Abstract
Subacute ruminal acidosis (SARA) is a metabolic disease in dairy cattle that occurs during early and mid-lactation and has traditionally been characterized by low rumen pH, but lactic acid does not accumulate as in acute lactic acid acidosis. It is hypothesized that factors such as increased gut permeability, bacterial lipopolysaccharides, and inflammatory responses may have a role in the etiology of SARA. However, little is known about the nature of the rumen microbiome during SARA. In this study, we analyzed the microbiome of 64 rumen samples taken from eight lactating Holstein dairy cattle using terminal restriction fragment length polymorphisms (TRFLP) of 16S rRNA genes and real-time PCR. We used rumen samples from two published experiments in which SARA had been induced with either grain or alfalfa pellets. The results of TRFLP analysis indicated that the most predominant shift during SARA was a decline in gram-negative Bacteroidetes organisms. However, the proportion of Bacteroidetes organisms was greater in alfalfa pellet-induced SARA than in mild or severe grain-induced SARA (35.4% versus 26.0% and 16.6%, respectively). This shift was also evident from the real-time PCR data for Prevotella albensis, Prevotella brevis, and Prevotella ruminicola, which are members of the Bacteroidetes. The real-time PCR data also indicated that severe grain-induced SARA was dominated by Streptococcus bovis and Escherichia coli, whereas mild grain-induced SARA was dominated by Megasphaera elsdenii and alfalfa pellet-induced SARA was dominated by P. albensis. Using discriminant analysis, the severity of SARA and degree of inflammation were highly correlated with the abundance of E. coli and not with lipopolysaccharide in the rumen. We thus suspect that E. coli may be a contributing factor in disease onset.
The bovine rumen is a classical host-microbe symbiotic system, and disturbances in this exquisitely balanced ecosystem may lead to disease in the host. An example is subacute ruminal acidosis (SARA), or non-lactic acid acidosis, which has a disease etiology distinct from that of acute lactic acid acidosis because there is no accumulation of lactic acid (35). Field studies in the United States estimated that 19% of early lactating cows and 26% of mid-lactation cows suffered from SARA (11). In Germany and The Netherlands, approximately 11% of early lactation and 18% of mid-lactation cows suffered from this disease (22). In the acute form, lactic acid accumulates in the rumen, causing metabolic acidosis, and it usually occurs when animals are abruptly transitioned to a high-grain diet from a predominantly forage diet (38). If, however, the adaptation is gradual, slower-growing lactic acid-consuming bacteria, like Megasphaera elsdenii, convert the lactic acid to propionic acid (29). In SARA, lactic acid does not accumulate during low-pH conditions and other factors, like microbial population shifts and immune responses, appear to be associated with the disease etiology (35).
In both acute and subacute acidosis, there is an increase in lipopolysaccharide (LPS) concentrations in the rumen (8, 14, 16). LPS and/or the low-pH rumen conditions may increase the permeability of the gut to LPS, which could trigger systemic inflammation (4). We previously developed two animal models of SARA, one based on grain and one based on alfalfa pellets (20, 21). Even though both models resulted in substantial reductions in rumen pH and an accumulation of LPS, only the grain induction model resulted in inflammation and the appearance of LPS in the peripheral blood (20, 21).
In contrast to the rumen microbiome during lactic acid acidosis, the rumen microbiome during SARA has not been evaluated (13, 28). Even in acute acidosis, studies are largely culture based, and the uncultured members of the community have not been extensively assessed (31, 46, 49). In this article, we describe the rumen microbiome when two SARA induction models were used. The shifts in microbial community structure were assessed using terminal restriction fragment length polymorphism (TRFLP) analysis and real-time PCR of key microbial populations.
MATERIALS AND METHODS
Animal experiments were conducted at the Glenlea Dairy Research Unit at the University of Manitoba (Winnipeg, MB, Canada) in accordance with the guidelines of the Canadian Council on Animal Care (3). Data for rumen pH, feed intake, milk production and composition, and rumen and blood metabolites for these experiments have been reported previously (20, 21). In this article, we reinterpret the previous data (20, 21) in the context of the microbial changes evaluated in this research.
Animal models and sampling.
As described previously (20, 21), SARA was induced in eight rumen-cannulated lactating dairy cows using two different induction models (total of eight animals from both experiments), one with grain, and the other with alfalfa pellets. In both experiments, animals were fed once daily at 9:00 a.m. and feed was always provided in an amount that met or exceeded the animal's nutritional requirements. In the first experiment (grain-induced SARA), cows received a basal diet with a forage-to-concentrate ratio of 50:50. The ration consisted of 25% as alfalfa silage, 25% as barley silage, 40% as an energy supplement, and 10% as a protein supplement. SARA was induced by replacing 21% of the dietary dry matter (DM) with wheat-barley pellets. In the second experiment (alfalfa pellet-induced SARA), cows received a basal diet that consisted of 50% of the DM as concentrate and 50% as chopped alfalfa hay. The concentrate fraction consisted of 39% as an energy supplement, 4.5% as a protein supplement, and 6.5% as roasted soybeans. SARA was induced by replacing 42% of the alfalfa hay DM with alfalfa pellets without changing the forage-to-concentrate ratio or the dietary starch content.
In both experiments, the severity of SARA in individual animals was determined based on objective criteria that included the duration of rumen pH below 5.6, free rumen LPS, and serum haptoglobin as an inflammatory marker (14). Rumen pH was monitored continuously using indwelling pH probes (21). Rumen fluids were collected from the ventral sac of the rumen 15 min before feeding (0 h) and at 6 h after feeding both during the control period and 4 days after induction of SARA during the SARA periods. Ruminal contents were strained through four layers of sterile cheesecloth, transferred to 50-ml sterile tubes, immediately frozen in liquid nitrogen, and stored at −20°C. The free rumen LPS content was determined with a chromogenic Limulus amebocyte lysate end-point assay (QCL-1000; Lonza group Ltd., Basel, Switzerland) (21). The serum concentration of haptoglobin was determined using an enzyme-linked immunosorbent assay kit (TP-801; Tri-Delta Diagnostics, Inc., Morris Plains, NJ) (21).
DNA extraction.
Rumen fluid samples (n = 64) were thawed at 32°C for 15 min and immediately centrifuged at 10,000 × g for 10 min. Supernatants were discarded, and pellets were resuspended in phosphate-buffered saline (1×) in new sterile tubes. Approximately 150 mg of microbial pellet was washed in 1 ml of phosphate-buffered saline (1×) and centrifuged at 10,000 × g for 2 min. The washing step was repeated twice. DNA was extracted from the pellets using a ZR fecal DNA kit (D6010; Zymo Research Corp., Orange, CA) which included a bead-beating step for the mechanical lysis of the microbial cells. DNA concentration and purity were determined spectrophotometrically by measuring the A260/280 (Beckman DU/800; Beckman Coulter, Inc., Fullerton, CA). DNA quality was also evaluated by gel electrophoresis.
PCR amplification and TRFLP.
The microbial composition in the rumen fluid was assessed using TRFLP as described by Sepehri et al. (41), with some modifications. In brief, the V1 and V2 regions of the 16S rRNA genes were PCR amplified using universal bacterial primers 27f (5′-GAAGAGTTTGATCATGGCTCAG-3′) and 342r (5′-CTGCTGCCTCCCGTAG-3′) (21). The forward primer was fluorescently labeled (WellRED D4dye; Sigma-Proligo, St. Louis, MO) to allow detection of the fragments by capillary electrophoresis. The PCRs were as follows: 37 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min, and a final extension at 72°C for 5 min. The first cycle used a denaturation step of 5 min instead of 1 min. To produce terminal restriction fragments (T-RFs), the PCR products were digested with HhaI (15 μl of PCR product, 10 units of HhaI, 1× HhaI buffer, and 20 μg of bovine serum; New England Biolabs, Ipswich, MA) at 37°C for 3 h. The precise lengths of the T-RF amplicons were determined on a CEQ 8800 genetic analysis system (Beckman Coulter, Inc., Fullerton, CA). A 2-μl volume of fluorescently labeled fragments, 29.5 μl of sample loading solution, and 1 μl of 600-bp DNA size standard (Beckman Coulter, Inc., Fullerton, CA) was mixed and applied to the capillaries. An electropherogram with peaks of different sizes was obtained for each rumen fluid sample, and each peak of unique size represents an operational taxonomic unit (OTU). All rumen fluid samples were run in duplicate, and results were accepted when the variability in the sizes of the T-RFs in the duplicate electropherograms were in the range of ±2 bp.
Fragment analysis.
CEQ software (version 9.0; Beckman Coulter, Inc., Fullerton, CA) with a binning parameter of 2 bp was used to analyze the fragment data. Signals were detected from electronic noise using a proportional threshold approach (40). In brief, the relative abundance of each T-RF within a profile was calculated as its peak height divided by the total peak height of all T-RFs in that profile. Only T-RFs with relative abundances higher than 1% of the total were included in the analysis. The incidence (presence/absence) data derived from the OTU profiles were grouped based on sampling times (0 h and 6 h) and dietary treatments and used for numerical analysis.
Richness, diversity, and similarity.
Chao2 incidence-based index of richness and Shannon incidence-based diversity estimators were calculated using EstimateS (version 7.5; http://purl.oclc.org/estimates) to determine the richness and diversity of microbial communities in each time/treatment group. An upper abundance limit of 5 was used to determine rare or infrequent species. The order of the samples was randomized 500 times for each run to reduce the effect of sample order. Similarities among microbial communities were also determined using SPADE (version 2.1; http://chao.stat.nthu.edu.tw/softwareCE.html), based on the Jaccard multiple incidence-based index.
Bioinformatic analysis of TRFLP data.
T-RFs alone do not allow for unequivocal identification of OTU to the genus or species level, and for high-resolution analysis, sequencing reactions need to be done. To partially circumvent this problem, we have previously described a bioinformatic approach in which we mined the Ribosomal Database Project (RDP-II) (6) for all sequences that were found in the digestive tract (1, 41). Essentially, a database was developed that only contained sequences that occur in the mammalian gut (42). This approach vastly improved data analysis because one does not have to account for taxa not found in the gut. We frequently update this database and it currently consists of over 200,000 curated near-full-length sequences from the RDP that only come from the mammalian and avian gut.
As described by Sepehri et al. (41), an in silico reference database based on 27f and 342r plus HhaI restriction digestion was created and submitted to the Phylogenetic Assignment Tool (PAT) (19) and compared to the experimentally produced data from capillary electrophoresis. The resultant libraries were then entered into the hierarchical browser of the RDP and converted to GenBank format. T-RFs with multiple accession numbers were assigned to taxonomic rank according to phylum, class, order, and family. Based on this analysis, reported values were expressed as the proportion of phylogenetic lineage for each library.
Primers and real-time PCR.
The PCR primers used are listed in Table 1. Primers were assembled from the literature or newly designed (Primer Express, version 3.0; Applied Biosystems, Foster City, CA) and tested for specificity in silico. Those primers that did not meet our selection criteria for specificity and performance were redesigned from sequence alignments. The oligonucleotides were synthesized by University Core DNA Services (University of Calgary, Calgary, AB, Canada).
TABLE 1.
Primers used for real-time PCR quantification
| Target organism(s) | Primer | Sequence (5′ → 3′)a | Tm (°C) | G+C (%) | Amplicon size (bp) | Source of primer |
|---|---|---|---|---|---|---|
| Eubacteria | 341-357F | CCTACGGGAGGCAGCAG | 55.2 | 70.6 | 189 | 25 |
| 518-534R | ATTACCGCGGCTGCTGG | 56.2 | 64.7 | |||
| Prevotella albensis | ProAlb4F | GCGCCACTGACGCTGAAG | 58.3 | 66.7 | 110 | This study |
| ProAlb4R | CCCCAAATCCAAAAGGACTCAG | 56.6 | 50.0 | |||
| Prevotella brevis | PreBre2F | GCGAACTGGTTTCCTTGAGTGTATT | 58.8 | 50.0 | 153 | This study |
| PreBre2R | ACCTTCGAGCTTTAGCGTCAGTTAT | 57.6 | 40.0 | |||
| Prevotella bryantii | ProBry4F | GAAGGCAGCTCGCTGTAGTGTT | 60.6 | 54.5 | 145 | This study |
| ProBry4R | CTTAACGCTTTCGCTTAGCCACT | 59.4 | 47.8 | |||
| Prevotella ruminicola | PreRum92862F | GCGAAAGTCGGATTAATGCTCTATG | 58.5 | 44.0 | 78 | This study |
| PreRum92862R | CCCATCCTATAGCGGTAAACCTTTG | 59.3 | 48.0 | |||
| Succinimonas amylolytica | SucAmy2F | CGTTGGGCGGTCATTTGAAAC | 55.2 | 52.4 | 139 | This study |
| SucAmy2R | CCTGAGCGTCAGTTACTATCCAGA | 56.2 | 50.0 | |||
| Succinivibrio dextrinisolvens | SucDex1F | TAGGAGCTTGTGCGATAGTATGG | 57.4 | 47.8 | 174 | This study |
| SucDex1R | CTCACTATGTCAAGGTCAGGTAAGG | 58.4 | 48.0 | |||
| Ruminobacter amylophilus | RumAmy2F | CTGGGGAGCTGCCTGAAT | 55.3 | 61.1 | 100 | 43 |
| RumAmy2R | CATCTGAATGCGACTGGTTG | 54.2 | 50.0 | |||
| Escherichia coli | EcoliFimH2F | GCCGGTGGCGCTTTATTTG | 57.3 | 57.9 | 114 | This study |
| EcoliFimH2R | TCATCGCTGTTATAGTTGTTGGTCT | 58.4 | 40.0 | |||
| Fibrobacter succinogenes | FibSuc4F | GGAGCGTAGGCGGAGATTCA | 58.7 | 60.0 | 97 | This study |
| FibSuc4R | GCCTGCCCCTGAACTATCCA | 58.5 | 60.0 | |||
| Anaerovibrio lipolytica | AnaLip2F | TGGGTGTTAGAAATGGATTCTAGTG | 56.6 | 40.0 | 109 | This study |
| AnaLip2R | GCACGTCATTCGGTATTAGCAT | 56.7 | 45.5 | |||
| Megasphaera elsdenii | MegEls1F | GACCGAAACTGCGATGCTAGA | 57.7 | 52.4 | 129 | 32 |
| MegEls1R | CGCCTCAGCGTCAGTTGTC | 58.2 | 63.2 | |||
| Selenomonas ruminantium | SelRum1F | GGCGGGAAGGCAAGTCAGTC | 60.4 | 65.0 | 83 | This study |
| SelRum1R | CCTCTCCTGCACTCAAGAAAGACAG | 61.1 | 52.0 | |||
| Ruminococcus albus | RumAlb1F | CCCTAAAAGCAGTCTTAGTTCG | 54.3 | 45.5 | 176 | 48 |
| RumAlb1R | CCTCCTTGCGGTTAGAACA | 53.8 | 52.6 | |||
| Ruminococcus flavefaciens | RumFla1F | CGAACGGAGATAATTTGAGTTTACTTAGG | 57.5 | 34.5 | 132 | 7 |
| RumFla1R | CGGTCTCTGTATGTTATGAGGTATTACC | 59.3 | 42.9 | |||
| Butyrivibrio fibrisolvens | ButFib2F | ACCGCATAAGCGCACGGA | 58.8 | 61.1 | 65 | 43 |
| ButFib2R | CGGGTCCATCTTGTACCGATAAAT | |||||
| Lactobacillus spp. | Ulac16S1F | AGCAGTAGGGAATCTTCCA | 51.5 | 47.4 | 345 | 24, 47 |
| Ulac16S1R | ATTCCACCGCTACACATG | 51.1 | 50.0 | |||
| Streptococcus bovis | SBovis1F | TTCCTAGAGATAGGAAGTTTCTTCGG | 57.9 | 42.3 | 127 | 43 |
| SBovis1R | ATGATGGCAACTAACAATAGGGGT | 57.9 | 41.7 | |||
| Treponema bryantii | TrpBry1F | GAGAAACGCTTTGTGGTGACTGT | 59.5 | 47.8 | 122 | This study |
| TrpBry1R | CCTACATGCCCTTTACGCTCAAT | 58.7 | 47.8 | |||
| Methanogenic archaea | MB1174f | GAGGAAGGAGTGGACGACGGTA | 60.6 | 59.1 | 232 | 30 |
| Arch1406-1389r | ACGGGCGGTGTGTGCAAG | 60.0 | 66.7 | |||
| Ciliate protozoa | UPorCil1F | GCTTTCGWTGGTAGTGTATT | 50.2 | 20.0 | 234 | 45 |
| UPorCil1R | CTTGCCCTCYAATCGTWCT | 50.4 | 47.4 |
Primers were based on 18S rRNA genes for ciliate protozoa, 16S rRNA genes for bacterial groups except for E. coli, and the fimH gene for E. coli.
Real-time PCR was carried out using an AB 7300 system (Applied Biosystems, Foster City, CA) and sequence detection software (version 1.3; Applied Biosystems, Foster City, CA). Each reaction mixture was run in triplicate in a volume of 25 μl in optical reaction plates (Applied Biosystems, Foster City, CA) sealed with optical adhesive film (Applied Biosystems, Foster City, CA). Amplification reactions were carried out with Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA) mixed with the selected primer set (Table 1) at a concentration of 0.5 μM for each primer and 2 μl (∼12 ng) of genomic DNA. Different concentrations of forward and reverse primers were only used for Prevotella brevis (0.5 and 0.9 μM, respectively) and Ruminobacter amylophilus (0.3 μM each). Amplification consisted of one cycle of 95°C (10 min), 40 cycles of denaturation at 95°C (15 s), and annealing/extension at 60°C (1 min). The only exceptions were for methanogenic archaea and ciliate protozoa, where annealing/extension steps of 63°C (30 s)/72°C (30 s) and 54°C (30 s)/72°C (1 min) were applied, respectively. Final melting analysis was obtained by slow heating from 65°C to 95°C. To evaluate the efficiency (E) of the amplification of each primer set, DNA templates were pooled (50 ng/reaction mixture) and serially diluted eightfold. Amplification efficiency was calculated from the slope of the standard curve generated by plotting the threshold cycle (CT) versus logarithmic values of different DNA concentrations using the following equation (7): E = 10−1/slope. Relative quantitation was accomplished using the following mathematical model (34): Ri = [(Etarget)ΔCTtarget (controli − SARAi)]/[(Eref)ΔCTref (controli − SARAi)], where target is the specific microbial group of interest, ref is the 16S rRNA gene of the Eubacteria, ΔCT is the CT deviation of SARA versus control period, i is sampling time (0 h or 6 h after feeding), and Ri is the relative expression ratio of a target gene compared to a reference gene at a specific time point.
Statistical analysis.
Discriminant multivariate analysis was conducted using JMP IN (version 5.1; SAS Institute, Inc., Cary, NC) to examine possible relationships among models of SARA induction and duration of rumen pH below 5.6, free rumen LPS, rumen microbial community dynamics, and serum haptoglobin as an inflammatory marker. Statistical significance (P < 0.05) was calculated using the least significant difference multiple comparison test to detect significant differences among times/treatments groups (39).
RESULTS
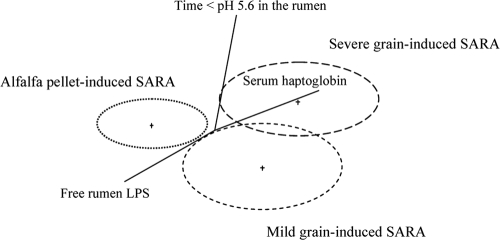
Significant variations in the animals' responses to SARA induction were observed with the grain-induced versus alfalfa-induced model (Tables 2 and 3) (20, 21). Multivariate discriminant analysis (Fig. 1) of time below pH 5.6, serum haptoglobin, and the concentration of free rumen LPS divided cows into three groups: severe grain-induced SARA, mild grain-induced SARA, and alfalfa pellet-induced SARA. The major differentiator between groups was the haptoglobin inflammatory marker.
TABLE 2.
Time spent below pH 5.6, rumen LPS, plasma LPS, and serum haptoglobin level of dairy cows during control period and grain-induced SARA
| Period | Mean valuea for indicated variable and disease severity |
|||||
|---|---|---|---|---|---|---|
| Time below pH 5.6 (min/day) |
Rumen LPS (EU/ml) |
Serum haptoglobin (μg/ml) |
||||
| Mild | Severe | Mild | Severe | Mild | Severe | |
| Control | 75 b | 161 b | 32,413 Bb | 29,933 b | 0.0 c | 0.0 c |
| Grain-induced SARA | 217 a | 337 a | 100,175 Ab | 179,762 a | 343.3 b | 608.0 a |
EU, endotoxin unit. a, b, and c, within each variable, mean values in the same row or column with different symbols differ (P < 0.05). A and B, within each variable, mean values in the same row or column with different symbols differ (P < 0.1). Standard errors of the means are 72 for time below pH 5.6, 30,072 for rumen LPS, and 35.8 for serum haptoglobin.
TABLE 3.
Time spent below pH 5.6, rumen LPS, plasma LPS, and serum haptoglobin of dairy cows during control period and alfalfa pellet-induced SARA
| Period | Mean resulta for indicated variable |
||
|---|---|---|---|
| Time below pH 5.6 (min/day) | Rumen LPS (EU/ml) | Serum haptoglobin (μg/ml) | |
| Control | 112 b | 42,122 b | 56 a |
| Alfalfa pellet-induced SARA | 510 a | 169,266 a | 21 b |
EU, endotoxin unit. a and b, mean values within the same column with different symbols differ (P < 0.05). Standard errors of the means are 109.5 for time below pH 5.6, 16,780 for rumen LPS, and 6 for serum haptoglobin.
FIG. 1.
Results of discriminant analysis of the duration of time below pH 5.6 in the rumen, free rumen LPS, and serum haptoglobin in response to SARA induction model. The circles are independent variables, and the distances between them reflect their dissimilarity. The straight lines are dependent variables, and their lengths and the angles between them are functions of relative effects of independent variables.
The feeding method (grain or alfalfa pellet) and the amplitudes of inflammatory markers were used as criteria to group TRFLP data (Tables 4 and 5). In other words, we used the SARA induction method and the inflammatory markers to decide how to group animals. We conducted TRFLP analysis with primers 27f and 342r or 1100r, but the larger amplicon appeared to be less efficiently produced and resulted in fewer T-RFs. Nine predominant phyla were detected: Bacteroidetes, Proteobacteria, Firmicutes, Spirochaetes, Actinobacteria, Fusobacteria, TM7, Tenericutes, and Deinococcus-Thermus (Tables 4 and 5). However, more than 98% of rumen bacteria were assigned to only three phyla (Bacteroidetes, Proteobacteria, and Firmicutes). Fibrobacteres was not detected with universal primers. We have determined that this is an artifact that has arisen because Fibrobacter succinogenes sequences in NCBI and RDP have not had the region upstream of the 27f site sequenced. Therefore, F. succinogenes is not included in the construction of our in silico database.
TABLE 4.
Comparison of putative microbial distribution generated from T-RF libraries of rumen fluid samples collected at 0 h
| Taxonomic rank | % of T-RFs with taxonomic rank at 0 h duringa: |
|||||
|---|---|---|---|---|---|---|
| Control period |
SARA |
|||||
| Grain-induced SARA |
Alfalfa pellet-induced SARA | Grain-induced SARA |
Alfalfa pellet-induced SARA | |||
| Mild | Severe | Mild | Severe | |||
| Phylum Bacteroidetes | 28.18 bc | 28.56 bc | 85.50 a | 25.78 bc | 17.75 c | 36.19 b |
| Class Bacteroidetes | 27.81 b | 28.25 b | 85.28 a | 25.53 bc | 17.66 c | 36.06 b |
| Order Bacteroidales | 27.81 b | 28.25 b | 85.28 a | 25.53 bc | 17.66 c | 36.06 b |
| Family Prevotellaceae | 2.71 c | 2.82 c | 25.49 a | 2.47 c | 2.24 c | 10.70 b |
| Phylum Proteobacteria | 4.99 | 3.83 | 1.14 | 4.02 | 1.97 | 0.78 |
| Class Epsilonproteobacteria | 0.39 | 0.28 | 0.35 | 0.33 | 0.03 | 0.15 |
| Order Campylobacterales | 0.39 | 0.28 | 0.35 | 0.33 | 0.03 | 0.15 |
| Class Gammaproteobacteria | 2.21 | 2.10 | 0.51 | 1.93 | 1.37 | 0.23 |
| Order Aeromonadales | 0.02 | 0.02 | 0.03 | 0.02 | 0.00 | 0.01 |
| Family Succinivibrionaceae | 0.02 | 0.02 | 0.03 | 0.02 | 0.00 | 0.01 |
| Order Enterobacteriales | 1.47 | 1.53 | 0.03 | 1.35 | 1.21 | 0.01 |
| Family Enterobacteriaceae | 1.47 | 1.53 | 0.03 | 1.35 | 1.21 | 0.01 |
| Phylum Firmicutes | 64.74 b | 66.03 b | 12.44 c | 68.53 b | 79.74 a | 62.34 b |
| Class Clostridia | 59.53 b | 60.91 b | 11.02 c | 64.08 ab | 75.89 a | 57.24 b |
| Order Clostridiales | 57.49 b | 58.84 b | 10.11 c | 63.08 ab | 75.17 a | 56.31 b |
| Family Clostridiaceae | 0.69 | 0.72 | 0.00 | 0.64 | 1.11 | 0.01 |
| Family Eubacteriaceae | 0.03 | 0.03 | 0.00 | 0.03 | 0.03 | 0.03 |
| Family Lachnospiraceae | 35.86 b | 36.23 b | 1.60 c | 43.60 b | 60.14 a | 39.12 b |
| Family Peptostreptococcaceae | 0.03 | 0.02 | 0.05 | 0.02 | 0.00 | 0.02 |
| Family Ruminococcaceae | 15.55 a | 16.29 a | 3.70 b | 14.15 a | 10.83 a | 11.72 a |
| Family Veillonellaceae | 3.83 | 3.98 | 3.80 | 3.46 | 2.31 | 7.70 |
| Class Bacilli | 2.54 | 2.33 | 1.12 | 2.01 | 1.61 | 2.65 |
| Order Lactobacillales | 2.39 | 2.17 | 0.84 | 1.87 | 1.49 | 2.53 |
| Family Streptococcaceae | 1.93 | 2.00 | 0.56 | 1.77 | 1.42 | 1.81 |
| Phylum Spirochaetes | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 |
| Phylum Actinobacteria | 1.43 | 1.29 | 0.53 | 1.12 | 0.64 | 0.29 |
| Phylum Fusobacteria | 0.02 | 0.02 | 0.00 | 0.02 | 0.00 | 0.00 |
| Phylum TM7 | 0.01 | 0.01 | 0.00 | 0.01 | 0.01 | 0.00 |
| Phylum Tenericutes | 0.10 | 0.02 | 0.00 | 0.07 | 0.04 | 0.03 |
| Phylum Deinococcus-Thermus | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 |
| Phylum-level unclassified bacteria | 0.50 | 0.21 | 0.38 | 0.42 | 0.03 | 0.29 |
Rumen fluid samples were collected 4 days after SARA induction during SARA periods. a, b, and c, mean values within a row with different symbols differ (P < 0.05).
TABLE 5.
Comparison of putative microbial distribution generated from T-RF libraries of rumen fluid samples collected at 6 h
| Taxonomic rank | % of T-RFs with taxonomic rank at 6 h duringa: |
|||||
|---|---|---|---|---|---|---|
| Control period |
SARA |
|||||
| Grain-induced SARA |
Alfalfa pellet-induced SARA | Grain-induced SARA |
Alfalfa pellet-induced SARA | |||
| Mild | Severe | Mild | Severe | |||
| Phylum Bacteroidetes | 24.58 bc | 20.04 bc | 47.41 a | 26.27 bc | 15.34 c | 34.65 ab |
| Class Bacteroidetes | 24.07 bc | 19.81 bc | 47.14 a | 26.09 bc | 15.29 c | 34.46 ab |
| Order Bacteroidales | 24.07 bc | 19.81 bc | 47.14 a | 26.09 bc | 15.29 c | 34.46 ab |
| Family Prevotellaceae | 2.41 b | 2.03 b | 13.53 a | 2.60 b | 7.65 ab | 9.99 a |
| Phylum Proteobacteria | 4.49 | 3.04 | 1.98 | 1.94 | 3.66 | 2.99 |
| Class Epsilonproteobacteria | 0.26 | 0.23 | 0.19 | 0.35 | 0.00 | 0.14 |
| Order Campylobacterales | 0.26 | 0.23 | 0.19 | 0.35 | 0.00 | 0.14 |
| Class Gammaproteobacteria | 2.03 | 1.45 | 0.62 | 0.40 | 2.96 | 1.92 |
| Order Aeromonadales | 0.02 | 0.02 | 0.03 | 0.00 | 0.00 | 0.02 |
| Family Succinivibrionaceae | 0.02 | 0.02 | 0.03 | 0.00 | 0.00 | 0.02 |
| Order Enterobacteriales | 1.31 | 1.12 | 0.03 | 0.37 | 1.22 | 1.28 |
| Family Enterobacteriaceae | 1.31 | 1.12 | 0.03 | 0.37 | 1.22 | 1.28 |
| Phylum Firmicutes | 69.33 b | 75.65 ab | 49.52 c | 70.72 ab | 79.58 a | 60.68 bc |
| Class Clostridia | 64.03 bc | 71.83 ab | 43.76 cd | 66.11 abc | 78.03 a | 56.44 c |
| Order Clostridiales | 62.23 bc | 70.34 ab | 42.78 cd | 65.01 abc | 78.19 a | 55.46 c |
| Family Clostridiaceae | 0.61 | 0.53 | 1.10 | 0.66 | 1.13 | 0.60 |
| Family Eubacteriaceae | 0.03 | 0.03 | 0.00 | 0.03 | 0.03 | 0.03 |
| Family Lachnospiraceae | 42.16 bc | 54.85 ab | 32.76 c | 45.73 bc | 62.60 a | 37.00 c |
| Family Peptostreptococcaceae | 0.08 | 0.00 | 0.03 | 0.02 | 0.00 | 0.07 |
| Family Ruminococcaceae | 13.90 a | 11.69 ab | 5.62 b | 13.70 a | 11.26 ab | 13.30 ab |
| Family Veillonellaceae | 4.09 | 2.14 | 2.66 | 3.69 | 2.34 | 3.37 |
| Class Bacilli | 2.90 | 1.67 | 2.46 | 2.11 | 0.41 | 1.93 |
| Order Lactobacillales | 2.77 | 1.56 | 2.29 | 1.97 | 0.29 | 1.80 |
| Family Streptococcaceae | 1.71 | 1.46 | 2.13 | 1.84 | 0.17 | 1.68 |
| Phylum Spirochaetes | 0.01 | 0.01 | 0.11 | 0.00 | 0.00 | 0.01 |
| Phylum Actinobacteria | 1.22 | 0.87 | 0.63 | 0.84 | 1.01 | 1.14 |
| Phylum Fusobacteria | 0.02 | 0.02 | 0.00 | 0.02 | 0.02 | 0.00 |
| Phylum TM7 | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 |
| Phylum Tenericutes | 0.10 | 0.02 | 0.12 | 0.00 | 0.08 | 0.11 |
| Phylum Deinococcus-Thermus | 0.01 | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 |
| Phylum-level unclassified bacteria | 0.23 | 0.34 | 0.30 | 0.17 | 0.21 | 0.40 |
Rumen fluid samples were collected 4 days after SARA induction during SARA periods. a, b, and c, mean values within a row with different symbols differ (P < 0.05).
Mild and severe grain-induced SARA and alfalfa pellet-induced SARA produced different TRFLP profiles at the phylum level (Tables 4 and 5). The major differences between mild and severe grain-induced SARA were a significant increase in the phylum Firmicutes (79.7% versus 69.6%) and a significant decrease in the phylum Bacteroidetes (16.6% versus 26%) in severe versus mild disease (Tables 4 and 5). Within the Firmicutes, the family most affected by the severe form was the Lachnospiraceae (61.4% versus 44.7%), which is richly populated with known rumen bacteria. Alfalfa pellet-induced SARA also showed a significant increase in the phylum Firmicutes and a decrease in the phylum Bacteroidetes (Tables 4 and 5). However, in the alfalfa pellet-induced model, Bacteroidetes were 35.4% and Firmicutes were 61.5% of the population (Tables 4 and 5). Microbial composition did not change over time, with the exception of the control period during alfalfa pellet-induced SARA, where a significant shift from Bacteroidetes to Firmicutes was observed at 6 h postfeeding (Table 5).
Species richness was not affected by the SARA induction model but was numerically lower (in both the grain and the alfalfa pellet model) than in the control period (Table 6). Species diversity was significantly lower during mild grain-induced SARA and tended to be lower in severe grain-induced SARA but was not different for alfalfa pellet-induced SARA (Table 6). The similarity between microbial communities was greater for the control period and alfalfa pellet-induced SARA and mild grain-induced SARA than for severe grain-induced SARA (Table 7).
TABLE 6.
Richness and diversity indices calculated from TRFLP incidence profiles of rumen fluida
| Time | Period | Mean valueb under indicated conditions for: |
|||||
|---|---|---|---|---|---|---|---|
| Richnessc |
Diversityd |
||||||
| Grain-induced SARA |
Alfalfa pellet-induced SARA | Grain-induced SARA |
Alfalfa pellet-induced SARA | ||||
| Mild | Severe | Mild | Severe | ||||
| 0 h | Control | 69.9 | 46.8 | 41.3 | 3.1 a | 2.8 | 2.8 |
| SARA | 48.5 | 29.6 | 31.7 | 2.4 b | 2 | 2.2 | |
| 6 h | Control | 93.6 | 48.1 | 25.2 | 3.2 a | 2.8 ab | 2.4 b |
| SARA | 45 | 49.2 | 42.5 | 2.3 b | 2.4 | 2.8 | |
Rumen fluid samples were collected 4 days after SARA induction during SARA period.
a and b, mean values within the same row or column with different symbols differ (P < 0.05). Standard errors of the means were 35.3 for richness and 0.2 for diversity.
Based on Chao-2 richness estimator.
Based on Shannon diversity estimator.
TABLE 7.
Multiple-incidence-based similarities of rumen bacterial communities between control period and grain-induced and alfalfa pellet-induced SARA calculated from TRFLP incidence profiles of rumen samples amplified using 27f and 342r primers
| Induction model | Period, disease severity | Time (h) | % Similaritya (mean ± SE) of bacterial community under indicated conditions |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grain induction |
Alfalfa pellet induction |
|||||||||||||
| Control |
SARA |
Control |
SARA |
|||||||||||
| Mild |
Severe |
Mild |
Severe |
|||||||||||
| 0 h | 6 h | 0 h | 6 h | 0 h | 6 h | 0 h | 6 h | 0 h | 6 h | 0 h | 6 h | |||
| Grain induced | Control, mild | 0 | 100 | |||||||||||
| 6 | 77 ± 8 | 100 | ||||||||||||
| Control, severe | 0 | 72 ± 8 | 66 ± 9 | 100 | ||||||||||
| 6 | 73 ± 8 | 61 ± 9 | 74 ± 10 | 100 | ||||||||||
| SARA, mild | 0 | 69 ± 9 | 58 ± 9 | 77 ± 8 | 71 ± 10 | 100 | ||||||||
| 6 | 59 ± 9 | 55 ± 8 | 73 ± 9 | 60 ± 9 | 86 ± 8 | 100 | ||||||||
| SARA, severe | 0 | 31 ± 8 | 30 ± 8 | 38 ± 9 | 46 ± 10 | 42 ± 9 | 50 ± 10 | 100 | ||||||
| 6 | 29 ± 7 | 28 ± 7 | 29 ± 8 | 40 ± 10 | 41 ± 9 | 43 ± 10 | 67 ± 10 | 100 | ||||||
| Alfalfa pellet induced | Control | 0 | 28 ± 8 | 28 ± 8 | 41 ± 9 | 31 ± 9 | 32 ± 8 | 39 ± 10 | 42 ± 11 | 42 ± 9 | 100 | |||
| 6 | 41 ± 9 | 44 ± 9 | 57 ± 10 | 46 ± 10 | 54 ± 10 | 57 ± 6 | 35 ± 10 | 36 ± 9 | 50 ± 11 | 100 | ||||
| SARA | 0 | 40 ± 10 | 52 ± 9 | 54 ± 10 | 44 ± 9 | 46 ± 10 | 54 ± 10 | 33 ± 10 | 34 ± 9 | 48 ± 10 | 52 ± 10 | 100 | ||
| 6 | 54 ± 8 | 64 ± 8 | 44 ± 8 | 46 ± 9 | 47 ± 8 | 48 ± 9 | 38 ± 9 | 46 ± 8 | 30 ± 8 | 42 ± 9 | 45 ± 9 | 100 | ||
Based on Jaccard index of similarity.
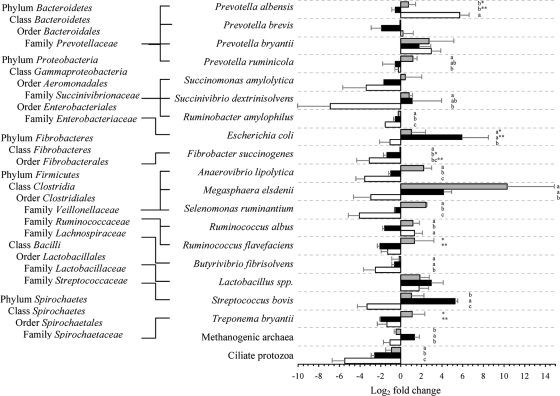
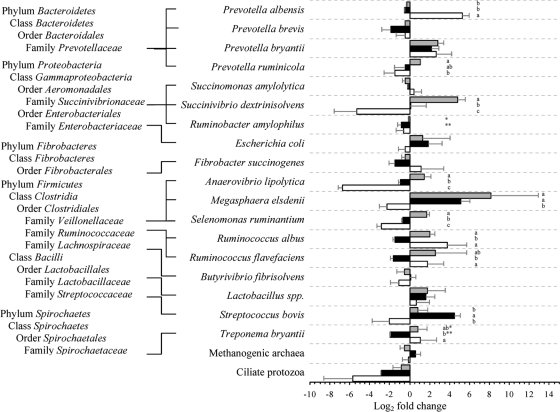
Relative quantification of major rumen bacteria, methanogenic archaea, and ciliate protozoa during SARA compared to the control period is shown in Fig. 2 and 3. At 0 h, severe grain-induced SARA was dominated by Escherichia coli, Streptococcus bovis, Megasphaera elsdenii, and Lactobacillus spp.; mild grain-induced SARA was dominated by M. elsdenii, Selenomonas ruminantium, Prevotella bryantii, and Anaerovibrio lipolytica; and alfalfa pellet-induced SARA was dominated only by Prevotella albensis. At 6 h after feeding, the severe grain-induced SARA group was dominated by M. elsdenii and S. bovis, while the mild group was dominated by M. elsdenii, Succinivibrio dextrinisolvens, P. bryantii, and Ruminococcus flavefaciens. In contrast, the alfalfa pellet-induced SARA group was dominated by P. albensis, P. bryantii, and Ruminococcus albus. Multivariate discriminant analysis of real-time PCR data indicated that the best predictor of severe grain-induced SARA was E. coli.
FIG. 2.
Changes (log2) in predominant rumen microorganisms at 0 h during mild (gray bars) and severe (black bars) grain-induced and alfalfa pellet-induced (white bars) SARA compared to the control period, determined with real-time PCR. The letters a, b, and c indicate statistical differences within species at a P value of <0.05; * and **, P < 0.1. Error bars show standard errors.
FIG. 3.
Changes (log2) in predominant rumen microorganisms at 6 h during mild (gray bars) and severe (black bars) grain-induced and alfalfa pellet-induced (white bars) SARA compared to the control period, determined with real-time PCR. The letters a, b, and c indicate statistical differences within species at a P value of <0.05; * and **, P < 0.1. Error bars show standard errors.
DISCUSSION
Current definitions of SARA are based on the low rumen pH typically generated on high-starch diets (35). However, there is no general agreement on the pH threshold that defines SARA, and moreover, rumen pH may not even be highly correlated with disease symptoms (2, 9). To refine the definition of SARA, Gozho et al. (14) suggested that the free rumen LPS concentration should be considered. In our previous work (20, 21), we demonstrated that the feed type and physical form of the diet had an effect on the induction of SARA. When we induced low rumen pH with highly fermentable alfalfa-pellets, inflammatory markers were absent even though the free rumen LPS concentrations were high (20). Thus, rumen fermentation conditions by themselves are not sufficient to explain SARA. In fact, we speculate that SARA is a historical artifact derived from the pH-related definition of acute ruminal acidosis.
To understand the underlying causes of SARA, we examined the microbial changes that occur with grain- and alfalfa pellet-induced SARA using rumen samples from our previous experiments (20, 21). A critical part of our analysis was to separate animals into mild and severe SARA groups based on objective criteria using multivariate statistical techniques (Table 2; Fig. 1). Clustering of cows into mild and severe groups was conducted using the animal as the independent variable and the free rumen LPS concentration, time below pH 5.6, and serum haptoglobin as dependent variables. Variation in animal response is one of the diagnostic criteria used to identify SARA on a herd basis. The coefficient of variation among animals is often used as an objective criterion to determine whether SARA is present (12). At this time, it is not clear in the literature whether the various SARA symptoms are the result of feed intake variation among individuals, natural variations in saliva buffering capacity, or rumen epithelial potential for uptake of short-chain fatty acids (10, 33). Differences in endotoxin tolerance and immune responses that predispose certain animals to the disease may also be a factor contributing to the variation in disease etiology seen in the field (17). Although the number of animals used in this study was small, which is typical of this type of experiment in dairy cattle (22), we were still able to detect a significant difference among individual responses to SARA, leading us to believe that our SARA induction models were characteristic of a field situation. We are confident that our experiment replicated the variation in disease etiology seen in the field.
We used TRFLP to evaluate the structure, dynamics, and diversity of the rumen microbiome in mild and severe grain-induced SARA and alfalfa pellet-induced SARA. We previously demonstrated (1, 41) that this analysis was not able to unambiguously identify peaks to the genus or family levels, but over 95% of T-RFs were unambiguous at the order level (41). The general population shifts we noted in our analysis were similar to those obtained with the 16S rRNA gene library studies of Tajima et al. (46) and Whitford et al. (49). However, these two studies (46, 49) evaluated typical high-grain diets but did not evaluate the possibility of reducing ruminal pH with highly fermentable nonstarch substrates like alfalfa pellets. By changing the particle size of the alfalfa (hay versus pellets), we could change the rate of fermentation without changing the chemical composition of the diet. Thus, fermentation substrate was not a confounding factor in the analysis.
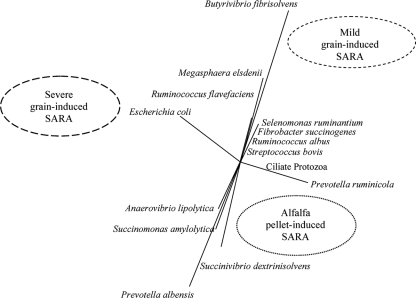
The real-time PCR data (Fig. 2 and 3) evaluated with multivariate statistics (Fig. 4) indicated that the major predictor of severe grain-induced SARA was E. coli. The TRFLP prevalence analysis also indicated changes in the Proteobacteria (contains E. coli), although the differences between treatments were not significant. Given that the rumen LPS concentration was high in both grain- and alfalfa pellet-induced SARA and that only grain-fed animals exhibited inflammatory responses, this excludes the possibility that LPS alone is the cause of inflammation.
FIG. 4.
Results of discriminant analysis of major rumen bacteria and ciliate protozoa in response to SARA induction model. The circles are independent variables, and the distances between them reflect their dissimilarity. The straight lines are dependent variables, and their lengths and the angles between them are functions of the relative effects of independent variables.
The most predominant shifts in microbial populations were among the gram-negative Bacteroidetes. These shifts were evident from the TRFLP prevalence data, as well as the real-time PCR data, for P. albensis, P. brevis, and Prevotella ruminicola, which are members of the Bacteroidetes. We suspect that a majority of the LPS produced came from these bacteria. Bacteroides spp. LPS is much less toxic than LPS from E. coli or Salmonella spp. (18, 44). A human study indicated that LPS from Salmonella typhi, E. coli, and Pseudomonas aeruginosa reached toxicity thresholds at ∼0.1, 1.0, and 60 ng/kg of body weight, respectively (15). In an earlier rumen study, LPS was extracted from mixed rumen bacteria from either grain-fed or hay-fed animals and compared to E. coli LPS in a rodent endotoxin model. The most-toxic LPS came from E. coli, followed by LPS from grain-fed and then hay-fed animals (26). M. elsdenii is a gram-negative bacterium and almost always in high abundance in grain-fed animals. To test the possibility that the LPS from this bacterium was toxic, its LPS was compared to that of E. coli in a rodent model and found to be much less potent (27).
The classical view of acidosis in the rumen is that as grain increases, so does the prevalence of starch-fermenting bacteria like S. bovis (38). Thus, there should be a high population ratio of lactate-consuming bacteria like M. elsdenii to prevent lactic acid acidosis (37). Our real-time PCR data indicated that the amylolytic bacterium whose abundance most closely mirrored the severity of SARA was S. bovis, and other major amylolytic bacteria were less affected. M. elsdenii populations were synchronized with the S. bovis numbers, indicating that this bacterium was effectively eliminating rumen lactate. In contrast, even though the rumen pH conditions created by the alfalfa pellet diet were typical of SARA, the levels of S. bovis and, consequently, M. elsdenii were low.
The results of multivariate analysis (Fig. 1 and 4) indicated that there was a significant association between alfalfa-fed animals, microbial populations, and the absence of inflammation. The microbial species most closely associated with this diet were P. ruminicola, A. lipolytica, Succinimonas amylolytica, P. albensis, and S. dextrinosolvens. In particular, P. albensis was in high abundance in this diet. Thus, Prevotella spp. may potentially be used as probiotics in SARA. Rodríguez (36) selected strains of Prevotella spp. that grew rapidly on starch and produced succinate and propionate as major end products. When this strain was inoculated into three lactic acid-challenged goats, it was found that lactic acid levels declined. Chiquette et al. (5) dosed Prevotella spp. into dairy cattle and observed small changes in lactic acid levels, but appropriate acidotic rumen conditions were not produced, so the effect of the Prevotella was equivocal. Our study is the first to our knowledge to demonstrate a clear association between SARA and particular microbial populations that might be protective, and it provides objective evidence for the use of Prevotella spp. as a probiotic in SARA.
In conclusion, we have demonstrated that grain- and alfalfa pellet-induced SARA result in different rumen microbial population structures even though rumen fermentation conditions are similar. We hypothesized that LPS concentrations in the rumen were not a predictor of SARA because high LPS levels could be created with an alfalfa pellet diet but no inflammation occurred. As the inflammatory response may have been due in part to translocation of LPS from the gut into the bloodstream, we conclude that LPS is indicative of a barrier function defect and that the key to understanding SARA is to identify potential microbial and physiological factors that result in increased epithelial permeability. Because E. coli is a major contributor to the rumen LPS pool and its LPS is highly toxic (17), we hypothesize that E. coli may be a contributing factor to disease onset.
Acknowledgments
This study was supported by grants from Dairy Farmers of Canada (DFC), the Agri-Food Research Development Initiative (ARDI), and the Natural Sciences and Engineering Research Council of Canada (NSERC).
We thank Roman Kotlowski and Shadi Sepehri for their technical assistance in the TRFLP and real-time PCR analysis.
Footnotes
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Bhandari, S. K., B. Xu, C. M. Nyachoti, D. W. Giesting, and D. O. Krause. 2008. Evaluation of alternatives to antibiotics using an Escherichia coli K88+ model of piglet diarrhea: effects on gut microbial ecology. J. Anim. Sci. 86:836-847. [DOI] [PubMed] [Google Scholar]
- 2.Bramley, E., I. J. Lean, W. J. Fulkerson, M. A. Stevenson, A. R. Rabiee, and N. D. Costa. 2008. The definition of acidosis in dairy herds predominantly fed on pasture and concentrates. J. Dairy Sci. 91:308-321. [DOI] [PubMed] [Google Scholar]
- 3.CCAC. 1993. Guide to the care and use of experimental animals, vol. 1. Canadian Council on Animal Care, Ottawa, ON, Canada.
- 4.Chin, A. C., A. N. Flynn, J. P. Fedwick, and A. G. Buret. 2006. The role of caspase-3 in lipopolysaccharide-mediated disruption of intestinal epithelial tight junctions. Can. J. Physiol. Pharmacol. 84:1043-1050. [DOI] [PubMed] [Google Scholar]
- 5.Chiquette, J., M. J. Allison, and M. A. Rasmussen. 2008. Prevotella bryantii 25A used as a probiotic in early-lactation dairy cows: effect on ruminal fermentation characteristics, milk production, and milk composition. J. Dairy Sci. 91:3536-3543. [DOI] [PubMed] [Google Scholar]
- 6.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denman, S. E., and C. S. McSweeney. 2006. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 58:572-582. [DOI] [PubMed] [Google Scholar]
- 8.Emmanuel, D. G. V., S. M. Dunn, and B. N. Ametaj. 2008. Feeding high proportions of barley grain stimulates an inflammatory response in dairy cows. J. Dairy Sci. 91:606-614. [DOI] [PubMed] [Google Scholar]
- 9.Enemark, J. M., R. J. Jorgensen, and N. B. Kristensen. 2004. An evaluation of parameters for the detection of subclinical rumen acidosis in dairy herds. Vet. Res. Commun. 28:687-709. [DOI] [PubMed] [Google Scholar]
- 10.Erickson, G. E., C. T. Milton, K. C. Fanning, R. J. Cooper, R. S. Swingle, J. C. Parrott, G. Vogel, and T. J. Klopfenstein. 2003. Interaction between bunk management and monensin concentration on finishing performance, feeding behavior, and ruminal metabolism during an acidosis challenge with feedlot cattle. J. Anim. Sci. 81:2869-2879. [DOI] [PubMed] [Google Scholar]
- 11.Garrett, E. F., K. V. Nordlund, W. J. Goodger, and G. R. Oetzel. 1997. A cross-sectional field study investigating the effect of preparturient dietary management on ruminal pH in early lactation dairy cows. J. Dairy Sci. 80:169. [Google Scholar]
- 12.Garrett, E. F., M. N. Pereira, K. V. Nordlund, L. E. Armentano, W. J. Goodger, and G. R. Oetzel. 1999. Diagnostic methods for the detection of subacute ruminal acidosis in dairy cows. J. Dairy Sci. 82:1170-1178. [DOI] [PubMed] [Google Scholar]
- 13.Goad, D. W., C. L. Goad, and T. G. Nagaraja. 1998. Ruminal microbial and fermentative changes associated with experimentally induced subacute acidosis in steers. J. Anim. Sci. 76:234-241. [DOI] [PubMed] [Google Scholar]
- 14.Gozho, G. N., J. C. Plaizier, D. O. Krause, A. D. Kennedy, and K. M. Wittenberg. 2005. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an inflammatory response. J. Dairy Sci. 88:1399-1403. [DOI] [PubMed] [Google Scholar]
- 15.Greisman, S. E., and R. B. Hornick. 1969. Comparative pyrogenic reactivity of rabbit and man to bacterial endotoxin. Proc. Soc. Exp. Biol. Med. 131:1154. [DOI] [PubMed] [Google Scholar]
- 16.Haubro Andersen, P., and N. Jarlov. 1990. Investigation of the possible role of endotoxin, TXA2, PGI2 and PGE2 in experimentally induced rumen acidosis in cattle. Acta Vet. Scand. 31:27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurley, J. C. 1995. Endotoxemia: methods of detection and clinical correlates. Clin. Microbiol. Rev. 8:268-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasper, D. L. 1976. Chemical and biological characterization of the lipopolysaccharide of Bacteroides fragilis subspecies fragilis. J. Infect. Dis. 134:59. [DOI] [PubMed] [Google Scholar]
- 19.Kent, A. D., D. J. Smith, B. J. Benson, and E. W. Triplett. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khafipour, E., D. O. Krause, and J. C. Plaizier. 2009. Alfalfa pellet-induced subacute ruminal acidosis in dairy cows increases bacterial endotoxin in the rumen without causing inflammation. J. Dairy Sci. 92:1712-1724. [DOI] [PubMed] [Google Scholar]
- 21.Khafipour, E., D. O. Krause, and J. C. Plaizier. 2009. A grain-based subacute ruminal acidosis challenge causes translocation of lipopolysaccharide and triggers inflammation. J. Dairy Sci. 92:1060-1070. [DOI] [PubMed] [Google Scholar]
- 22.Kleen, J. L., G. A. Hooijer, J. Rehage, and J. P. T. M. Noordhuizen. 2004. Rumenocentesis (rumen puncture): a viable instrument in herd health diagnosis. Dtsch. Tierarztl. Wochenschr. 111:458-462. [PubMed] [Google Scholar]
- 23.Krause, K. M., and G. R. Oetzel. 2005. Inducing subacute ruminal acidosis in lactating dairy cows. J. Dairy Sci. 88:3633-3639. [DOI] [PubMed] [Google Scholar]
- 24.Lan, Y., S. Xun, S. Tamminga, B. A. Williams, M. W. Verstegen, and G. Erdi. 2004. Real-time PCR detection of lactic acid bacteria in cecal contents of Eimeria tenella-infected broilers fed soybean oligosaccharides and soluble soybean polysaccharides. Poult. Sci. 83:1696-1702. [DOI] [PubMed] [Google Scholar]
- 25.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaraja, T. G., E. E. Bartley, L. R. Fina, H. D. Anthony, B. E. Brent, and D. A. Sapienza. 1979. Chemical characteristics of rumen bacterial endotoxin. J. Anim. Sci. 48:1250-1256. [DOI] [PubMed] [Google Scholar]
- 27.Nagaraja, T. G., L. R. Fina, B. A. Lassman, E. E. Bartley, H. D. Anthony, D. A. Sapienza, and B. E. Brent. 1979. Characterization of endotoxin from the rumen bacterium Megasphaera elsdenii. Am. J. Vet. Res. 40:35-39. [PubMed] [Google Scholar]
- 28.Nagaraja, T. G., and E. C. Titgemeyer. 2007. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J. Dairy Sci. 90(Suppl. 1):E17-E38. [DOI] [PubMed] [Google Scholar]
- 29.Nocek, J. E. 1997. Bovine acidosis: implications on laminitis. J. Dairy Sci. 80:1005-1028. [DOI] [PubMed] [Google Scholar]
- 30.Ohene-Adjei, S., A. V. Chaves, T. A. McAllister, C. Benchaar, R. M. Teather, and R. J. Forster. 2008. Evidence of increased diversity of methanogenic archaea with plant extract supplementation. Microb. Ecol. 56:234-242. [DOI] [PubMed] [Google Scholar]
- 31.Owens, F. N., D. S. Secrist, W. J. Hill, and D. R. Gill. 1998. Acidosis in cattle: a review. J. Anim. Sci. 76:275-286. [DOI] [PubMed] [Google Scholar]
- 32.Ozutsumi, Y., K. Tajima, A. Takenaka, and H. Itabashi. 2006. Real-time PCR detection of the effects of protozoa on rumen bacteria in cattle. Curr. Microbiol. 52:158-162. [DOI] [PubMed] [Google Scholar]
- 33.Penner, G. B., J. R. Aschenbach, G. Gabel, R. Rackwitz, and M. Oba. 2009. Epithelial capacity for apical uptake of short chain fatty acids is a key determinant for intraruminal pH and the susceptibility to subacute ruminal acidosis in sheep. J. Nutr. 139:1714-1720. [DOI] [PubMed] [Google Scholar]
- 34.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plaizier, J. C., D. O. Krause, G. N. Gozho, and B. W. McBride. 2008. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet. J. 176:21-31. [DOI] [PubMed] [Google Scholar]
- 36.Rodríguez, F. 2003. Control of lactate accumulation in ruminants using Prevotella bryantii. Ph.D. thesis. Iowa State University, Ames.
- 37.Russell, J. B., M. A. Cotta, and D. B. Dombrowski. 1981. Rumen bacterial competition in continuous culture: Streptococcus bovis versus Megasphaera elsdenii. Appl. Environ. Microbiol. 41:1394-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell, J. B., and T. Hino. 1985. Regulation of lactate production in Streptococcus bovis: a spiraling effect that contributes to rumen acidosis. J. Dairy Sci. 68:1712-1721. [DOI] [PubMed] [Google Scholar]
- 39.SAS. 2004. SAS/STAT user's guide, release 9.1.2. SAS Institute, Inc., Cary, NC.
- 40.Schutte, U. M. E., Z. Abdo, S. J. Bent, C. Shyu, C. J. Williams, J. D. Pierson, and L. J. Forney. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80:365-380. [DOI] [PubMed] [Google Scholar]
- 41.Sepehri, S., R. Kotlowski, C. N. Bernstein, and D. O. Krause. 2007. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm. Bowel Dis. 13:675-683. [DOI] [PubMed] [Google Scholar]
- 42.Shyu, C., T. Soule, S. J. Bent, J. A. Foster, and L. J. Forney. 2007. MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms of 16S and 18S rRNA genes. Microb. Ecol. 53:562-570. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson, D. M., and P. J. Weimer. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75:165-174. [DOI] [PubMed] [Google Scholar]
- 44.Stewart, D. J. 1977. Biochemical and biological studies on lipopolysaccharide of bacteroides-nodosus. Res. Vet. Sci. 23:319-325. [PubMed] [Google Scholar]
- 45.Sylvester, J. T., S. K. Karnati, Z. Yu, M. Morrison, and J. L. Firkins. 2004. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J. Nutr. 134:3378-3384. [DOI] [PubMed] [Google Scholar]
- 46.Tajima, K., R. I. Aminov, T. Nagamine, K. Ogata, M. Nakamura, H. Matsui, and Y. Benno. 1999. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol. Ecol. 29:159-169. [Google Scholar]
- 47.Walter, J., C. Hertel, G. W. Tannock, C. M. Lis, K. Munro, and W. P. Hammes. 2001. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:2578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1997. PCR detection of Ruminococcus spp. in human and animal faecal samples. Mol. Cell. Probes 11:259-265. [DOI] [PubMed] [Google Scholar]
- 49.Whitford, M. F., R. J. Forster, C. E. Beard, J. H. Gong, and R. M. Teather. 1998. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe 4:153-163. [DOI] [PubMed] [Google Scholar]