Abstract
Parasites play important roles in local population dynamics and genetic structure. However, due to insufficient diagnostic tools, detailed host-parasite interactions may remain concealed by hidden parasite diversity in natural systems. Microscopic examination of 19 European lake Daphnia populations revealed the presence of three groups of parasites: fungi, microsporidia, and oomycetes. For most of these parasites no genetic markers have been described so far. Based on sequence similarities of the nuclear small-subunit and internal transcribed spacer (ITS) rRNA gene regions, one fungus, four microsporidian, and nine oomycete taxa were discovered in 147 infected Daphnia (and/or three other zooplankton crustaceans). Additionally, cloning of rRNA gene regions revealed parasite sequence variation within host individuals. This was most pronounced in the ITS region of one microsporidian taxon, where the within-host sequence variation ranged from 1.7% to 5.3% polymorphic sites for parasite isolates from 14 different geographical locations. Interestingly, the parasite isolates from close locations grouped together based on sequence similarities, suggesting that there was parasite dispersal. Taken together, the data obtained in this study revealed hidden diversity of parasite communities in Daphnia lake populations. Moreover, a higher level of resolution for identifying parasite strains makes it possible to test new hypotheses with respect to parasite dispersal, transmission routes, and coinfection.
During the last decade, microparasites of Daphnia species, which are small zooplankton crustaceans, have become a popular study system in ecological and evolutionary research (for a review, see reference 15). It has been shown both in the field and under controlled laboratory conditions that parasites have a substantial impact on Daphnia fitness (7, 21, 52). Parasite-induced reductions in Daphnia population density (11, 12) or even population crashes (17) might result in disruptions of aquatic food webs, as daphnids play important roles as main phytoplankton grazers and as a major food of planktivorous fish (27). Moreover, as infections are often genotype specific (6, 8), they can lead to changes in the gene pool of a Daphnia population (7, 14), sometimes significantly increasing the genetic diversity of the host population (12, 54). Thus, Daphnia parasites cause not only ecological but also evolutionary changes in aquatic systems.
Conclusions regarding the importance of parasites in natural systems require powerful tools to detect and properly identify parasite taxa. Thus far, few species-specific molecular markers have been developed for Daphnia parasites (33, 38, 39, 41) and then used in experimental studies (3). In surveys of natural Daphnia populations, parasite identification has been based primarily on microscopic examination (4, 5, 29, 52), with only one exception (32). The parasites recorded in natural populations of Daphnia are thus considered members of certain taxa, or even species, without genetic confirmation. The fact that molecular markers are not used to characterize Daphnia infections makes it difficult to compare epidemic patterns across different habitats and/or various field surveys, as parasites cannot be unambiguously identified by microscopic examination alone. Even if microscopic identification is theoretically possible (for example, by examining ultrastructural morphology by electron microscopy [37]), this approach is not feasible for routine analysis. Consequently, classification of parasites that actually belong to different taxa in the same group might introduce noise into field surveys, as parasite taxa differ widely in virulence and host range (for a review, see reference 15).
Most of the known Daphnia parasites that have been described were obtained from small temporary ponds and rock pools (4, 16, 43). In permanent lakes, lower parasite diversity was assumed, mainly because increased fish predation reduces the population density of potential hosts (18), whereas high host density is a crucial determinant of epidemic spread (1, 2, 45). In addition, infected Daphnia spp. are more vulnerable to fish that hunt visually due to loss of their transparent appearance (11, 13). On the other hand, it was recently shown that even if Daphnia host density was reduced by selective fish predation, the prevalence of infection did not decline, probably due to the very high rates of transmission of the parasite that was observed (11). In contrast, we expected that the highly heterogeneous biotic and abiotic conditions in permanent lakes (27) would provide a variety of niches (for a review, see reference 47), which also favor a high level of parasite diversity. Therefore, Czech canyon-shaped reservoirs were chosen as our main study systems, because in these lakes environmental gradients are particularly pronounced in both the horizontal and vertical dimensions (42). Moreover, the Daphnia communities of these reservoirs are dominated by members of the Daphnia longispina complex (35), taxa which have previously been shown to be infected by a variety of parasites (52).
The results of our study revealed a high level of diversity of Daphnia parasites in permanent lakes. Fourteen different parasite taxa were detected using nuclear small-subunit (SSU) and internal transcribed spacer (ITS) rRNA gene sequence information. In addition, a high level of sequence variation was observed in the ITS region of one microsporidian taxon. Thus, molecular markers are now available which allow discrimination with high resolution among and within parasite taxa and provide tools to address more detailed questions concerning lake Daphnia-microparasite systems.
MATERIALS AND METHODS
Parasite origin and morphological examination.
Infected Daphnia spp. were isolated from freshly collected zooplankton samples from four small quarry lakes in Munich, Germany, and three larger, natural lakes near this city (all screened for infections in May to September 2008) and from ethanol-preserved zooplankton samples from 12 canyon-shaped reservoirs in the Czech Republic (collected for another purpose in July and October of 2004 and 2005 [35, 42]) (Table 1). Further, ethanol-preserved infected Daphnia spp. from three small lakes in the United States served as reference samples for the parasite taxon Metschnikowia bicuspidata. All of the lake Daphnia studied belong to European or North American members of the D. longispina complex. Additionally, parasite isolates obtained from Daphnia magna (either collected from an experimental basin or incidentally infected in laboratory cultures) and from infected individuals of other zooplankton crustaceans (Bosmina, Diaphonosoma, and Diaptomus) cooccurring in the field samples were analyzed. As some parasites infecting other crustaceans had a phenotype strikingly similar to that of Daphnia parasites, we wanted to clarify whether this was the result of cross-infection or whether these similar phenotypes represented genetically different taxa.
TABLE 1.
Numbers of molecularly identified parasite isolatesa
Within each taxon, at least one isolate from each location and host species was completely sequenced in the SSU and ITS regions (METS isolates were also sequenced in the D1/D2 domain). For all other isolates, partial sequence information was sufficient to unambiguously identify the parasite taxon.
METS, Metschnikowia sp.; BERW, Berwaldia sp.; MIC1, MIC2, and MIC3, Microsporidium sp. (with distinct identities); APH1, APH2, and APH3, Aphanomyces sp. (with distinct identities); PHY1 and PHY2, Pythiaceae (with distinct identities); SAP1, SAP2, SAP3, and SAP4, Saprolegnia sp. (with distinct identities).
All Daphnia hosts belong to the D. longispina complex, unless indicated otherwise.
Daphnia dentifera.
D. magna.
Experimental basin belonging to Ludwig-Maximilians University (Martinsried).
incidental lab infection.
In one case both parasite taxa were detected in the same host.
All zooplankton samples were screened for signs of parasite infection using a stereomicroscope at a magnification of ×250 (Zeiss, Göttingen, Germany). At least 200 Daphnia adults were screened for each location and sampling date. The parasites were classified into functional groups based on pathology and morphological traits (15).
Genomic DNA extraction.
Genomic DNA was extracted by overnight incubation of a single infected individual with 100 μg/ml proteinase K (Merck KGaA, Darmstadt, Germany) and 0.1% sodium dodecyl sulfate in proteinase K buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 25 mM EDTA [pH 8.0]) at 55°C. After heat inactivation of proteinase K for 12 min at 95°C, DNA was precipitated using isopropanol. DNA was dissolved in 50 to 100 μl of PCR-grade sterile water.
PCR amplification, cloning, and sequencing.
To initially determine the nucleotide sequences of rRNA gene regions of fungal or oomycete parasites, DNA was amplified using universal primers for eukaryotic SSU and ITS regions (30, 40). For microspordian parasites, primers for the SSU region were designed based on rRNA gene sequences of a variety of microsporidian parasites of Daphnia obtained from GenBank (for primer information see Table S1 in the supplemental material). Each PCR was carried out with 2 to 3 μl of genomic DNA, 0.05 U/μl DreamTaq DNA polymerase, 0.02 U/μl Pfu DNA polymerase, 1× DreamTaq buffer, 0.25 mM deoxynucleoside triphosphates (all obtained from Fermentas GmbH, St. Leon-Rot, Germany), and 0.5 μM of each primer. PCR amplicons of the correct size were gel eluted using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). The SSU and ITS regions were cloned using a TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA) and a CloneJET PCR cloning kit (Fermentas GmbH, St. Leon-Rot, Germany), respectively. Between three and six clones were sequenced for each parasite isolate using the BigDye v1.1 sequencing mixture (Applied Biosystems Inc., Foster City, CA). All sequencing reaction mixtures were resolved using an ABI 3730 capillary sequencer (Applied Biosystems Inc., Foster City, CA). Based on the sequences obtained and alignments of representative rRNA gene sequences from GenBank, primers specific for each group of parasites or single parasite taxa were designed in order to avoid amplification of Daphnia rRNA genes together with parasite rRNA genes (see Table S1 in the supplemental material). PCR with parasite-specific primers was carried out as described above, and amplicons were directly sequenced. If direct sequencing was not possible due to polymorphisms in rRNA gene copies, products were cloned as described above and four to six clones were sequenced for each amplicon. In addition, for the fungal parasite part of the large-subunit (LSU) rRNA gene (D1/D2 domain of the LSU rRNA gene) was amplified (for primer information see Table S1 in the supplemental material), and all PCR products were directly sequenced.
Molecular identification, phylogenetic analyses, and genetic diversity.
Similarity searches with the sequences obtained were conducted using the NCBI nucleotide database, and sequences that produced significant scores were selected. The outgroup taxa for each phylogenetic tree were chosen based on previously published phylogenies. The sequences were aligned using ClustalW; minor manual corrections were made using BioEdit v7.0.9 (22). Sequence similarities were displayed in neighbor-joining trees based on uncorrected Kimura two-parameter distances and the complete deletion option using MEGA 4.0 (46). Phylogenetic relationships among sequences were corroborated by performing maximum parsimony analyses with the same data sets (heuristic search, random stepwise additions) using PAUP* 4.0b10 (44). The percentages of 1,000 replicate trees in which the associated sequences clustered together were obtained by performing bootstrap tests.
Parasite sequence variation within a host.
All parasite isolates that were cloned were analyzed to determine the percentage of single-nucleotide polymorphisms (SNPs), including insertions and deletions (indels). Indels larger than 1 bp were counted as a single SNP since they might have resulted from a single event. In addition, mean pairwise differences between cloned rRNA gene copies were calculated using uncorrected Kimura two-parameter distances (MEGA 4.0). The SNP frequency distribution for the cloned ITS copies of 14 isolates of the microsporidian taxon BERW (each isolate was obtained from a distinct location) was analyzed by principal component analysis (PCA) in order to test for a phylogeographical pattern (26). To corroborate the results, a similar PCA was performed for the directly sequenced SSU region of the same 14 isolates, but the presence of polymorphic sites was inferred from visible double peaks in the sequencing chromatograms. For both analyses, only SNPs that were detected in at least 2 of 14 screened isolates were included to minimize the chance of relying on misincorporated nucleotides caused by DNA polymerase errors. The PCA were performed using STATISTICA for Windows, release 8.0 (StatSoft, Inc.).
Nucleotide sequence accession numbers.
The SSU, ITS, and partial LSU rRNA gene sequences determined have been deposited in the GenBank database under accession numbers FJ763440 to FJ763572 and FJ794855 to FJ794943 (see Table S2 in the supplemental material).
RESULTS
Morphological and molecular identification.
In 19 European lakes and five other locations (lakes in the United States, an experimental basin, and a laboratory infection), the following three groups of Daphnia crustacean parasites were observed by microscopic examination: fungi, microsporidia, and oomycetes. The fungi formed needle-like structures, and massive amounts of spores filled the entire host body cavity. Microsporidia were also observed in the body cavity of Daphnia and appeared to be whitish when dark-field microscopy was used. Oomycete parasites filled either the brood pouch or the entire body cavity with hypha-like structures. It was not possible to discriminate distinct taxa in each group of parasites using light microscopy and morphological traits.
Sequencing of rRNA gene regions detected one, four, and nine different taxa in the groups of fungi, microsporidia, and oomycetes, respectively, based on sequence divergence (Table 1 shows all parasite taxa and their abbreviations). Complete or partial rRNA gene sequences were obtained for a total of 147 parasite isolates (Table 1). Specifically, at least one isolate for each parasite taxon, location, and host species was completely sequenced in both the SSU and ITS regions (59 isolates) or in only the SSU region if amplification of the ITS was unsuccessful (seven isolates) (see Table S2 in the supplemental material). For the remaining 81 isolates, partial SSU sequences sufficient to unambiguously identify each parasite taxon were obtained. For the majority of parasite isolates belonging to a taxon, an identical main sequence variant was detected for the SSU, ITS, and partial LSU rRNA gene regions (the latter was sequenced only for the fungal parasite) by direct sequencing. When polymorphic sites were identified by two distinct peaks overlapping at the same nucleotide position, one of the peaks was always consistent with the nucleotide observed in all other isolates of the corresponding taxon. If sequences were poorly resolved (e.g., due to indels), the rRNA gene region was cloned to elucidate the main sequence variants.
Phylogenetic analyses and genetic diversity.
The phylogenetic analyses were in most cases based on rRNA gene sequences obtained without cloning, as the main sequence variant obtained by direct sequencing was the same for the majority of isolates from a particular parasite taxon. Otherwise, the PCR product was cloned, and the most abundant sequence variant was chosen. For taxon identification we used membership in clusters in neighbor-joining trees based on the genetic distance between sequences and grouped the taxa with previously described taxa. Phylogenetic relationships among sequences were corroborated with parsimony analyses.
(i) Fungi.
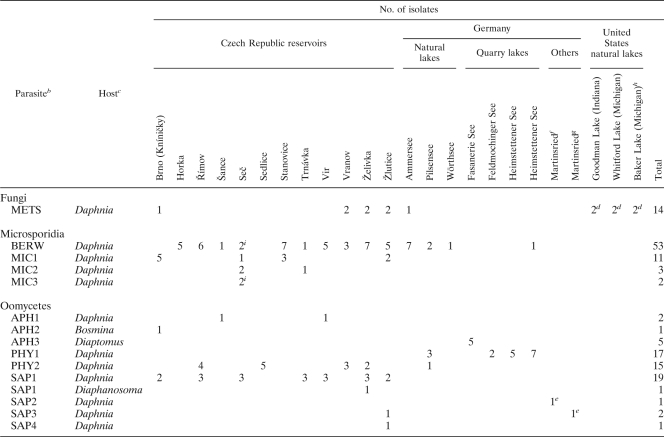
For 14 parasite isolates classified by morphology as fungi and originating from five locations in Europe and three locations in the United States (Table 1), only one sequence variant (referred to below as METS) was detected by direct sequencing of SSU, ITS, and partial LSU regions. Because the reference parasite isolates from the United States had been described previously as isolates belonging to Metschnikowia bicuspidata (a yeast in the family Hemiascomycetes [12, 21]), the sequences obtained were aligned with several published sequences of Hemiascomycetes. Neighbor-joining and parsimony analyses showed that the SSU, ITS, and partial LSU sequences of the isolated fungal parasite taxon METS are clearly distinct from the SSU, ITS, and partial LSU sequences of several M. bicuspidata isolates that have been published previously (bootstrap support, >90%) (Fig. 1A and B; see Fig. S1 in the supplemental material). However, the highest levels of sequence similarity were obtained with uncultured fungal isolates (99.0% and 91.4% sequence similarity), which is also shown in the grouping of sequences in the trees (Fig. 1A and B).
FIG. 1.
Neighbor-joining phylogeny of Hemiascomycetes (yeasts), including the METS taxon (in bold type), based on (A) SSU and (B) ITS rRNA gene sequences. The numbers on branches are bootstrap values (1,000 replicates; values less than 50% are not shown). Branch lengths are based on the expected numbers of nucleotide substitutions per site. In addition, the levels of bootstrap support for nodes obtained by parsimony analyses (trees with nearly identical topologies, with only some internal branches collapsed) are indicated by bold type. GenBank accession numbers are indicated after the species names.
(ii) Microsporidia.
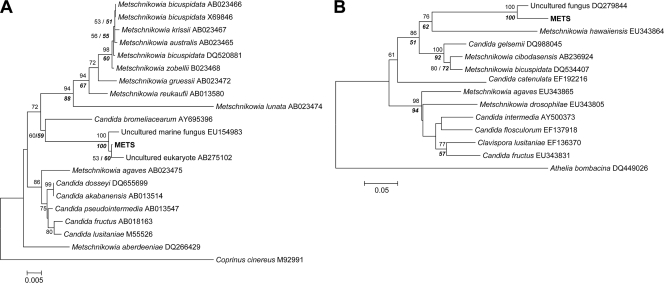
For 69 isolates classified as microsporidia and originating from 15 locations, four distinct microsporidian taxa were detected (BERW, MIC1, MIC2, and MIC3). BERW was especially common, as it was found in 14 different lakes (Table 1). Notably, in the Czech reservoir Seč, all four taxa cooccurred in the same year. In addition, in the same reservoir a single Daphnia was found to be coinfected by BERW and MIC3. The phylogenetic analysis performed for the SSU rRNA gene region included data for other previously described Daphnia microsporidia (Fig. 2). In the resulting tree, each taxon was located in a separate branch (with 100% bootstrap support). Thus, the BERW taxon was almost identical to Berwaldia schaefernai (99.9% sequence similarity) and the unidentified microsporidium Angskar 21, whereas MIC1 clustered with two other unidentified microsporidia. Since cloning, as well as direct sequencing of the MIC2 and MIC3 taxa, detected two variants in the SSU rRNA gene region, which were always present in all isolates of the corresponding parasite taxa, both sequence variants (variants a and b) were included in the phylogenetic analyses. In particular, variant b of MIC2 had 26 single-nucleotide exchanges and a 1-bp deletion in a 58-bp region, whereas variant b of MIC3 had two larger deletions (29 bp and 4 bp) compared to the sequence of variant a. Both MIC2 variants were similar to the unidentified microsporidium DP-1-19, whereas the MIC3 variants resembled Ordospora colligata. The ITS tree had a topology similar to that of the SSU tree, but since there was no published ITS sequence of B. schaefernai or microsporidian isolate DP-1-19, the tree is not shown. Nevertheless, in contrast to the high levels of sequence similarity between O. colligata and MIC3 in the SSU region, O. colligata had three deletions (8 bp, 26 bp, and 26 bp) in the ITS region compared to both MIC3 variants.
FIG. 2.
Neighbor-joining phylogeny of microsporidia, including the BERW, MIC1, MIC2, and MIC3 taxa (in bold type), based on SSU rRNA gene sequences. The numbers on branches are bootstrap values (1,000 replicates; values less than 50% are not shown). Branch lengths are based on the expected numbers of nucleotide substitutions per site. In addition, the levels of bootstrap support for nodes obtained by parsimony analysis (tree with identical topology) are indicated by bold type. GenBank accession numbers are indicated after the species names. *, MIC2 and MIC3 had a second underlying sequence, which was the same in all isolates in the corresponding parasite taxon (for details see text).
(iii) Oomycetes.
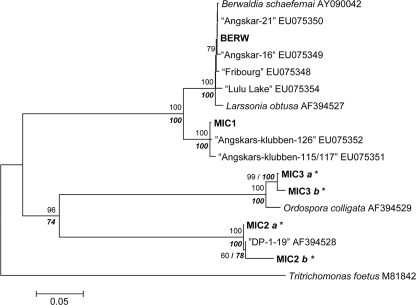
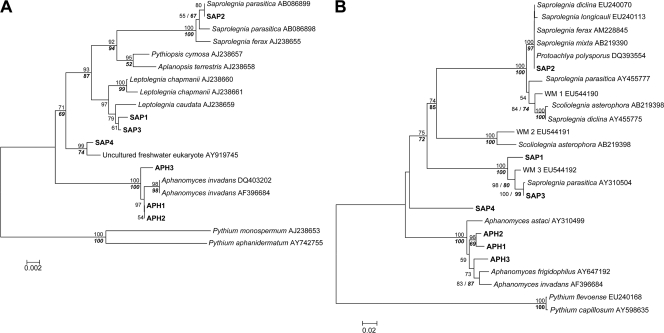
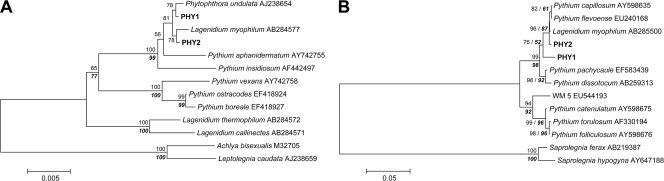
For 64 isolates tested originating from 18 locations, nine different oomycete taxa were detected (SAP1, SAP2, SAP3, SAP4, APH1, APH2, APH3, PHY1, and PHY2). In addition to Daphnia, three other zooplankton crustaceans were infected: Bosmina, Diaphanosoma, and Diaptomus (Table 1). Phylogenetic analyses were performed using two sets, because seven taxa belong to the family Saprolegniaceae, whereas two taxa belong to the family Pythiaceae (based on sequence similarities to previously described taxa). Within the Saprolegniaceae, the SAP1, SAP2, SAP3, and SAP4 taxa clustered in the genus Saprolegnia, whereas the APH1, APH2, and APH3 taxa clustered in the genus Aphanomyces on both SSU and ITS trees (Fig. 3A and B). In the SSU tree, both the SAP1 and SAP3 taxa clustered with Leptolegnia caudata, whereas in the ITS tree, as no Leptolegnia sequence was available for comparison, SAP3 was almost identical to a Saprolegnia parasitica strain of unknown origin. The SSU tree showed that the SAP2 taxon was identical to an S. parasitica strain isolated from eggs of cultured salmon, whereas the ITS tree revealed sequence identity of SAP2 with three different Saprolegnia species, all of which are well-known pathogens of fish. The SAP4 taxon clustered in the SSU tree closely with an uncultured eukaryote originating from lake water (99.8% sequence similarity). Otherwise, the position of this taxon was poorly resolved (Fig. 3A and B). In the SSU and ITS trees, the APH1, APH2, and APH3 taxa clustered with Aphanomyces invadans or Aphanomyces frigidophilus strains isolated from infected fish. The APH1 (obtained from a Daphnia host) and APH2 (obtained from a Diaptomus host) taxa were identical in the SSU region but differed in the ITS region (Fig. 3A and B). Within the Pythiaceae, the PHY1 and PHY2 taxa were closely related to each other, but the PHY1 taxon was present only in German lakes, whereas PHY2 predominated in the Czech Republic (Table 1). In both SSU and ITS trees, the PHY2 taxon clustered closely with Lagenidium myophilum strains isolated from infected shrimp (Fig. 4A and B).
FIG. 3.
Neighbor-joining phylogeny of oomycetes, including the APH1, APH2, APH3, SAP1, SAP2, SAP3, and SAP4 taxa (in bold type), based on the (A) SSU and (B) ITS rRNA gene sequences. The numbers on branches are bootstrap values (1,000 replicates; values less than 50% are not shown). Branch lengths are based on expected numbers of nucleotide substitutions per site. In addition, the levels of bootstrap support for nodes obtained by parsimony analyses (trees with nearly identical topologies, with only some internal branches collapsed) are indicated by bold type. GenBank accession numbers are indicated after the species names. WM1, WM2, and WM3 correspond to oomycete taxa isolated from Daphnia in a previous study (53).
FIG. 4.
Neighbor-joining phylogeny of oomycetes, including the PHY1 and PHY2 taxa (in bold type), based on (A) SSU and (B) ITS rRNA gene sequences. The numbers on branches are bootstrap values (1,000 replicates; values less than 50% are not shown). Branch lengths are based on expected numbers of nucleotide substitutions per site. In addition, the levels of bootstrap support for nodes obtained by parsimony analyses (trees with identical topologies) are indicated by bold type. GenBank accession numbers are indicated after the species names. WM5 corresponds to the oomycete taxon isolated from Daphnia in a previous study (53).
Parasite sequence variation within a host.
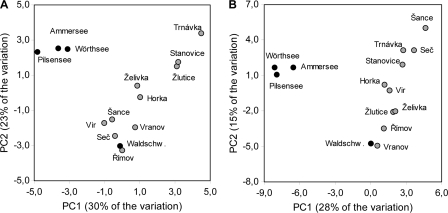
Within-host nucleotide polymorphism in different rRNA gene regions was observed for all cloned parasite isolates, which represented nine distinct taxa (Table 2). In most taxa the polymorphism was low (<2%). Differences between clonal variants were characterized mostly by single base pair substitutions, whereas nucleotide indels were rare and short (1 to 2 bp). The only exception was the SSU region of the microsporidian taxon MIC3, which contained 29-bp and 4-bp deletions in one sequence variant. The 14 cloned isolates of another microsporidian taxon, BERW, differed in the amount of polymorphism detected in the ITS region. For example, the isolate from Ammersee exhibited the greatest sequence divergence (5.27% polymorphic sites and 1.85% mean pairwise differences), whereas the isolate from Stanovice showed the least divergence (1.69% polymorphic sites and 0.60% mean pairwise differences) (Table 2). Moreover, the PCA based on cloned rRNA gene copies of all BERW isolates revealed a clear phylogeographic pattern, with some polymorphic sites shared by individuals from Ammersee, Wörthsee, and Pilsensee (Fig. 5A), which are lakes that are only ∼3 km apart. The fourth German lake, which did not cluster with the other lakes, Waldschwaig See, is located 28 to 34 km from the other lakes. A similar cluster including the same three German lakes was formed when polymorphic sites were inferred from double peaks in the chromatograms of directly sequenced SSU regions (Fig. 5B).
TABLE 2.
Summary of the extent of within-host parasite nucleotide polymorphism detected by cloning of the SSU and ITS rRNA gene regions of different parasites of Daphnia
| Sequence | Parasitea |
Study site | No. of clones | % of polymorphic sitesc | Mean pairwise difference (%) | |
|---|---|---|---|---|---|---|
| Group | Taxon | |||||
| ITS | Fungi | METS | Žlutice | 4 | 0.58 | 0.29 |
| Microsporidia | BERW | Ammersee | 6 | 5.27 | 1.85 | |
| BERW | Horka | 6 | 2.07 | 0.51 | ||
| BERW | Pilsensee | 6 | 3.01 | 1.15 | ||
| BERW | Římov | 6 | 2.64 | 1.18 | ||
| BERW | Šance | 6 | 1.69 | 0.67 | ||
| BERW | Seč | 6 | 3.77 | 1.44 | ||
| BERW | Stanovice | 6 | 1.69 | 0.60 | ||
| BERW | Trnávka | 6 | 2.64 | 1.11 | ||
| BERW | Vir | 6 | 1.88 | 0.65 | ||
| BERW | Vranov | 6 | 2.26 | 0.87 | ||
| BERW | Waldschwaig See | 6 | 2.26 | 0.70 | ||
| BERW | Wörthsee | 6 | 4.90 | 1.78 | ||
| BERW | Želivka | 6 | 2.45 | 0.70 | ||
| BERW | Žlutice | 6 | 2.64 | 0.85 | ||
| Oomycetes | APH1 | Šance | 6 | 1.16 | 0.30 | |
| APH2 | Vir | 5 | 1.29 | 0.47 | ||
| PHY1 | Brno | 3 | 0.12 | 0.08 | ||
| PHY2 | Vranov | 3 | 0.35 | 0.14 | ||
| SAP1 | Želivka | 3 | 0.41 | 0.27 | ||
| SSU | Microsporidia | BERW | Sečb | 4 | 1.37 | 0.70 |
| BERW | Stanovice | 2 | 0.77 | 0.77 | ||
| MIC2 | Seč | 6 | 3.05 | 0.79 | ||
| MIC3 | Seč | 6 | 1.51 | 0.43 | ||
| MIC3 | Sečb | 2 | 0.89 | 0.83 | ||
| Oomycetes | PHY2 | Brno | 3 | 0.17 | 0.11 | |
| PHY2 | Vranov | 3 | 0.17 | 0.11 | ||
| SAP1 | Želivka | 2 | 0.06 | 0.06 | ||
For an explanation of the abbreviations of parasite taxa see Table 1, footnote b.
Sequences were obtained from one Daphnia host infected with two different parasite taxa.
Including indels.
FIG. 5.
Distribution of 14 isolates of the microsporidian taxon BERW, plotted using two principal components. The plots were based on SNPs inferred from (A) variation among clonal variants of the ITS region and (B) double peaks in the chromatograms of directly sequenced SSU regions. Each symbol represents an individual Daphnia isolate from the lake indicated. Black circles, lakes around Munich, Germany; gray circles lakes in the Czech Republic.
Theoretically, Taq DNA polymerase errors and/or cloning artifacts may have contributed to the within-host parasite sequence variation observed in this study. Therefore, three BERW clones representing different ITS sequences were subjected to a second round of PCR. All PCR products were directly sequenced and cloned again. As determined by direct sequencing of the reamplified ITS regions, no single-nucleotide exchange was detected. Of the 14 clones examined, 13 were identical to the original sequences, whereas one clone contained a single SNP. This represents a rate of 1.3 × 10−4 error per site, which is 100-fold lower than the sequence polymorphism detected in this study.
DISCUSSION
Fourteen different parasite taxa infecting Daphnia were detected in this survey. Particularly high diversities of microsporidian and oomycete parasites were observed (four and nine taxa, respectively). Microsporidia have already received a lot of attention as parasites of Daphnia (36, 37) and other zooplankton species (28, 50). Despite attempts, researchers have failed repeatedly to maintain many microsporidia in laboratory cultures (49, 50). One proposed reason for the inability to culture these microsporidian parasites is the assumption that they have a complex life cycle requiring a second host (36). So far, the existence of a second host has been confirmed only for microsporidia infecting copepods, which are transmitted by mosquitoes (51). Nevertheless, our phylogenetic trees showed that the BERW and MIC1 microsporidian taxa described in this study cluster with microsporidia with a proposed complex life cycle (36, 37). On the other hand, the MIC3 taxon clusters with O. colligata, a microsporidium with direct, horizontal transmission from host to host (19), whereas the transmission route of the microsporidium DP-1-19, a relative of the MIC2 taxon, has not been tested so far. Interestingly, both O. colligata and DP-1-19 are gut parasites (36) that are reported to be practically invisible without dissection of the host (15), whereas the MIC2 and MIC3 taxa were found to appear as clear body cavity infections.
In contrast to the importance of microsporidia, the importance of oomycete parasites has been underestimated in Daphnia populations. It has been shown recently that some oomycete parasites are highly virulent to Daphnia (53). Our phylogenetic analyses demonstrate that the SAP1 and SAP3 taxa are closely related to these recently recognized virulent isolates (e.g., isolate WM3 [Fig. 3B]). Furthermore, based on our results we propose that oomycetes infecting Daphnia are generalist parasites. First, the SAP3 taxon was detected in different Daphnia species (D. longispina group and D. magna). Also, the oomycete taxa isolated from Daphnia and Bosmina (APH1 and APH2) or from Daphnia and Diaphanosoma (SAP1) had identical SSU sequences (Fig. 3A). In addition, there was 100% sequence identity for SAP2 (SSU region) and only one single-nucleotide difference for SAP3 (ITS region) with a known parasite of fish. This clearly shows that a single oomycete taxon is able to parasitize species at various trophic levels. So far, fish predation has usually been thought to dampen epidemics of zooplankton (13, 25). In contrast, our results suggest that the presence of fish might introduce and facilitate the spread of some oomycete infections in zooplankton communities, but this hypothesis needs to be tested further (10).
The only fungal parasite detected, METS, was morphologically identified as M. bicuspidata, which was first described by Metschnikoff (31) as a yeast-like organism infecting D. magna. Later, this parasite species was frequently reported in literature from the United States (5, 12) and Europe (43). Surprisingly, phylogenetic analyses of the METS taxon (including reference isolates from the United States) showed that this parasite is clearly distinct from several M. bicuspidata isolates deposited in GenBank (however, none of them was isolated from a Daphnia host). This unexpected finding highlights the fact that morphological identification of parasites may have low discriminatory power because genetically divergent taxa are pooled by phenotype. Although the METS taxon described in our study clusters with members of the Hemiascomycetes, it is rather distinct from any Metschnikowia or Candida species sequenced so far (Fig. 1A and B; see Fig. S1 in the supplemental material). The phylogenetic analyses showed that the closest relatives of the METS isolates were three different uncultured eukaryote clones, which were not collected from Daphnia habitats but were isolated from deep-sea environmental samples. Together with the fact that identical sequences were obtained from hosts belonging to the D. longispina group from Europe and North America, this finding supports the conclusion that the METS taxon is a generalist parasite that is able to infect different hosts occupying very different habitats.
Cloning of rRNA genes from several parasite taxa revealed various levels of within-host parasite sequence variation, especially in the ITS region. This indicates either that units of the rRNA gene multigene family comprise multiple genetic variants (i.e., intragenomic variation), which is common across various species (20, 24, 26), or that Daphnia spp. were infected by multiple parasite strains, or both. In our study, the level of within-host polymorphism was particularly high for the microsporidian taxon BERW (up to 5.3% polymorphic sites). A similar level was detected in another microsporidium, Nosema bombi, within a single spore or a single nucleus (34). On the other hand, a low level of polymorphism was observed across various oomycete taxa in our study, suggesting that genetic diversity within populations of different parasite taxa is taxon specific and spans a wide range. While a less variable genetic makeup may be successful in cases of generalist parasites (allowing them to switch between different hosts), high genetic variability of specialist parasites may be essential for successful arms races (23) in very specific host-parasite relationships (6).
Regardless of the cause of the within-host parasite rRNA variation, the observed pattern may reflect the population structure (for example, the extent of gene flow). Thus, individuals originating from lakes that were close to each other had more similar rRNA gene copies than isolates from localities that were more distant (≥28 km apart), and they clustered together in the PCA plots (Fig. 5A and B). This is in agreement with a recent study exploring variable-number-of-tandem-repeat markers in the bacterium Pasteuria ramosa, another parasite of Daphnia, which revealed strong parasite clustering by location (32). The genetic similarities of parasite isolates from populations that were close to each other detected in the present study indicate that either Daphnia populations of the lakes were originally colonized by genetically similar parasite pools or there is ongoing parasite dispersal among the populations. In principle, the microsporidium BERW may disperse via its Daphnia host, transported either by water currents or by avian vectors (9). However, if the BERW taxon requires passage through a secondary host (such as insects [36, 51]), these vectors could also transmit the infection to nearby Daphnia populations.
Overall, this study shows that the parasite diversity in lake Daphnia populations is high, despite the presence of selective predators. As parasites play a fundamental role in natural ecosystems because they regulate host population density, influence diversity, and stabilize food webs (48) but species differ substantially in the strength of the selection pressure that they can exert, it is important to correctly determine their identities. Our results highlight the significance of using molecular markers to assess parasite identity in natural populations. Many Daphnia infections, although morphologically indistinguishable, are actually caused by different parasite taxa that often coexist in the same habitat. Moreover, we found that identical oomycete parasites were shared by various zooplankton species, as well as by zooplankton and fish. This suggests that oomycetes, although previously ignored, may play a complex role in the dynamics of aquatic ecosystems, and this possibility certainly deserves further investigation.
Supplementary Material
Acknowledgments
We thank Adam Petrusek and Jaromir Seda for providing zooplankton samples from their field study of 12 Czech reservoirs and Spencer Hall for providing reference samples from the United States. We especially thank Rita Jaenichen for doing a large part of the molecular work. Dominik Refardt, Adam Petrusek, Tommy Leung, and an anonymous reviewer provided valuable comments which helped us improve the manuscript. We thank Tommy Leung for linguistic help.
This work was supported in part by DFG grant WO 1587/2-1 and by department research money provided by Wilfried Gabriel.
Footnotes
Published ahead of print on 18 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anderson, R. M., and R. M. May. 1978. Regulation and stability of host-parasite population interactions. 1. Regulatory processes. J. Anim. Ecol. 47:219-247. [Google Scholar]
- 2.Arneberg, P., A. Skorping, B. Grenfell, and A. F. Read. 1998. Host densities as determinants of abundance in parasite communities. Proc. R. Soc. Ser. B 265:1283-1289. [Google Scholar]
- 3.Ben-Ami, F., L. Mouton, and D. Ebert. 2008. The effects of multiple infections on the expression and evolution of virulence in a Daphnia-endoparasite system. Evolution 62:1700-1711. [DOI] [PubMed] [Google Scholar]
- 4.Bengtsson, J., and D. Ebert. 1998. Distributions and impacts of microparasites on Daphnia in a rockpool metapopulation. Oecologia 115:213-221. [DOI] [PubMed] [Google Scholar]
- 5.Caceres, C. E., S. R. Hall, M. A. Duffy, A. J. Tessier, C. Helmle, and S. MacIntyre. 2006. Physical structure of lakes constrains epidemics in Daphnia populations. Ecology 87:1438-1444. [DOI] [PubMed] [Google Scholar]
- 6.Carius, H. J., T. J. Little, and D. Ebert. 2001. Genetic variation in a host-parasite association: potential for coevolution and frequency-dependent selection. Evolution 55:1136-1145. [DOI] [PubMed] [Google Scholar]
- 7.Decaestecker, E., S. Gaba, J. A. M. Raeymaekers, R. Stoks, L. Van Kerckhoven, D. Ebert, and L. De Meester. 2007. Host-parasite “Red Queen” dynamics archived in pond sediment. Nature 400:870-873. [DOI] [PubMed] [Google Scholar]
- 8.Decaestecker, E., A. Vergote, D. Ebert, and L. De Meester. 2003. Evidence for strong host clone-parasite species interactions in the Daphnia microparasite system. Evolution 57:784-792. [DOI] [PubMed] [Google Scholar]
- 9.De Meester, L., A. Gomez, B. Okamura, and K. Schwenk. 2002. The monopolization hypothesis and the dispersal-gene flow paradox in aquatic organisms. Acta Oecol. 23:121-135. [Google Scholar]
- 10.Duffy, M. 2009. Staying alive: the post-consumption fate of parasite spores and its implications for disease dynamics. Limnol. Oceanogr. 54:770-773. [Google Scholar]
- 11.Duffy, M. A. 2007. Selective predation, parasitism, and trophic cascades in a bluegill-Daphnia-parasite system. Oecologia 153:453-460. [DOI] [PubMed] [Google Scholar]
- 12.Duffy, M. A., C. E. Brassil, S. R. Hall, A. J. Tessier, C. E. Caceres, and J. K. Conner. 2008. Parasite-mediated disruptive selection in a natural Daphnia population. BMC Evol. Biol. 8:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duffy, M. A., S. R. Hall, A. J. Tessier, and M. Huebner. 2005. Selective predators and their parasitized prey: are epidemics in zooplankton under top-down control? Limnol. Oceanogr. 50:412-420. [Google Scholar]
- 14.Duncan, A., and T. J. Little. 2007. Parasite-driven genetic change in a natural population of Daphnia. Evolution 64:796-803. [DOI] [PubMed] [Google Scholar]
- 15.Ebert, D. 2005. Ecology, epidemiology, and evolution of parasitism in Daphnia. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD.
- 16.Ebert, D., J. W. Hottinger, and V. I. Pajunen. 2001. Temporal and spatial dynamics of parasite richness in a Daphnia metapopulation. Ecology 82:3417-3434. [Google Scholar]
- 17.Ebert, D., M. Lipsitch, and K. L. Mangin. 2000. The effect of parasites on host population density and extinction: experimental epidemiology with Daphnia and six microparasites. Am. Nat. 156:459-477. [DOI] [PubMed] [Google Scholar]
- 18.Ebert, D., R. Payne, and W. Weisser. 1997. The epidemiology of parasitic diseases in Daphnia, p. 91-111. In K. Dettner, G. Bauer, and W. Völkl (ed.), Vertical food web interactions evolutionary patterns and driving forces. Ecological studies, vol. 130. Springer, Berlin, Germany. [Google Scholar]
- 19.Ebert, D., C. D. Zschokke Rohringer, and H. J. Carius. 2000. Dose effects and density-dependent regulation of two microparasites of Daphnia magna. Oecologia 122:200-209. [DOI] [PubMed] [Google Scholar]
- 20.Gribble, K. E., and D. M. Anderson. 2007. High intraindividual, intraspecific, and interspecific variability in large-subunit ribosomal DNA in the heterotrophic dinoflagellates Protoperidinium, Diplopsalis, and Preperidinium (Dinophyceae). Phycologia 46:315-324. [Google Scholar]
- 21.Hall, S. R., A. J. Tessier, M. A. Duffy, M. Huebner, and C. E. Caceres. 2006. Warmer does not have to mean sicker: temperature and predators can jointly drive timing of epidemics. Ecology 87:1684-1695. [DOI] [PubMed] [Google Scholar]
- 22.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 23.Hamilton, W. D. 1980. Sex versus non-sex versus parasite. Oikos 35:282-290. [Google Scholar]
- 24.Hartmann, S., J. D. Nason, and D. Bhattacharya. 2001. Extensive ribosomal DNA genic variation in the columnar cactus Lophocereus. J. Mol. Evol. 53:124-134. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, P. T. J., D. E. Stanton, E. R. Preu, K. J. Forshay, and S. R. Carpenter. 2006. Dining on disease: how interactions between infection and environment affect predation risk. Ecology 87:1973-1980. [DOI] [PubMed] [Google Scholar]
- 26.Keller, I., P. Veltsos, and R. A. Nichols. 2008. The frequency of rDNA variants within individuals provides evidence of population history and gene flow across a grasshopper hybrid zone. Evolution 62:833-844. [DOI] [PubMed] [Google Scholar]
- 27.Lampert, W., and U. Sommer. 1999. Limnoökologie, 2nd ed. Georg Thieme Verlag, Suttgart, Germany.
- 28.Larsson, R. 1981. A new microsporidium Berwaldia singularis gen. et sp. nov. from Daphnia pulex and a survey of microsporidia described from Cladocera. Parasitology 83:325-342. [Google Scholar]
- 29.Little, T. J., and D. Ebert. 1999. Associations between parasitism and host genotype in natural populations of Daphnia (Crustacea: Cladocera). J. Anim. Ecol. 68:134-149. [Google Scholar]
- 30.Medlin, L., H. J. Elwood, S. Stickel, and M. L. Sogin. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA coding regions. Gene 71:491-499. [DOI] [PubMed] [Google Scholar]
- 31.Metschnikoff, E. 1884. A disease of Daphnia caused by a yeast. A contribution to the theory of phagocytes as agents for attack on disease-causing organisms. Archiv. Pathol. Anat. Physiol. Klin. Med. 96:177-195. (In German.) [Google Scholar]
- 32.Mouton, L., and D. Ebert. 2008. Variable-number-of-tandem-repeats analysis of genetic diversity in Pasteuria ramosa. Curr. Microbiol. 56:447-452. [DOI] [PubMed] [Google Scholar]
- 33.Mouton, L., G. Nong, J. F. Preston, and D. Ebert. 2007. Variable-number tandem repeats as molecular markers for biotypes of Pasteuria ramosa in Daphnia spp. Appl. Environ. Microbiol. 73:3715-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Mahony, E. M., W. T. Tay, and R. J. Paxton. 2007. Multiple rRNA variants in a single spore of the microsporidian Nosema bombi. J. Eukaryot. Microbiol. 54:103-109. [DOI] [PubMed] [Google Scholar]
- 35.Petrusek, A., J. Seda, J. Machacek, S. Ruthova, and P. Smilauer. 2008. Daphnia hybridization along ecological gradients in pelagic environments: the potential for the presence of hybrid zones in plankton. Philos. Trans. R. Soc. Lond. B 363:2931-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Refardt, D., E. U. Canning, A. Mathis, S. A. Cheney, N. J. Lafranchi-Tristem, and D. Ebert. 2002. Small subunit ribosomal DNA phylogeny of microsporidia that infect Daphnia (Crustacea: Cladocera). Parasitology 124:381-389. [DOI] [PubMed] [Google Scholar]
- 37.Refardt, D., E. Decaestecker, P. T. J. Johnson, and J. Vavra. 2008. Morphology, molecular phylogeny, and ecology of Binucleata daphniae n. g., n. sp. (fungi: microsporidia), a parasite of Daphnia magna Straus, 1820 (Crustacea: Branchiopoda). J. Eukaryot. Microbiol. 55:393-408. [DOI] [PubMed] [Google Scholar]
- 38.Refardt, D., and D. Ebert. 2006. Quantitative PCR to detect, discriminate and quantify intracellular parasites in their host: an example from three microsporidians in Daphnia. Parasitology 133:11-18. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigues, J. L. M., M. A. Duffy, A. J. Tessier, D. Ebert, L. Mouton, and T. M. Schmidt. 2008. Phylogenetic characterization and prevalence of “Spirobacillus cienkowskii,” a red-pigmented, spiral-shaped bacterial pathogen of freshwater Daphnia species. Appl. Environ. Microbiol. 74:1575-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlotterer, C., M. T. Hauser, A. Vonhaeseler, and D. Tautz. 1994. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol. Biol. Evol. 11:513-522. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, L. M., L. Mouton, G. Nong, D. Ebert, and J. F. Preston. 2008. Genetic and immunological comparison of the cladoceran parasite Pasteuria ramosa with the nematode parasite Pasteuria penetrans. Appl. Environ. Microbiol. 74:259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seda, J., A. Petrusek, J. Machacek, and P. Smilauer. 2007. Spatial distribution of the Daphnia longispina species complex and other planktonic crustaceans in the heterogeneous environment of canyon-shaped reservoirs. J. Plankton Res. 29:619-628. [Google Scholar]
- 43.Stirnadel, H. A., and D. Ebert. 1997. Prevalence, host specificity and impact on host fecundity of microparasites and epibionts in three sympatric Daphnia species. J. Anim. Ecol. 66:212-222. [Google Scholar]
- 44.Swofford, D. L. 2001. PAUP: phylogenetic analysis using parsimony (and other methods), 4.0b10 ed. Sinauer Associates, Sunderland, MA.
- 45.Takemoto, R. M., G. C. Pavanelli, M. A. P. Lizama, J. L. Luque, and R. Poulin. 2005. Host population density as the major determinant of endoparasite species richness in floodplain fishes of the upper Parana River, Brazil. J. Helminthol. 79:75-84. [DOI] [PubMed] [Google Scholar]
- 46.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 47.Tews, J., U. Brose, V. Grimm, K. Tielborger, M. C. Wichmann, M. Schwager, and F. Jeltsch. 2004. Animal species diversity driven by habitat heterogeneity/diversity: the importance of keystone structures. J. Biogeogr. 31:79-92. [Google Scholar]
- 48.Thieltges, D. W., B. L. Fredensborg, and R. Poulin. 2009. Geographical variation in metacercarial infection levels in marine invertebrate hosts: parasite species character versus local factors. Mar. Biol. 156:983-990. [Google Scholar]
- 49.Vavra, J. 1964. A failure to produce artificial infection in cladoceran microsporidia. J. Protozool. 11:35. [Google Scholar]
- 50.Vavra, J., M. Hylis, M. Obornik, and C. R. Vossbrinck. 2005. Microsporidia in aquatic microcrustacea: the copepod microsporidium Marssoniella elegans Lemmermann, 1900 revisited. Folia Parasitol. 52:163-172. [PubMed] [Google Scholar]
- 51.Vossbrinck, C. R., T. G. Andreadis, J. Vavra, and J. J. Becnel. 2004. Molecular phylogeny and evolution of mosquito parasitic microsporidia (Microsporidia: Amblyosporidae). J. Eukaryot. Microbiol. 51:259-261. [DOI] [PubMed] [Google Scholar]
- 52.Wolinska, J., B. Keller, M. Manca, and P. Spaak. 2007. Parasite survey of a Daphnia hybrid complex: host-specificity and environment determine infection. J. Anim. Ecol. 76:191-200. [DOI] [PubMed] [Google Scholar]
- 53.Wolinska, J., K. C. King, F. Vigneux, and C. M. Lively. 2008. Virulence, cultivating conditions, and phylogenetic analyses of oomycete parasites in Daphnia. Parasitology 135:1667-1678. [DOI] [PubMed] [Google Scholar]
- 54.Wolinska, J., and P. Spaak. 2009. The cost of being common: evidence from natural Daphnia populations. Evolution 63:1893-1901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.