with primers that were specific for the bclB gene of B. anthracis (bclB F2 and bclB R4) or with control primers specific for the bclB gene of non-B. anthracis organisms (bclB F3 and bclB R5) belonging to the B. cereus group (Table 2). PCR amplification using B. anthracis-specific primers and ∼104 spores of B. anthracis Sterne in the reaction mixture yielded the expected DNA product, while PCR amplification using spores of B. cereus, B. thuringiensis, and B. mycoides did not (Fig. 4, upper panel). Importantly, all of the latter spores yielded DNA products in control reactions with the non-B. anthracis-specific primers, whereas B. anthracis spores did not (Fig. 4, bottom panel). These data demonstrate that amplification of the bclB gene can specifically differentiate B. anthracis spores from spores of other members of the B. cereus group; however, experiments using a large panel of spores obtained from various Bacillus strains are necessary to validate bclB-based detection performed directly in the field.
FIG. 4.
Amplification of the bclB gene using Bacillus sp. spores. Spores were prepared from B. anthracis Sterne (Ba), B. cereus ATCC 13061 (Bc), B. thuringiensis ATCC 33679 (Bt), and B. mycoides ATCC 6462 (Bm). Specific detection of B. anthracis spores (∼104 spores per reaction) by PCR was performed with B. anthracis-specific bclB primers bclB F2 and bclB R4 (upper panel). A control positive PCR amplification was performed with primers bclB F3 and bclB R5, which amplify the bclB gene from the non-B. anthracis Bacillus species B. cereus, B. thuringiensis, and B. mycoides (lower panel). PCR products were analyzed by 2% agarose gel electrophoresis. Lane M, 2-log DNA ladder.
Fingerprinting of B. anthracis strains based on bclABCDE length polymorphism.
Significant sequence length polymorphism was observed in the CL regions of BclA to BclE. The variability in the length of the CL region encoded by various bclA alleles of several B. anthracis strains has previously been used for strain differentiation (6, 42). Here, we significantly improved the discriminatory power by simultaneous analysis of the lengths of the CL regions of all five bcl genes examined (bclABCDE). First, PCR amplifications were performed with genomic DNAs from B. anthracis strains Sterne and Ames using primers flanking the bclABCDE CL regions (Table 2). PCR products were found in 2% agarose gels as single DNA bands at the predicted sizes deduced from sequence data (Fig. 5, left panel). We next loaded side by side combined samples containing bclABCDE gene products obtained from each strain into single wells, and band patterns were resolved by agarose gel electrophoresis (Fig. 5, middle panel). The results show that the fingerprints generated for B. anthracis strain Sterne and strain Ames were significantly different and consisted of five and four bands, respectively. The bclABCDE amplification products of strain Sterne were separated from each other, but the amplification products of bclA (728 bp) and bclC (743 bp) were not resolved in a sample from strain Ames.
FIG. 5.
bcl-based fingerprinting of B. anthracis strains. Fragments of individual bclABCDE genes from B. anthracis strains Sterne and Ames were PCR amplified using primers flanking the CL regions (left panel). Expected sizes of PCR products are indicated below the gel. The individual PCR samples of bclABCDE for either strain Sterne or Ames were combined, and the sets were each loaded into single wells (middle panel). The solid squares indicate PCR bands in the gels. Multiplex PCR was performed with combined bclB primers using DNA from strain Sterne as the template (right panel). All PCR products were analyzed by 2% agarose gel electrophoresis. Lane M, 2-log DNA ladder.
Finally, multiplex PCR with all five primer pairs was attempted with DNA of the Sterne strain as the template by using a temperature gradient from 50 to 65°C for primer annealing and an Mg2+ concentration range of 1.5 to 6.5 mM in the buffer (Fig. 5, right panel). The bclABCDE genes were all amplified with an annealing temperature of 50°C and an Mg2+ concentration of 1.8 mM, although the intensities of the bclA and bclE bands were relatively low. Together, these data demonstrate that significant length variation in the CL regions of the bclABCDE genes that are present in the genomes of all available B. anthracis strains can be a valuable tool in strain fingerprinting.
Discriminating among B. anthracis strains using bclABCDE-based fingerprinting.
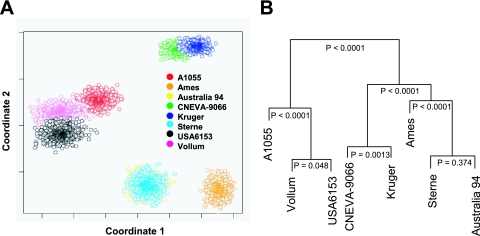
A computational approach was used to establish the feasibility of discriminating among B. anthracis strains using length polymorphism in the CL regions of the bclABCDE genes. The first step was to develop and calibrate a quantitative relationship between experimentally measured gel migration patterns of the bclABCDE gene products PCR amplified from B. anthracis strains Sterne and Ames, and the theoretical fragment lengths that were inferred from the sequence data (see Fig. S1 in the supplemental material). The calibrated model was next used to predict the fragment sizes amplified by PCR for each of the bclABCDE genes present in the genomes of six additional B. anthracis strains. The uncertainty associated with strain fingerprinting using multivariate measurement of the amplified fragments derived from the bclABCDE genes was estimated using bootstrap resampling (10). Bootstrap resampling was used to create a population of synthetic replicates. The ability to distinguish among the strains using the bclABCDE genes was represented in two dimensions using multidimensional scaling (Fig. 6A). The levels of confidence associated with distinguishing among these strains are shown in an annotated dendrogram in Fig. 6B. The eight strains clustered into four distinct groups. The Kruger and CNEVA-9066 strains clustered together, while the A1055, Vollum, and USA6153 strains formed a separate group (P < 0.0001). The Ames strain appeared to be distinct from the other strains (P < 0.0001). Only the Sterne and Australia 94 strains, which exhibited the highest level of similarity, could not be distinguished from each other (P = 0.377). Hence, we predicted that under the experimental conditions used here, we would be able to differentiate with confidence strains of B. anthracis, with the exception of the Sterne and Australia 94 strains, using a multilocus typing approach based on bclABCDE length polymorphism.
FIG. 6.
Mathematical modeling of bclABCDE-based fingerprinting of B. anthracis strains. (A) Multidimensional scaling representation of the similarity among the bootstrap replicates of eight B. anthracis strains based on multivariate PCR measurement of bclABCDE gene fragments. (B) Dendrogram with confidence levels associated with correct classification of the eight B. anthracis strains.
bcl gene-based fingerprinting of the B. cereus group organisms.
Determination of the origin of certain spores may also be important for non-B. anthracis Bacillus species in the event of a hoax, a blunder by a perpetrator, or psychological terrorism. Primer pairs that were optimized for the bclABCDE genes of B. anthracis were used to generate fingerprints using DNA templates from three B. cereus strains, one B. thuringiensis strain, and one B. mycoides strain (Fig. 7). Not all primer pairs yielded bclABCDE gene products with all DNA templates (see Fig. S4 in the supplemental material). Despite partial amplification of three or four bands, the combined PCR samples generated unique fingerprint patterns for the strains analyzed. Inclusion of the bclF and bclG genes in the fingerprint analysis, as well as primer optimization, should significantly improve the discriminating power. This test demonstrates that bcl-based fingerprinting could also be employed in forensic applications for differentiation of strains of all members of the B. cereus group.
FIG. 7.
bcl-based fingerprinting of non-B. anthracis Bacillus species. PCR amplification of the bclABCDE genes was performed using DNA templates from the following strains: B. cereus (Bc) strains ATCC 14579, ATCC 4342, and ATCC 13061; B. thuringiensis (Bt) ATCC 33679; and B. mycoides (Bm) ATCC 6462. Individually amplified bclABCDE gene products were combined and analyzed by 2% agarose gel electrophoresis. The estimated products sizes are indicated below the gel. Some primer pairs did not yield a detectable DNA product (ND). Lane M, 2-log DNA ladder.
DISCUSSION
The dissemination of B. anthracis spores to government offices and media outlets in the United States in late 2001 heightened public awareness of the potential for biological attacks, and since that time much emphasis has been placed on microbial forensic techniques that could determine the identity and origin of organisms used as biological weapons (3, 7); however, sequencing of B. anthracis genomes was required for these analyses (11). This is because the chromosomes of B. cereus group organisms are virtually identical and interspecies differences are largely determined by plasmid contents. The present work was initiated to identify and characterize potential markers specific to B. anthracis, as well as to individual B. anthracis strains. Here, we describe significant diversity and sequence polymorphisms in the Bacillus bcl genes and use of these genes for B. anthracis detection and strain fingerprinting.
The CLPs identified in our searches yielded proteins that were classified into 10 clans based on CLANS (see Fig. S2 in the supplemental material). The Bcl proteins of B. anthracis belong to clans 2a (BclB), 4a (BclC), 6a (BclD and BclE), and 7 (BclA). In addition to B. anthracis, the BclA, BclB, BclC, BclD, and BclE proteins are found in many other members of the B. cereus group. Despite the uniqueness of proteins that accounts for clan groups, several of the clans share protein fold predictions. Clans 4, 6, 7, 9, and 10 all have a predicted C-terminal TNF/C1q-like domain, although the level of predicted homology varies for each clan. The crystal structure of the BclA CTD (clan 7) was recently solved, and the authors reported that this domain is strikingly similar to the C-terminal globular domain of C1q (35). The mammalian proteins belonging to the TNF/C1q superfamily are involved in many diverse functions, including inflammation, autoimmunity, host defense, and apoptosis (21). The BclA protein is found in the exosporium and affects the hydrophobicity and adhesive properties of B. anthracis spores (5). A recent study showed that BclA interacts with the integrin receptor Mac-1 present on phagocytic and nonphagocytic cells and that this interaction affects spore uptake and infectivity in mice (31). Here, we predict that a TNF/C1q-like fold may be widespread in Bcl proteins. Other Bcl CTDs, such as those in clan 1, are predicted to contain a cupredoxin-like fold. The cupredoxins are a group of copper-containing proteins that are found in numerous organisms and function in a wide variety of cellular processes, including many enzymatic reactions and aerobic and anaerobic respiration (36). Additionally, Bcl proteins in clan 5 organisms contain a predicted WD40 domain, which is also found in the eukaryotic cell cycle protein CDC20 and in several G-protein β subunits, where it functions in protein-protein interactions (26, 49). Until this work, cupredoxin-like and WD40 domains in Bcl proteins had been not reported or investigated, and their biological significance is not known. Nonetheless, our analyses indicate that Bcl proteins are common in the members of the B. cereus group and can be classified into distinct clan groups based on the predicted protein folds, which are often shared with protein folds found in mammalian proteins.
Both BclA and BclB were identified as components of the outermost spore layer called the exosporium, which is characteristic of the spores of B. cereus group organisms but not the spores of B. subtilis group organisms (16). Sequences of bcl genes and Bcl proteins were found in the genomes and proteomes of B. cereus group organisms, such as B. anthracis, B. cereus, and B. thuringiensis, as well as two related species, B. mycoides and B. weihenstephanensis. In contrast, they were not present in B. subtilis group organisms. Considering the fact that Bcl proteins have a common architecture, it is tempting to speculate that all of them are associated with the exosporium. Recently, an exosporium-targeting sequence motif was identified in the N-terminal domain of BclA and BclB (44), as well as two other proteins designated BAS3290 and BAS4623, which we refer to here as a protein belonging to clan 10 (BclF) and BclE, respectively. The N-terminal domain of BclA is proteolytically cleaved, and the processed mature protein is inserted into the exosporium. However, the BclC and BclD proteins, as well as several other Bcl proteins, lack the consensus targeting sequence, and their association with the exosporium remains to be verified experimentally.
In addition to variation within the noncollagenous domains of Bcl proteins, there are both length and sequence polymorphisms in their collagenous domains. The CL regions of proteins BclA to BclE in B. anthracis Sterne consist of 28 distinct GXY triplets (Fig. S5). However, there is a strong preponderance of GXT triplets that account for about 97, 92, 49, 98, and 96% of all CL repeats in BclA to BclE, respectively. It has been shown that both BclA (40, 41) and BclB (47) are glycoproteins and that threonine residues are O glycosylated (8), which may explain the observed high GXT repeat content. Bcl CL glycosylation is not unique to B. anthracis; rather, it is an intrinsic property of the Bcl proteins in bacilli (43). Glycosylation of other Bcl proteins has yet to be confirmed. The BclC CL region is unique and is characterized by a lower frequency of GXT repeats (∼49%), which is accompanied by a high frequency of GXQ triplets (∼32%), which are not found in other Bcls. These differences provide an additional basis for differentiating Bcl variants. For example, BclD and BclE have high CTD sequence identity and are both grouped in clan 6a; however, the BclE CL region contains several triplets (GST, GET, GNT, GTT, GGT, GMT, GSA, GSI, GSM, GNM, GPM, GDT, and GVS) that are not present in the BclD CL region (see Fig. S5 in the supplemental material).
Here, we identified sequence polymorphisms that occur in the bclB alleles as a way to discriminate between B. anthracis and other members of the B. cereus group. Primers were designed that specifically amplified the bclB gene product when chromosomal DNA of B. anthracis was used as a template for PCR but not when DNAs of the closely related species B. cereus, B. thuringiensis, and B. mycoides, which contain different bclB alleles, were used as templates. Importantly, the same results were obtained when spores were used as the PCR templates. While we demonstrated the feasibility of our approach, the bclB-based detection system could be developed to identify B. anthracis in the field using portable PCR devices with speed and sensitivity.
Although bclB alone serves as a B. anthracis genetic identifier, in aggregate, the bcl genes exhibit significant diversity, which could be used to generate B. anthracis strain “fingerprints” (Fig. 5 and 6). It has been shown that length polymorphism in the CL region of bclA could differentiate some B. anthracis strains (42), while length variation in the bclB CL region in B. anthracis has not been explored (47). Since bclA is present in the genomes of other B. cereus group members (B. anthracis, B. cereus, and B. thuringiensis), PCR amplification of bclA was coupled with various electrophoretic separation methods to discriminate among B. cereus members at the strain level (6). The present work employed three additional bcl genes (bclC, bclD, and bclE) that are present in the genomes of B. anthracis strains and are characterized by significant length variation in their CL regions. This method should allow even greater discrimination between strains because it employs five variables (bclA, bclB, bclC, bclD, and bclE) rather than a single variable (bclA), as proposed previously. Other methods have been proposed that leverage the previously used multiple-locus VNTR analysis (MLVA) method to differentiate strains of B. anthracis (20, 23, 27). Using eight marker loci, Keim et al. analyzed a large worldwide collection of B. anthracis strains that clustered into six major genetic groups (20). A subsequent study by Lista et al., employing 25-locus MLVA for typing of large French and Italian collections, as well as reference B. anthracis strains, showed increased discriminatory power (27). Interestingly, some of the repeats used in that study (Bams13 and Bams30) are associated with bcl genes. Our bootstrap resampling analysis of a limited number of B. anthracis reference strains, which was based on bclABCDE length polymorphism, predicted strain clusters that had some differences from and some similarities to clusters obtained using the 25-locus MLVA criteria. For example, B. anthracis Volum (cluster A4), Sterne (cluster A3b), and Ames (cluster A3b) were on the same main branch as determined by both methods, although our analysis discriminated strains Sterne and Ames better. On the other hand, the Australia 94 and Sterne strains clustered separately in clusters A3a and A3b, respectively, based on the 25-locus MLVA criteria, while we could not discriminate between them based on bclABCDE polymorphism.
Here we used agarose gel electrophoresis to separate the bcl PCR products; however, this technique may not be optimal considering the relatively large sizes of the DNA fragments analyzed. Alternative separation techniques could significantly improve our results (6). The size and number of DNA bands could also be altered by incorporating endonuclease digestion of PCR products. An increase in the number of bands accompanied by a decrease in the band sizes would likely result in increased discriminatory power. This alternative protocol, including the digestion of PCR products, can be readily incorporated into the mathematical approach employed in this study. Together, our “proof of principle” experiments determined the feasibility of bcl fingerprinting, but further work is needed to validate our model.
In conclusion, we describe use of the bcl genes of B. cereus group organisms as chromosomal genetic identifiers of the species, as well as a means of discrimination at the strain level. Incorporation of bcl-based identification into existing systems may improve B. anthracis detection, and it may also improve microbial forensic techniques used to identify individuals responsible for biological attacks. In addition to using bcl gene polymorphisms as species- and strain-identifying markers, it is important that the Bcl proteins be assessed in terms of their biology. Additional research is needed to determine the role of Bcl in the pathogenesis of B. cereus group organisms. The present work proposes the first comprehensive classification of the CLPs of bacilli, which is based on the predicted structural characteristics of these proteins. This study increases our understanding of the diversity of this unique family of prokaryotic proteins that have been found in many pathogenic bacteria.
Supplementary Material
Acknowledgments
We are grateful to Baochuan Lin for providing the genomic DNA of B. anthracis strain Ames. We thank Chris Garton and Stephanie Boblett for assistance with some experiments.
Chris Garton and Stephanie Boblett were supported by NIH grant 5P20RR016477 to the West Virginia IDeA Network for Biomedical Research Excellence. This work was supported in part by NIH grants AI50666 (S.L.) and CA132124 (D.J.K.), by the PhRMA Foundation (D.J.K.), and by a West Virginia University Health Science Center internal grant from the Office of Research and Graduate Education (S.L.). J.M.B. and M.P. were supported by NIH grant GM081680-01.
Footnotes
Published ahead of print on 18 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharjee, Y., and M. Enserink. 2008. Anthrax investigation. FBI discusses microbial forensics—but key questions remain unanswered. Science 321:1026-1027. [DOI] [PubMed] [Google Scholar]
- 4.Boydston, J. A., P. Chen, C. T. Steichen, and C. L. Turnbough, Jr. 2005. Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J. Bacteriol. 187:5310-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmbhatt, T. N., B. K. Janes, E. S. Stibitz, S. C. Darnell, P. Sanz, S. B. Rasmussen, and A. D. O'Brien. 2007. Bacillus anthracis exosporium protein BclA affects spore germination, interaction with extracellular matrix proteins, and hydrophobicity. Infect. Immun. 75:5233-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castanha, E. R., R. R. Swiger, B. Senior, A. Fox, L. N. Waller, and K. F. Fox. 2006. Strain discrimination among B. anthracis and related organisms by characterization of bclA polymorphisms using PCR coupled with agarose gel or microchannel fluidics electrophoresis. J. Microbiol. Methods 64:27-45. [DOI] [PubMed] [Google Scholar]
- 7.Dance, A. 2008. Anthrax case ignites new forensics field. Nature 454:813. [DOI] [PubMed] [Google Scholar]
- 8.Daubenspeck, J., H. Zeng, P. Chen, S. Dong, C. Steichen, N. Krishna, D. Pritchard, and C. Turnbough. 2004. Novel oligosaccharide side-chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J. Biol. Chem. 279:30945-30953. [DOI] [PubMed] [Google Scholar]
- 9.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efron, B., and R. Tibshirani. 1986. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1:54-77. [Google Scholar]
- 11.Enserink, M. 2008. Anthrax investigation. Full-genome sequencing paved the way from spores to a suspect. Science 321:898-899. [DOI] [PubMed] [Google Scholar]
- 12.Frickey, T., and A. Lupas. 2004. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702-3704. [DOI] [PubMed] [Google Scholar]
- 13.Hall, P., and M. P. Wand. 1988. On nonparametric discrimination using density differences. Biometrica 75:541-547. [Google Scholar]
- 14.Han, R., A. Zwiefka, C. C. Caswell, Y. Xu, D. R. Keene, E. Lukomska, Z. Zhao, M. Hook, and S. Lukomski. 2006. Assessment of prokaryotic collagen-like sequences derived from streptococcal Scl1 and Scl2 proteins as a source of recombinant GXY polymers. Appl. Microbiol. Biotechnol. 72:109-115. [DOI] [PubMed] [Google Scholar]
- 15.Hart, S. J., A. Terray, T. A. Leski, J. Arnold, and R. Stroud. 2006. Discovery of a significant optical chromatographic difference between spores of Bacillus anthracis and its close relative, Bacillus thuringiensis. Anal. Chem. 78:3221-3225. [DOI] [PubMed] [Google Scholar]
- 16.Henriques, A. O., J. Moran, and P. Charles. 2007. Structure, assembly, and function of the spore surface layers. Annu. Rev. Microbiol. 61:555-588. [DOI] [PubMed] [Google Scholar]
- 17.Inglesby, T. V., D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Hauer, J. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, K. Tonat, and the Working Group on Civilian Biodefense. 1999. Anthrax as a biological weapon: medical and public health management. JAMA 281:1735-1745. [DOI] [PubMed] [Google Scholar]
- 18.Katoh, K., K. Misawa, K.-I. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keim, P., A. M. Klevytska, L. B. Price, J. M. Schupp, G. Zinser, K. L. Smith, M. E. Hugh-Jones, R. Okinaka, K. K. Hill, and P. J. Jackson. 1999. Molecular diversity in Bacillus anthracis. J. Appl. Microbiol. 87:215-217. [DOI] [PubMed] [Google Scholar]
- 20.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishore, U., C. Gaboriaud, P. Waters, A. K. Shrive, T. J. Greenhough, K. B. M. Reid, R. B. Sim, and G. J. Arlaud. 2004. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 25:551-561. [DOI] [PubMed] [Google Scholar]
- 22.Kurowski, M. A., and J. M. Bujnicki. 2003. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 31:3305-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Fleche, P., Y. Hauck, L. Onteniente, A. Prieur, F. Denoeud, V. Ramisse, P. Sylvestre, G. Benson, F. Ramisse, and G. Vergnaud. 2001. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leighton, T. J., and R. H. Doi. 1971. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J. Biol. Chem. 246:3189-3195. [PubMed] [Google Scholar]
- 25.Lew, D. P. 2000. Bacillus anthracis (anthrax), p. 2215-2220. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious diseases, vol. 2. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 26.Li, D., and R. Roberts. 2001. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58:2085-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lista, F., G. Faggioni, S. Valjevac, A. Ciammaruconi, J. Vaissaire, C. le Doujet, O. Gorge, R. De Santis, A. Carattoli, A. Ciervo, A. Fasanella, F. Orsini, R. D'Amelio, C. Pourcel, A. Cassone, and G. Vergnaud. 2006. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 6:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundstrom, J., L. Rychlewski, J. Bujnicki, and A. Elofsson. 2001. Pcons: a neural-network-based consensus predictor that improves fold recognition. Protein Sci. 10:2354-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muddiman, D. C., D. S. Wunschel, C. Liu, L. Pasa-Tolic, K. F. Fox, A. Fox, G. A. Anderson, and R. D. Smith. 1996. Characterization of PCR products from bacilli using electrospray ionization FTICR mass spectrometry. Anal. Chem. 68:3705-3712. [DOI] [PubMed] [Google Scholar]
- 30.Mueller, O., K. Hahnenberger, M. Dittmann, H. Yee, R. Dubrow, R. Nagle, and D. Ilsley. 2000. A microfluidic system for high-speed reproducible DNA sizing and quantitation. Electrophoresis 21:128-134. [DOI] [PubMed] [Google Scholar]
- 31.Oliva, C. R., M. K. Swiecki, C. E. Griguer, M. W. Lisanby, D. C. Bullard, C. L. Turnbough, Jr., and J. F. Kearney. 2008. The integrin Mac-1 (CR3) mediates internalization and directs Bacillus anthracis spores into professional phagocytes. Proc. Natl. Acad. Sci. USA 105:1261-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips, A. P., and J. W. Ezzell. 1989. Identification of Bacillus anthracis by polyclonal antibodies against extracted vegetative cell antigens. J. Appl. Bacteriol. 66:419-432. [DOI] [PubMed] [Google Scholar]
- 33.Rasko, D. A., M. R. Altherr, C. S. Han, and J. Ravel. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29:303-329. [DOI] [PubMed] [Google Scholar]
- 34.Rasmussen, M., M. Jacobsson, and L. Bjorck. 2003. Genome-based identification and analysis of collagen-related structural motifs in bacterial and viral proteins. J. Biol. Chem. 278:32313-32316. [DOI] [PubMed] [Google Scholar]
- 35.Rety, S., S. Salamitou, I. Garcia-Verdugo, D. J. S. Hulmes, F. Le Hegarat, R. Chaby, and A. Lewit-Bentley. 2005. The crystal structure of the Bacillus anthracis spore surface protein BclA shows remarkable similarity to mammalian proteins. J. Biol. Chem. 280:43073-43078. [DOI] [PubMed] [Google Scholar]
- 36.Savelieff, M. G., T. D. Wilson, Y. Elias, M. J. Nilges, D. K. Garner, and Y. Lu. 2008. Experimental evidence for a link among cupredoxins: red, blue, and purple copper transformations in nitrous oxide reductase. Proc. Natl. Acad. Sci. USA 105:7919-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 40.Steichen, C., P. Chen, J. F. Kearney, and C. L. Turnbough, Jr. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J. Bacteriol. 185:1903-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol. Microbiol. 45:169-178. [DOI] [PubMed] [Google Scholar]
- 42.Sylvestre, P., E. Couture-Tosi, and M. Mock. 2003. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J. Bacteriol. 185:1555-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamborrini, M., M. A. Oberli, D. B. Werz, N. Schurch, J. Frey, P. H. Seeberger, and G. Pluschke. 2009. Immuno-detection of anthrose containing tetrasaccharide in the exosporium of Bacillus anthracis and Bacillus cereus strains. J. Appl. Microbiol. 106:1618-1628. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, B., and G. Stewart. 2008. Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol. Microbiol. 70:421-434. [DOI] [PubMed] [Google Scholar]
- 45.Todd, S. J., A. J. G. Moir, M. J. Johnson, and A. Moir. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J. Bacteriol. 185:3373-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valjevac, S., V. Hilaire, O. Lisanti, F. Ramisse, E. Hernandez, J.-D. Cavallo, C. Pourcel, and G. Vergnaud. 2005. Comparison of minisatellite polymorphisms in the Bacillus cereus complex: a simple assay for large-scale screening and identification of strains most closely related to Bacillus anthracis. Appl. Environ. Microbiol. 71:6613-6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waller, L., M. Stump, K. Fox, W. Harley, A. Fox, G. Stewart, and M. Shahgholi. 2005. Identification of a second collagen-like glycoprotein produced by Bacillus anthracis and demonstration of associated spore-specific sugars. J. Bacteriol. 187:4592-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, Y., D. R. Keene, J. M. Bujnicki, M. Hook, and S. Lukomski. 2002. Streptococcal Scl1 and Scl2 proteins form collagen-like triple helices. J. Biol. Chem. 277:27312-27318. [DOI] [PubMed] [Google Scholar]
- 49.Yu, H. 2007. Cdc20: a WD40 activator for a cell cycle degradation machine. Mol. Cell 27:3-16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.