Abstract
Sulfate-reducing prokaryotes (SRP) cause severe problems like microbial corrosion and reservoir souring in seawater-injected oil production systems. One strategy to control SRP activity is the addition of nitrate to the injection water. Production waters from two adjacent, hot (80°C) oil reservoirs, one with and one without nitrate treatment, were compared for prokaryotic community structure and activity of SRP. Bacterial and archaeal 16S rRNA gene analyses revealed higher prokaryotic abundance but lower diversity for the nitrate-treated field. The 16S rRNA gene clone libraries from both fields were dominated by sequences affiliated with Firmicutes (Bacteria) and Thermococcales (Archaea). Potential heterotrophic nitrate reducers (Deferribacterales) were exclusively found at the nitrate-treated field, possibly stimulated by nitrate addition. Quantitative PCR of dsrAB genes revealed that archaeal SRP (Archaeoglobus) dominated the SRP communities, but with lower relative abundance at the nitrate-treated site. Bacterial SRP were found in only low abundance at both sites and were nearly exclusively affiliated with thermophilic genera (Desulfacinum and Desulfotomaculum). Despite the high abundance of archaeal SRP, no archaeal SRP activity was detected in [35S]sulfate incubations at 80°C. Sulfate reduction was found at 60°C in samples from the untreated field and accompanied by the growth of thermophilic bacterial SRP in batch cultures. Samples from the nitrate-treated field generally lacked SRP activity. These results indicate that (i) Archaeoglobus can be a major player in hot oil reservoirs, and (ii) nitrate may act in souring control—not only by inhibiting SRP, but also by changing the overall community structure, including the stimulation of competitive nitrate reducers.
During the process of secondary oil recovery in offshore oil fields, most often sulfate-rich seawater is injected into the reservoir to increase pressure and enhance recovery. The supply of large amounts of sulfate as an electron acceptor and the presence of oil organics and their degradation products as electron donors facilitate the enrichment and growth of sulfate-reducing prokaryotes (SRP) in the reservoir, as well as in piping and topside installations (51, 54). The activity of SRP causes severe economic problems due to the reactivity and toxicity of the produced hydrogen sulfide (H2S). In addition to microbiologically influenced corrosion and reservoir souring, the efficiency of oil production is decreased due to plugging by SRP biomass and precipitated metal sulfides (12, 39). Besides the use of broad-spectrum biocides or inhibitors for sulfate reduction, the addition of nitrate effectively decreased the net production of H2S in model column studies (15, 20, 38) and field trials (7, 53). The mechanisms by which nitrate addition might affect souring control are (i) the stimulation of heterotrophic nitrate-reducing bacteria (hNRB) that outcompete SRP for electron donors, (ii) the activity of nitrate-reducing, sulfide-oxidizing bacteria (NR-SOB), and (iii) the inhibition of SRP by the production of nitrite and nitrous oxides (21, 51).
Identification and quantification of reservoir microorganisms, including NRB and SRP, has so far most frequently been assessed by cultivation-dependent methods (7, 12, 53), and cultivation-independent methods have only recently been introduced into the field of reservoir microbiology (11, 17, 28, 32). Considering the small number of these studies, information currently available on the microbial communities and especially on the abundance of nitrate and sulfate reducers present in oil reservoirs or production systems is sparse and, most notably, not quantitative.
The goal of this study was to compare the diversity, abundance, and activity of SRP in production water (PW) from a nitrate-treated and an untreated oil reservoir using a combination of 16S rRNA and dsrAB gene-based analyses, newly developed quantitative PCR (qPCR) assays, and 35SO42− radiotracer incubations. The two analyzed oil reservoirs (Dan and Halfdan) share similar physicochemical characteristics with regard to injection water composition and reservoir conditions, but nitrate has only been added at Halfdan since the start of production. It is hypothesized that the addition of nitrate to the injection water favored the growth of hNRB and/or NR-SOB, thereby inhibiting the activity of SRP and reducing the concentration of H2S, and is consequently reflected in a lower abundance of SRP and a more specialized prokaryotic community.
MATERIALS AND METHODS
Field site and sampling.
The Dan and Halfdan oil fields are located in the North Sea, ∼200 km west off Southern Jutland (Denmark). The crude oil contained in the Dan and Halfdan formations is situated about 2,000 m below the surface. Oil production at Dan commenced in 1972, and oil and natural gas are produced from low-permeability chalks of the latest Cretaceous as well as Paleocene (46). The Halfdan gas and oil accumulation was discovered in 1998 by drilling an ∼9-km-long horizontal well from Dan to Halfdan (26). Production at Halfdan commenced in 1999. Both production systems show a temperature gradient from the reservoir (∼80°C) through the tubing (∼50 to 60°C) to the separators (∼40 to 50°C) accompanied by depressurization through a cascade of separators. Three-phase separation is carried out, yielding gas, oil, and PW. PW from both reservoirs is a mixture of different types of formation water as well as seawater (26). The chemical composition of recovered PW is therefore strongly dependent on the mixing ratio of the different water types and the occurrence of seawater breakthrough in individual wells, as well as on chemical and microbial processes in the piping and the reservoirs. Seawater injection is applied in secondary oil recovery, and injection water is transported from the Dan platform to Halfdan via a 9-km subsea pipeline. Injection water facilities are typically treated weekly with the bactericide tetrakishydroxymethylphosphonium sulfate (THPS). Nitrate (65 mg liter−1) has been added to Halfdan injection water since 2001 in order to decrease the net production of H2S and prevent reservoir souring and corrosion (26). No nitrate is added at the Dan field.
PW was sampled after separation from Dan (HP separator V-3440) and Halfdan (test separator) in March 2008. Samples were kept cool (4°C) and anoxic without further preservation. They were transported to the laboratory within 24 h (Dan) or 48 h (Halfdan) and immediately processed for activity measurements and molecular analyses.
Activity of sulfate reducers and enrichment of active SRP.
PW samples (20 ml) were dispensed into 25-ml tubes and crimped with aluminum seals, and the remaining headspace was flushed with nitrogen and equilibrated at incubation temperatures, i.e., at either 60°C or 80°C, representing in situ temperatures in the production system and the reservoir, respectively. After 4 h of equilibration, samples were supplemented with ∼1 MBq carrier-free 35SO42− (Hartmann Analytic GmbH, Braunschweig, Germany), Na2SO4 (0.2 mM, final concentration), dithionite as the reducing agent, and resazurin as the redox indicator (0.5 mg liter−1, final concentration). All incubations were carried out in duplicates.
To estimate nonstimulated activities, incubations were terminated after 2 days of incubation by the addition of 2 ml zinc acetate solution (20%) and 0.5 ml of a 0.45 mM Na2S suspension (in 20% zinc acetate). Precipitated zinc sulfide was recovered by centrifugation and subjected to a single-step chromium distillation as described previously (1). Activities were stimulated by adding a substrate mixture of lactate, acetate, and glucose (0.5 mM each, final concentration) and yeast extract (0.01%, final concentration) and incubation for 2 days (Dan and Halfdan), 42 days (Dan), and up to 100 days (Halfdan). Samples without added substrates were incubated in parallel for 42 and 100 days. Incubations were terminated and further processed as described above. PW samples that were not amended with substrates and/or 35SO42− served as negative controls and to determine background activities.
To identify and enrich for potentially active SRP, nonradioactive parallel cultures were incubated at 60°C and 80°C for 42 days (Dan) to 100 days (Halfdan). Aliquots of these cultures were removed for DNA extraction and subsequent gene quantification via qPCR and cloning and sequencing of 16S rRNA and dsrAB genes.
DNA extraction and PCR amplification.
DNA extracts were obtained from PW samples (triplicate extracts) and from SRP enrichment cultures (single extracts). Samples of 100 to 250 ml PW and 5 to 12.5 ml enrichment culture were filtered onto white polycarbonate filters (0.2-μm pore size; Nuclepore, Whatman, Kent, United Kingdom). DNA was extracted by combining bead beating and enzymatic and chemical lysis with the FastDNA spin kit for soil (Q-Biogene, Carlsbad, CA) (16).
Archaeal and bacterial 16S rRNA genes were amplified using the universal primer 1492R (34), together with 8F for Bacteria (22) and 21F for Archaea (13). PCRs contained 20 μl of distilled water (dH2O) (Sigma-Aldrich, Copenhagen, Denmark), 25 μl of Taq DNA polymerase master mix red (Ampliqon, Copenhagen, Denmark), 1 μl of bovine serum albumin (10 mg ml−1, Amersham Biosciences, Uppsala, Sweden), 1 μl each of forward and reverse primers (10 pmol μl−1 each; MWG Biotech, Ebersberg, Germany), and 1 μl of template DNA. The thermal cycling conditions were as follows: initial denaturation at 96°C for 4 min, 25 to 28 cycles of 30 s of denaturation at 94°C, 45 s of annealing at 57°C (bacterial 16S rRNA genes) or 56°C (for archaeal 16S rRNA genes), and 2 min of elongation at 72°C. Cycling was completed by a final elongation step at 72°C for 10 min. PCR amplification of dsrAB genes was performed as described previously using primer mixtures of DSR1F and DSR4R (24, 34). The thermal cycling conditions were as follows: 5 min at 96°C, 35 cycles of 30 s at 48°C, 45 s at 48°C, and 2 min at 72°C, completed by a final elongation step of 10 min at 72°C.
Sequencing and phylogenetic analysis.
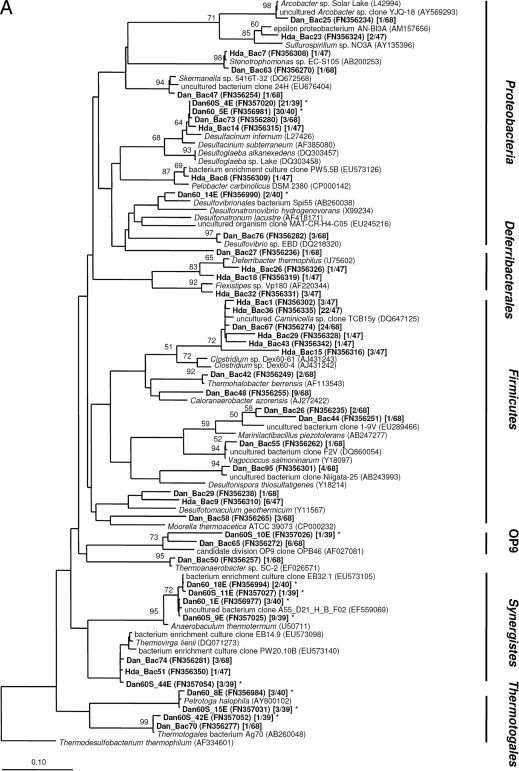
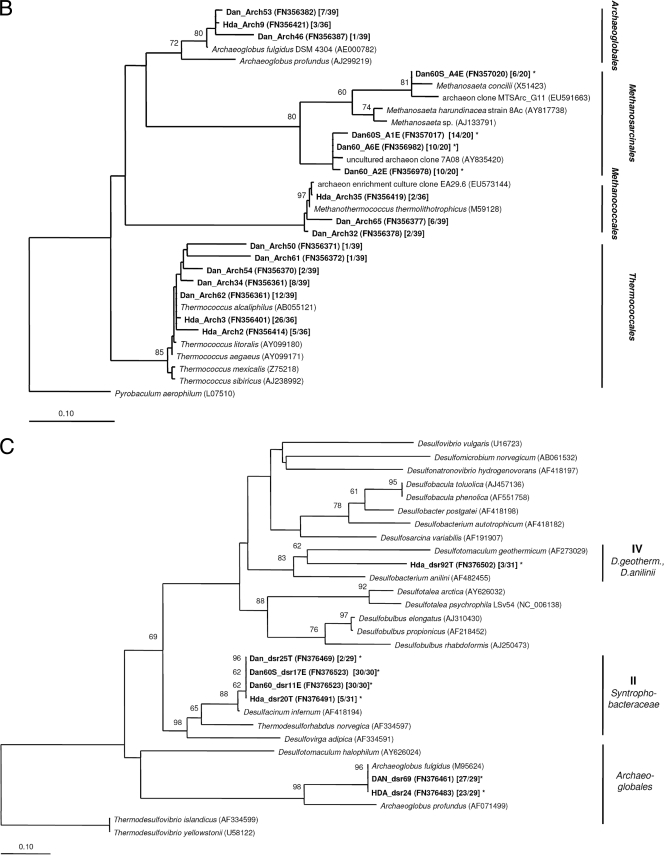
Amplicons of 16S rRNA genes and dsrAB genes were purified with the Gen Elute PCR clean up kit (Sigma, Copenhagen, Denmark) and cloned using the pGEM-T vector system (Promega Corp., Madison, WI) for 16S rRNA genes and the TOPO XL cloning kit (Invitrogen Corp., Carlsbad, CA) for dsrAB genes. Plasmids were isolated and purified with the QIAprep spin miniprep kit (Qiagen GmbH, Hilden, Germany). Inserts were sequenced by Macrogen (Korea) using the vector primers M13F (16S rRNA) and M13R (dsrAB). Sequences (750 to 850 nucleotides [nt]) were checked for possible chimeric origin using Bellerophon (19). Initial alignment of amplified sequences and close relatives identified with BLAST (2) was performed using the SILVA alignment tool (41) for the 16S rRNA gene sequences. dsrAB gene sequences were translated into amino acid sequences and manually aligned in ARB (35). Operational taxonomic units (OTU) were determined with DOTUR (45) and defined as clone sequences sharing >97% nucleotide sequence identity for 16S rRNA and >97% amino acid sequence identity for DsrAB. Coverage, species richness, and diversity indexes were calculated using SPADE (http://chao.stat.nthu.edu.tw). For each OTU of the 16S rRNA gene clone libraries generated from PW samples, at least one representative full-length sequence (>1,450 nt) was determined. The similarity of 16S rRNA clone libraries was statistically evaluated using LIBSHUFF (http://libshuff.mib.uga.edu/). Phylogenetic trees based on 16S rRNA sequences (>1,300 nt) and deduced DsrAB amino acid sequences (540 unambiguously aligned sequence positions) were created based on distance analysis using the neighbor-joining algorithm of the ARB program package (35). Different filters were applied to select sequence positions for the phylogenetic analyses (see the legend to Fig. 2A and B). The robustness of inferred topologies was tested by bootstrap resampling using the same distance model (1,000 replicates). Short sequences were added to the generated 16S rRNA- and DsrAB amino acid-based trees, applying maximum parsimony criteria without changing the overall tree topologies. A broad range of taxa were included in the phylogenetic analyses; some of the reference sequences were removed from the presented phylogenetic trees to enhance clarity.
FIG. 2.
Neighbor-joining distance trees showing the phylogenetic relationship of bacterial (A) and archaeal (B) 16S rRNA gene sequences and deduced DsrAB amino acid sequences (C) of representative clones derived from PW samples and enrichment cultures (all shown in bold). Sequences from this study are named according to the sampling sites (Dan and Halfdan [Hda]) and the enrichment conditions (Dan60 and Dan60S). Accession numbers are given in parentheses. Square brackets show the number of sequences out of the total number of sequences analyzed. 16S rRNA-based trees were constructed using either the Bacteria or the Archaea filter of the ssu_jan04_corr_optARB database (http://www.arb-home.de). Short sequences (16S rRNA, <1,300 nt; DsrAB, <540 amino acids), marked by asterisks, were added without changing the overall tree topology using maximum parsimony criteria. DsrAB gene group names refer to those used by Kaneko et al. (23). Bootstrap values are based on 1,000 replicates; only values greater 50% are indicated near the nodes. The scale bar represents 10% sequence divergence.
qPCR assays.
Bacterial and archaeal 16S rRNA genes were amplified with the primer set 8F/338R for Bacteria and 806F/958R for Archaea (see reference 33 and the references therein) (Table 1). For the quantification of bacterial and archaeal dsrAB gene copies, the novel primers Del1075R [5′-G(CT)TC(ACG)CGGTTCTT(GAT)C-3′] (Table 1) and Arch1830F [5′-TGCTGTC(ACGT)AACATG-3′] (Table 1) were designed with Primrose (4) based on 41 public domain sequences and 91 dsrAB sequences retrieved during an initial study of the sampling site (27). The primers were designed to specifically target sequences affiliated with either the Archaeoglobales (Arch1830F) or a range of bacterial orders, including Desulfovibrionales, Syntrophobacterales, Desulfobacterales, and Clostridiales (Del1075R). These target groups were previously shown to be present in PW samples from Dan and Halfdan (27). The newly designed primers were used in combination with DSR1F and DSR4R, respectively (55). Specificity of the primer sets DSR1F/Del1075R (bacterial dsrA) and Arch1830F/DSR4R (archaeal dsrB) was theoretically checked using the probe match function of ARB and by using standards of plasmids containing known dsrAB genes of bacterial or archaeal origin as templates in conventional PCR. Furthermore, clone libraries were generated from qPCR products, and 36 (archaeal dsrB) and 38 (bacterial dsrA) sequences were phylogenetically analyzed. Clones from each library were exclusively affiliated with the respective target group, confirming the specificity of the primer sets (data not shown).
TABLE 1.
Primers used for qPCR
| Primer | Sequence (5′→3′) | Temp (°C) |
Elongation time (s) | Specificity | Reference | |
|---|---|---|---|---|---|---|
| Annealing | Data acquisitiona | |||||
| 16S rRNA targeted | ||||||
| 8F | AGAGTTTGAT(CT)(AC)TGGCTC | 55 | 80 | 15 | Most Bacteria | 34 |
| 338R | GCTGCCTCCCGTAGGAGT | 55 | 80 | 15 | Most Bacteria | 3 |
| 806F | ATTAGATACCCSBGTAGTCC | 55 | 78 | 10 | Most Archaea | 42 |
| 958R | YCCGGCGTTGAMTCCAATT | 55 | 78 | 10 | Most Archaea | 13 |
| dsrAB targeted | ||||||
| DSR1F | AC(GC)CACTGGAAGCACG | 58 | 80 | 40 | Most SRP | 55 |
| Del1075R | G(CT)TC(ACG)CGGTTCTT(GAT)C | 58 | 80 | 40 | Deltaproteobacterial SRP | This study |
| DSR4R | GTGTAGCAGTTACCGCA | 56 | 80 | 15 | Most SRP | 55 |
| Arch1830F | TGCTGTC(ACGT)AACATG | 56 | 80 | 15 | Archaeal SRP | This study |
Fluorescence data were acquired at assay-specific temperatures to avoid the detection of fluorescence originating from primer dimers (see Materials and Methods).
Quantification of copy numbers of the different genes was performed with a Roche Lightcycler 480 instrument. Each reaction mixture (20 μl) contained 1 μl DNA template (∼3 ng DNA), 1 μl bovine serum albumin (10 mg/ml; Amersham Biosciences, Uppsala, Sweden), 1 μl each primer (100 pmol μl−1 each), 6 μl dH2O, and 10 μl Lightcycler 480 Sybr green I master mix (Roche Molecular Biochemicals, Mannheim, Germany). Cycling conditions varied depending on the primer set (see Table 1 for details). In general, initial denaturation (10 min at 95°C) was followed by 40 cycles of 30 s at 95°C, 30 s at 55 to 58°C, and 15 to 40 s at 72°C and completed by fluorescence data acquisition at an assay-specific temperature used for target quantification. Product specificity was confirmed by melting point analysis (55°C to 95°C with a plate read every 0.5°C).
Plasmids containing either 16S rRNA or dsrAB gene target sequences were used as standards. DNA concentrations for the initial standard as well as unknown samples were calculated from triplicate measurements with a Nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). The gene copy number in the initial standard was calculated from the DNA content, the length of the cloned gene fragment, and the mean weight of a base pair (1.1 × 10−21 g) and adjusted to 108 target molecules per 1 μl. Gene copy numbers in unknown samples were determined based on standard curves constructed from 10-fold serial dilutions of the standard. Amplification efficiencies were calculated from the slope of standard curves. The detection limits for the various assays (i.e., the lowest standard concentration that is significantly different from the nontemplate controls) were less than 50 gene copies for each assay. Samples, standards, and nontemplate controls were run in triplicate.
Nucleotide sequence accession numbers.
The gene sequences determined in this study are available from EMBL under accession no. FN356234 to FN356423 (bacterial and archaeal 16S rRNA and PW), FN356977 to FN357095 (bacterial and archaeal 16S rRNA and enrichments), and FN376454 to FN376573 (dsrAB, PW, and enrichments).
RESULTS
Physicochemical characterization of PW.
PW from Dan and Halfdan were rather similar in their chemical composition (Table 2), with high salinity (950 to 1,100 mM) and sulfate concentrations (>12 mM) and moderate to high concentrations of fatty acids (acetate, 3 to 6 mM; propionate and butyrate, <1 mM). The main differences were the absence of sulfide and the higher ammonium concentrations (2.8 mM) in PW from the nitrate-treated Halfdan field compared to Dan PW, suggesting that nitrate addition to the injection water resulted in a reduced net sulfide production and that at least some of the nitrate was converted to ammonium.
TABLE 2.
Chemical composition of PW, total cell counts, and gene copy numbers of 16S rRNA and dsrAB
| Parameter | Result for field: |
|
|---|---|---|
| Dan | Halfdan | |
| PW composition (mg liter−1)a | ||
| Chlorideb | 33,747 (952) | 38,652 (1,090) |
| Sulfateb | 1,174 (12.2) | 1,175 (12.3) |
| Sulfide | 5 (0.2) | 0.1 (0.003) |
| Ammoniumc | 33 (1.8) | 51 (2.83) |
| Nitratec | <0.5 (<0.01) | |
| Nitritec | <0.5 (<0.01) | |
| Acetate | 282 (4.7) | 340 (5.7) |
| Propionate | 46 (0.6) | 60 (0.8) |
| Butyrate | 3 (0.03) | <2 (<0.02) |
| DAPI countsd | 1.2 × 105-2.0 × 105 | 0.03 × 106-4.3 × 106 |
| Gene copy no./ml PW | ||
| Bacterial 16S rRNA | 1.95 × 104 ± 0.55 × 104 | 0.83 × 106 ± 0.10 × 106 |
| Archaeal 16S rRNA | 2.66 × 104 ± 0.62 × 104 | 0.28 × 106 ± 0.05 × 106 |
| Bacterial dsrA | 1.95 × 102 ± 1.1 × 102 | 3.12 × 103 ± 0.77 × 103 |
| Archaeal dsrB | 0.66 × 104 ± 0.37 × 104 | 3.19 × 104 ± 0.45 × 104 |
| Sum of bacterial and archaeal dsrAB | 0.68 × 104 ± 0.38 × 104 | 3.50 × 104 ± 0.51 × 104 |
All values are given in mg liter−1. Millimolar concentrations are given in parentheses. Data were provided by Maersk Oil (Copenhagen, Denmark) and are average values from several samplings.
Chloride and sulfate are routinely monitored to control seawater breakthrough.
Ammonium, nitrate, and nitrate are not routinely analyzed at Dan, as no nitrate is added to the injection water.
Range of DAPI counts from Dan and Halfdan PW (data provided by Maersk Oil).
Microbial diversity in production waters.
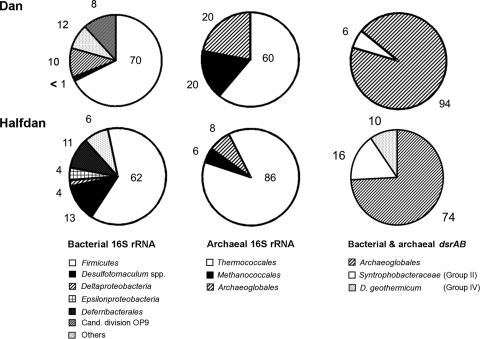
To assess microbial community structure and diversity at Dan and Halfdan, clone libraries for bacterial 16S rRNA, archaeal 16S rRNA, and dsrAB genes were generated from PW samples (Fig. 1).
FIG. 1.
Community composition of Bacteria and Archaea from 16S rRNA gene libraries and dsrAB gene libraries in PW from Dan and Halfdan. Numbers indicate the percentage of clones from each group. The dsrAB gene group names refer to those used by Kaneko et al. (23). Cand., candidate.
(i) Bacteria.
Both PW samples contained a diverse bacterial community whose members were affiliated with nine different phyla (Fig. 2A; see Table S1 in the supplemental material). Five of these phyla (Firmicutes; Gamma-, Delta-, and Epsilonproteobacteria; and Synergistes) were present in both clone libraries, whereas members of the Deferribacterales exclusively showed up in the clone library constructed from Halfdan PW. Clones affiliated with the candidate division OP9, the Alphaproteobacteria and the Thermotogales, were only recovered from Dan PW. Statistical analysis (LIBSHUFF) supported the significant difference of both communities (P value of 0.001). In general, diversity and species richness were higher at Dan than at Halfdan (Table 3) but were in the same range as reported for a hot oil reservoir previously (32). Good's coverage estimates indicated that further screening of clones would probably have resulted in more unique sequences, higher diversity indexes, and richness estimates. The major group of phylotypes recovered from both Dan and Halfdan PW were members of the Firmicutes (70% and 62%) (Fig. 1) that affiliated with clone sequences or isolates previously obtained from other high-temperature oil production systems (11, 43). Bacterial sulfate reducers accounted for the second-most-abundant group of phylotypes. For Dan PW, the majority of these sequences were assigned to deltaproteobacterial sulfate reducers (10%) (Fig. 1) affiliated with the genera Desulfacinum and Desulfovibrio (see Table S1 in the supplemental material). Only a minor fraction of the sequences (<1%) was assigned to Desulfotomaculum geothermicum. Desulfotomaculum-related SRB appeared to be more abundant at Halfdan (13%) (Fig. 1). Clone sequences representing members of the nitrate-reducing, sulfide-oxidizing Epsilonproteobacteria were recovered from both Dan and Halfdan PW in very low numbers, but showed a slightly higher relative abundance in Halfdan PW. In contrast, members of the Deferribacterales were only found at Halfdan and constituted a large fraction in the clone library (11%) (Fig. 1). Several of the described species within this phylum utilize nitrate as an electron acceptor and have been found to play a major role in nitrate-dependent souring control of oil field systems (18, 21).
TABLE 3.
Statistics on microbial diversity in Dan and Halfdan PW
| Field | Target domain or group | Target gene | No. of sequences | No. of OTUa | Good's coverage | Shannon diversity index (H′) | Species richness byb: |
|
|---|---|---|---|---|---|---|---|---|
| Chao1-bcc | ACEd | |||||||
| Dan | Bacteria | 16S rRNA | 68 | 19 | 0.868 | 2.557 | 31.0 (21.7, 73.7) | 31.5 (22.5, 63.5) |
| Archaea | 16S rRNA | 39 | 9 | 0.95 | 2.004 | 9.3 (9.0, 13.8) | 10.5 (9.2, 20.2) | |
| SRP | dsrAB | 29 | 2 | 1 | 0.334 | 2.0 | 2.0 | |
| Halfdan | Bacteria | 16S rRNA | 47 | 14 | 0.830 | 2.212 | 28.0 (16.9, 81.1) | 27.1 (17.2, 68.4) |
| Archaea | 16S rRNA | 36 | 4 | 1 | 0.919 | 4.0 | 4.0 | |
| SRP | dsrAB | 31 | 3 | 1 | 0.774 | 3.0 | 3.0 | |
OTU defined as <3% nucleotide sequence difference (16S rRNA) and <3% amino acid sequence difference (DsrAB).
Mean values are shown, with lower and upper 95% confidence intervals given in parentheses.
A bias-corrected form for the Chao1 (see reference 9).
ACE, abundance-based coverage estimator.
(ii) Archaea.
Compared to the bacterial communities, the archaeal 16S rRNA gene diversity and species richness recovered from clone libraries of Dan and Halfdan PW were quite low (Table 3). The level of diversity was comparable to previous studies on hot oil reservoirs (32). However, there are only a small number of studies assessing the archaeal diversity within these environments, and higher and even lower diversity estimates have been reported as well (11, 30). Both clone libraries included members of the three phyla Thermococcales, Methanothermococcales, and Archaeoglobales (Fig. 2B; see Table S2 in the supplemental material) and were not significantly different from each other (LIBSHUFF, P value of 0.026), even though the dominance of Thermococcales was more pronounced in the library from Halfdan PW than in that from Dan PW (Fig. 1).
(iii) Sulfate reducers.
Construction of dsrAB gene clone libraries yielded a detailed picture of the SRP communities present in Dan and Halfdan PW and confirmed the results obtained from the 16S rRNA-based approach (Fig. 2C; see Table S3 in the supplemental material). dsrAB gene diversity and species richness were low at Dan and only slightly higher at Halfdan (Table 3). Both dsrAB gene clone libraries were dominated by sequences closely affiliated with Archaeaoglobus fulgidus. In addition, representatives of the deltaproteobacterial dsrAB gene sequence groups II and IV (II, Syntrophobacteraceae, including Desulfacinum; IV, D. geothermicum/D. anilinii; groups named according to Kaneko et al. [23]) were recovered from Halfdan PW and accounted for ∼25% of all dsrAB sequences in the library (Fig. 1). At Dan, deltaproteobacterial SRP represented only a minor fraction of the clone library (6%) (Fig. 1) and were exclusively affiliated with the Syntrophobacteraceae. dsrAB sequences assigned to the genera Desulfovibrio and Desulfonatronum were not detected, although they were found in the 16S rRNA gene-based clone libraries constructed from Dan PW.
In situ abundance of Bacteria, Archaea, and sulfate reducers.
Quantification of bacterial and archaeal 16S rRNA genes yielded a total of 4.6 × 104 ± 0.33 × 104 prokaryotic 16S rRNA gene copies per ml PW for Dan and about 25-times-higher copy numbers for Halfdan PW (1.1 × 106 ± 0.15 × 106 ml−1) (Table 2). Archaea slightly dominated the prokaryotic community in Dan PW, as archaeal 16S rRNA gene copies accounted for 57.6% of all 16S rRNA gene copies detected. In contrast, Bacteria clearly dominated the prokaryotic community in PW from Halfdan, with 74.7% of all 16S rRNA gene copies detected. The sums of bacterial and archaeal 16S rRNA gene copies were in approximately the same order of magnitude as average DAPI (4′,6-diamidino-2-phenylindole) counts (Table 2). However, cell numbers calculated from 16S rRNA gene copies would be lower than the DAPI counts (16S rRNA gene copies per cell of 4.13 for Bacteria and 1.77 for Archaea; http://ribosome.mmg.msu.edu/rrndb/). Although DAPI counting was not performed on the same samples, this observation indicated that DNA was not quantitatively extracted from the PW samples.
Two specific qPCR assays for the quantification of deltaproteobacterial and archaeal dsrAB gene copies were developed to estimate the abundance of bacterial and archaeal sulfate reducers. About 5-times-higher bacterial and archaeal dsrAB gene copy numbers were detected in Halfdan PW than in Dan PW (Table 2). The qPCR results corroborated the cloning results as archaeal dsrB genes (i.e., Archaeoglobus relatives) were highly abundant and dominated the dsrAB gene-carrying community. Relative to the sum of prokaryotic 16S rRNA gene copies, SRP in total accounted for 14.5% of the total prokaryotic community at Dan, but only 3.2% at Halfdan (Table 2). The contribution of archaeal SRP to the total prokaryotic community was much lower at Halfdan (2.9%) than at Dan (14.1%), as already indicated from the archaeal 16S rRNA gene-based clone libraries (Fig. 1). Deltaproteobacterial sulfate reducers occurred at low abundance (0.3 to 0.4%) (Table 2) in both PW samples.
Potential activity of sulfate reducers.
Activity of sulfate reducers was only detectable in PW samples from Dan (Table 4). Incubations that were not amended with substrates showed very low activity at 60°C (Dan60) and no detectable activity at 80°C (Dan80) after 2 days of incubation. Activity at 60°C could be slightly stimulated by addition of a substrate mixture and further increased during an incubation period of 42 days in both unamended incubations (Dan60) and incubations amended with a substrate mixture (Dan60S; amended with lactate, acetate, and glucose at 0.5 mM each and 0.01% yeast extract). Substrate addition did not enhance activity in the 80°C incubations. Rates were still close to the detection limit after 42 days, indicating that archaeal sulfate reducers were not stimulated to grow under the given incubation conditions.
TABLE 4.
Sulfate reduction rates determined in Dan PW
| Incubation time (days) | Incubation condition(s) | Sulfate reduction rate (nmol ml−1 day−1) |
|---|---|---|
| 2 | 60°C | 0.07 ± 0.01 |
| 60°C + substrates | 0.2 ± 0.1 | |
| 80°C | BDa | |
| 80°C + substrates | BD | |
| 42 | 60°C | 20.9 ± 2.4 |
| 60°C + substrates | 31.8 ± 3.1 | |
| 80°C | BD | |
| 80°C + substrates | 0.02 ± 0.003 |
BD, below detection. The detection limit was defined as twice the amount of cpm measured for the sample blank (∼60 cpm, which equals 0.01 nmol ml−1 day−1).
Assuming that activity at 60°C was due to bacterial sulfate reducers only, bacterial dsrA gene copy numbers (Table 2) were used to estimate cell-specific sulfate reduction rates between 0.36 and 1.03 pmol cell−1 day−1. These values are in the lower range of cell-specific rates calculated from activity measurements and most probable number (MPN) counts in PW from the Marion Lake oil field, Canada (1.8 to 2.5 pmol cell−1 day−1) (12). However, the latter rates are most likely overestimates due to their dependence on cultivation-based MPN counts that generally underestimate the number of sulfate reducers compared to molecular approaches (fluorescence in situ hybridization and qPCR). In addition, activity of SRP in Dan PW was several orders of magnitude higher than rate estimates from pure culture experiments with thermophiles, e.g., a close relative to Desulfacinum infernum (8) and Desulfotomaculum geothermicum (14).
Identification of bacterial sulfate reducers in enrichments.
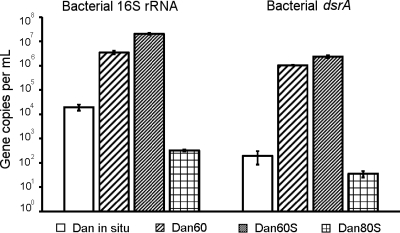
The activation of bacterial sulfate reducers in Dan PW was accompanied by a significant increase in copy numbers of bacterial 16S rRNA and dsrA genes (Dan60 and Dan60S) (Fig. 3), as evidenced from parallel incubations without radioactive tracer. Gene copies increased by 2 to 3 and 4 to 5 orders of magnitude, respectively. The ratio of bacterial dsrA genes to bacterial 16S rRNA genes increased from ca. 1% (Dan in situ) (Fig. 3) to 30% (Dan60) and 11.5% (Dan60S). The lower proportion of dsrAB gene-carrying organisms in the substrate-amended incubation indicated parallel stimulation of non-sulfate-reducing prokaryotes. Archaeal 16S rRNA increased less than 1 order of magnitude, whereas the amount of archaeal dsrB genes decreased compared to their in situ abundance (data not shown).
FIG. 3.
Abundance of bacterial 16S rRNA and dsrA gene copies in PW from Dan prior to incubation (Dan in situ) and after 42 days at 60°C without (Dan60) and with (Dan60S) substrate addition, as well as at 80°C with substrate addition (DAN80S). Mean values and standard deviations were calculated from triplicate (original PW) or single (PW after incubation) DNA extracts. Each extract was run in triplicates within a single qPCR setup.
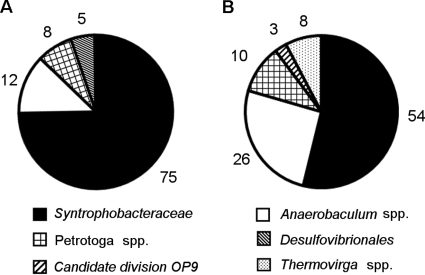
Bacterial 16S rRNA gene-based clone libraries from both Dan60 and Dan60S were dominated by sequences affiliated with thermophilic sulfate reducers of the Syntrophobacteraceae, accounting for 75% and 54% of all sequences detected (Fig. 4). The closest relatives were members of the genera Desulfacinum and Desulfoglaeba (97 to 98% sequence identity). All sequences from Dan60 and Dan60S that were affiliated with these two genera formed 1 distinct OTU within the Syntrophobacteraceae (Fig. 2A). dsrAB gene analysis (30 clone sequences each) supported this finding. All sequences were assigned to the Syntrophobacteraceae, with Desulfacinum infernum as the most closely related organism (Fig. 2C).
FIG. 4.
Community composition of Bacteria from 16S rRNA gene clone libraries in PW from Dan that were incubated for 42 days at 60°C without addition of any substrates (Dan60 [chart A]) and with addition of a substrate mixture (Dan60S [chart B]). Numbers indicate the percentage of clones from each group.
Representatives of non-sulfate-reducing genera recovered from the enrichments were affiliated with environmental clones and isolates of the thermophilic genera Anaerobaculum, Petrotoga, and Thermovirga, which were previously derived from high-temperature oil reservoirs (10, 11, 36). Members of these genera were described to be involved in sulfur cycling by producing sulfide from elemental sulfur and/or thiosulfate. They were more abundant in the clone library obtained from Dan60S than that from Dan60. Additionally, a small fraction of clone sequences in the Dan60S clone library (3%) were affiliated with the candidate division OP9, which lacks cultivated representatives. Archaeal 16S rRNA gene-based clone libraries (20 clones each) yielded only two distinct phylotypes within the Methanosarcinales. These were not detected in clone libraries from the original PW samples.
DISCUSSION
Presence of indigenous, thermophilic sulfate reducers.
Due to elevated temperatures and nutrient limitations in hot oil reservoirs, it has previously been argued that sulfide production in situ mainly occurred near the injection well bore, where the active zone is cooled by contact with injection water (51). It has been concluded that sulfide was produced by mesophilic SRP that were nonindigenous to the reservoir but introduced during exploitation of oil formations or water injection (11, 31). However, only a small fraction of the obtained 16S rRNA gene sequences from Dan and Halfdan PW were affiliated with mesophilic SRP that might have been introduced by seawater injection. In contrast, both thermophilic bacterial and archaeal SRP were present and, in the case of the enriched bacterial SRP from Dan, their activity was demonstrated at elevated temperatures (60°C). Therefore, bacterial SRP, although less abundant than archaeal SRP, might also be active in situ (Table 4) and might contribute significantly to the production of H2S and the concurrent economic problems. From the physiology of oil reservoir-derived prokaryotes (5, 43) and the temperature regimens present at Dan and Halfdan, it was inferred that archaeal SRP were restricted to the reservoirs (∼80°C), whereas bacterial SRP were more likely to be found in liquid phases and biofilms of piping and topside facilities (∼40 to 60°C). The decrease in bacterial 16S rRNA and dsrAB gene copies after 6 weeks of incubation at 80°C (Fig. 3) indicated that bacterial SRP could not survive these conditions. Therefore, they most likely did not originate from the injection, but from the production well-bore.
Potential impact of nitrate on the prokaryotic community and stimulation of nitrate reducers.
Higher absolute cell numbers in Halfdan PW indicated that growth of the prokaryotic community was stimulated by the addition of nitrate. As nitrate reduction is thermodynamically more favorable than sulfate reduction, it results in higher biomass yields by the oxidation of electron donors that can be utilized by both sulfate and nitrate reducers, e.g., volatile fatty acids. Higher biomass in Halfdan PW was accompanied by a lower bacterial species richness, which might be explained by interspecies competition or the direct inhibition by nitrite formed during nitrate reduction (21, 47). Although nitrite concentrations were only <0.01 mM in the PW samples, nitrite might accumulate in biofilms and reach higher concentrations in situ. As shown in field and laboratory studies, nitrite is a strong inhibitor of the dissimilatory sulfite reductase and therefore SRP activity (20, 38, 39).
The very low nitrate concentration in Halfdan PW (<0.5 mg liter−1) compared to the amount of nitrate added to the injection water (65 mg liter−1) indicated that nitrate was consumed near the injection well-bore and/or during water passage by the activity of hNRB and NR-SOB. Although the activity of hNRB and NR-SOB was not specifically assessed within the scope of the present study, sequences affiliated with representatives of both types of nitrate reducers were recovered from 16S rRNA gene clone libraries and included members of the Epsilonproteobacteria (Sulfurospirillum spp. and Arcobacter spp.) and the Deferribacterales (Deferribacter spp.). Interestingly, signatures of the Deferribacterales were exclusively recovered from Halfdan PW, indicating that their growth was favored due to the addition of nitrate. Several thermophilic pure cultures of the genus Deferribacter reduce nitrate to ammonium in the presence of various organic acids (18, 37, 52) and might therefore be potential heterotrophic competitors to SRP. Although whether these organisms were indigenous to the reservoir or part of the biofilms and water phase in the piping remained unknown, it can be inferred from our data that they were stimulated to grow in the nitrate-treated system and that their presence and activity—either by the production of nitrite as an intermediate or the direct competition for substrates—resulted in an inhibition of SRP. The underlying mechanism needs to be approached by, e.g., in situ activity measurements or studies of cocultures of the antagonistic organisms.
The second group of potential competitors, Sulfurospirillum spp. and other members of the Epsilonproteobacteria, have frequently been found in oil fields (7, 17, 53), but little is known about their metabolic features in hot environments. Pure cultures of Sulfurospirillum spp. from the Colville oil field (Saskatchewan, Canada) have been shown to possess both hNRB and NR-SOB activity, giving the first evidence of facultative chemolithotrophy among the Epsilonproteobacteria (21). Their presence at both fields leaves their role in sulfide mitigation unresolved.
Differences in community composition and abundance of SRP.
In contrast to previous molecular studies on seawater-flooded, high-temperature oil reservoirs (6, 32, 40), bacterial and archaeal SRP were identified to form a major fraction of the prokaryotic communities at Dan and Halfdan. Differences in the SRP community composition, a lower relative abundance, and the absence of detectable activity at Halfdan most likely resulted from the addition of nitrate. The lower abundance of SRP relative to the total prokaryotic community at Halfdan was mainly due to a smaller fraction of archaeal SRP, consistently inferred from clone libraries and dsrAB quantification. Apparently, the addition of nitrate primarily affected the archaeal SRP in the reservoir but had only a minor effect on the abundance of bacterial SRP. The abundance and activity of archaeal SRP in high-temperature reservoirs had not been specifically targeted earlier, and therefore they were most likely overlooked in previous studies. SRP in oil production systems were routinely monitored by applying the MPN technique, which requires cultivation and cannot mirror in situ conditions and the particular needs of different physiological groups of SRP (e.g., temperature, nutrients, and syntrophic interactions). Cultivation therefore often favored fast-growing, moderate SRP, e.g., Desulfovibrio spp. (7), that were not necessarily the numerically most abundant or in situ active SRP. To assess differences between fields and to target the effect of nitrate systematically, archaeal SRP need to be detected more specifically, e.g., by the amplification and quantification of archaeal dsrAB genes.
Another major difference between the two fields was the presence of SRP related to the spore-forming, thermophilic genus Desulfotomaculum, which formed a substantial component of the bacterial community in Halfdan PW and dominated the fraction of bacterial SRP. Oil field-derived Desulfotomaculum strains have previously been reported to grow at temperatures of up to 78°C (44), but due to the lack of detectable activity in Halfdan PW, it remained an open question whether these signatures were derived from vegetative cells or inactive spores that did not germinate during incubation.
Other sulfidogenic organisms.
In general, SRP have been considered the main suspects for H2S production in seawater-flooded oil fields. Our results showed that other sulfidogenes within the Clostridiales (genera Thermoanaerobacter and Desulfonispira), the Thermotogales, and the Thermococcales were prominent members of the prokaryotic communities in both fields; they may therefore additionally contribute to hydrogen sulfide production in the system. Hyperthermophilic archaeal sulfur reducers of the Thermococcales were previously shown to be global residents in subsurface oil reservoir environments and other marine hydrothermal systems (29, 40, 48). It has been argued that they possess the capability to desulfurize organosulfur compounds in crude oil generating hydrogen sulfide (49). Due to their extreme requirements in growth temperature and their capability to utilize complex organic substrates as electron donors, members of this phylum would probably not be inhibited by the stimulation of nitrate reducers and should therefore be considered in souring control strategies in the future.
Lack of activity of archaeal SRP.
Archaeoglobus-related SRP were the dominant SRP phylotypes at both Dan and Halfdan. However, their activity could not be demonstrated, although a very sensitive tracer incubation method was applied, which showed activity of bacterial SRP in Dan PW at an incubation temperature of 60°C. As the samples were depressurized during water passage from the reservoir to the separators, it might be argued that only genetic material, but no living archaeal cells from the reservoir, was present in the PW. However, other reservoir-derived Archaeoglobus strains have been previously cultivated from depressurized samples and tolerated atmospheric pressure conditions (5, 29, 50). It is therefore plausible that the archaeal cells remained intact in the PW, but either lacked an essential growth factor in the incubations or were inhibited by compounds in the PW that were less toxic to bacterial SRP. In situ, such inhibition might be coped with by the formation of biofilms that protect against toxic compounds and/or biocides (25). Future studies on the contribution of archaeal SRP to reservoir souring therefore need to target their physiological status in situ, e.g., by applying RNA-targeted techniques like catalyzed reporter deposition-fluorescence in situ hybridization and reverse transcription-PCR.
In conclusion, our comparative analyses of two high-temperature oil production systems showed that the addition of nitrate resulted in the inhibition of SRP activity, a decrease in the relative abundance of SRP, and a general shift of the microbial community, including the appearance of putative hNRB of the Deferribacterales. However, the increase in total microbial biomass and absolute numbers of SRP may aggravate plugging problems and cause resumption of sulfide if nitrate is omitted from the injection water. Further understanding of microbial populations and mechanisms is therefore needed before finally assessing the prospects in nitrate addition in souring control.
Supplementary Material
Acknowledgments
We thank Britta Poulsen, Trine Søgaard Thomsen, and Tove Wiegers for excellent assistance with molecular and analytical techniques. Adam Wieczorek is acknowledged for help in activity measurements and identification of enriched organisms. We thank Jan Larsen (Maersk Oil, Copenhagen, Denmark), who provided access to the sampling site, and the Petroleum Engineering Department for providing chemical data on formation and production water.
This work was funded by the Danish Agency for Science, Technology and Innovation and is part of the innovation consortium “At-Line Monitoring of Bacteria” (AMBA).
Footnotes
Published ahead of print on 2 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abildgaard, L., M. B. Nielsen, K. U. Kjeldsen, and K. Ingvorsen. 2006. Desulfovibrio alkalitolerans sp. nov., a novel alkalitolerant, sulphate-reducing bacterium isolated from district heating water. Int. J. Syst. Evol. Microbiol. 56:1019-1024. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Muyers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashelford, K. E., A. J. Weightman, and J. C. Fry. 2002. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 30:3481-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beeder, J., R. K. Nilsen, J. T. Rosnes, T. Torsvik, and T. Lien. 1994. Archaeoglobus fulgidus isolated from hot North Sea oil field waters. Appl. Environ. Microbiol. 60:1227-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bødtker, G., K. Lysnes, T. Torsvik, E. Ø. Bjørnestad, and E. Sunde. 2009. Microbial analysis of backflowed injection water from a nitrate-treated North Sea oil reservoir. J. Ind. Microbiol. Biotechnol. 36:439-450. [DOI] [PubMed] [Google Scholar]
- 7.Bødtker, G., T. Thorstenson, B.-L. P. Lillebø, B. E. Thorbjørnsen, R. H. Ulvøen, E. Sunde, and T. Torsvik. 2008. The effect of long-term nitrate treatment on SRB activity, corrosion rate and bacterial community composition in offshore water injection systems. J. Ind. Microbiol. Biotechnol. 35:1625-1636. [DOI] [PubMed] [Google Scholar]
- 8.Böttcher, M. E., S. M. Sievert, and J. Kuever. 1999. Fractionation of sulfur isotopes during dissimilatory reduction of sulfate by a thermophilic gram-negative bacterium at 60 degrees C. Arch. Microbiol. 172:125-128. [DOI] [PubMed] [Google Scholar]
- 9.Chao, A., R. L. Chazdon, R. K. Colwell, and T.-J. Shen. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 8:148-159. [Google Scholar]
- 10.Dahle, H., and N.-K. Birkeland. 2006. Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int. J. Syst. Evol. Microbiol. 56:1539-1545. [DOI] [PubMed] [Google Scholar]
- 11.Dahle, H., F. Garshol, M. Madsen, and N.-K. Birkeland. 2008. Microbial community structure analysis of produced water from a high-temperature North Sea oil-field. Antonie van Leeuwenhoek 93:37-49. [DOI] [PubMed] [Google Scholar]
- 12.Davidova, I., M. S. Hicks, P. M. Fedorak, and J. M. Suflita. 2001. The influence of nitrate on microbial processes in oil industry production waters. J. Ind. Microbiol. Biotechnol. 27:80-86. [DOI] [PubMed] [Google Scholar]
- 13.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Detmers, J., V. Brüchert, K. S. Habicht, and J. Kuever. 2001. Diversity of sulfur isotope fractionations by sulfate-reducing prokaryotes. Appl. Environ. Microbiol. 67:888-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunsmore, B. C., J. Youldon, D. R. Thrasher, and I. Vance. 2006. Effects of nitrate treatment on a mixed species, oil field microbial biofilm. J. Ind. Microbiol. Biotechnol. 33:454-462. [DOI] [PubMed] [Google Scholar]
- 16.Foesel, B. U., A. Gieseke, C. Schwermer, P. Stief, L. Koch, E. Cytryn, J. R. de la Torré, J. van Rijn, D. Minz, H. L. Drake, and A. Schramm. 2008. Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol. Ecol. 63:192-204. [DOI] [PubMed] [Google Scholar]
- 17.Grabowski, A., O. Nercessian, F. Fayolle, D. Blanchet, and C. Jeanthon. 2005. Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol. Ecol. 54:427-443. [DOI] [PubMed] [Google Scholar]
- 18.Greene, A. C., B. K. C. Patel, and A. J. Sheehy. 1997. Deferribacter thermophilus gen. nov., sp. nov., a novel thermophilic manganese- and iron-reducing bacterium isolated from a petroleum reservoir. Int. J. Syst. Bacteriol. 47:505-509. [DOI] [PubMed] [Google Scholar]
- 19.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 20.Hubert, C., M. Nemati, G. E. Jenneman, and G. Voordouw. 2005. Corrosion risk associated with microbial souring control using nitrate or nitrite. Appl. Microbiol. Biotechnol. 68:272-282. [DOI] [PubMed] [Google Scholar]
- 21.Hubert, C., and G. Voordouw. 2007. Oil field souring control by nitrate-reducing Sulfurospirillum spp. that outcompete sulfate-reducing bacteria for organic electron donors. Appl. Environ. Microbiol. 73:2644-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juretschko, S., G. Timmermann, M. Schmid, K.-H. Schleifer, A. Pommerening-Röser, H.-P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko, R., T. Hayashi, M. Tanahashi, and T. Naganuma. 2007. Phylogenetic diversity and distribution of dissimilatory sulfite reductase genes from deep-sea sediment cores. Mar. Biotechnol. 9:429-436. [DOI] [PubMed] [Google Scholar]
- 24.Kjeldsen, K. U., B. V. Kjellerup, K. Egli, B. Frølund, P. H. Nielsen, and K. Ingvorsen. 2007. Phylogenetic and functional diversity of bacteria in biofilms from metal surfaces of an alkaline district heating system. FEMS Microbiol. Ecol. 61:384-397. [DOI] [PubMed] [Google Scholar]
- 25.LaPaglia, C., and P. L. Hartzell. 1997. Stress-induced production of biofilm in the hyperthermophile Archaeoglobus fulgidus. Appl. Environ. Microbiol. 63:3158-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen, J., T. L. Skovhus, M. Agerbæk, T. R. Thomsen, and P. H. Nielsen. 2006. Bacterial diversity study applying novel molecular methods on Halfdan produced waters, paper 06668. Corrosion NACExpo 2006, San Diego, CA. NACE International, Houston, TX.
- 27.Larsen, J., T. L. Skovhus, A. M. Saunders, B. Højris, and M. Agerbæk. 2008. Molecular identification of MIC bacteria from scale and produced water: similarities and differences, paper 08652. Corrosion NACExpo 2008, New Orleans, LA. NACE International, Houston, TX.
- 28.Larsen, J., S. Zwolle, B. V. Kjellerup, B. Frølund, J. L. Nielsen, and P. H. Nielsen. 2005. Identification of bacteria causing souring and biocorrosion in the Halfdan Field by application of new molecular techniques, paper 05629. Corrosion NACExpo 2005, Houston, TX. NACE International, Houston, TX.
- 29.L'Haridon, S., A. L. Reysenbach, P. Glenat, D. Prieur, and C. Jeanthon. 1995. Hot subterranean biosphere in a continental oil reservoir. Nature 377:223-224. [PubMed] [Google Scholar]
- 30.Li, H., S.-Z. Yang, and B.-Z. Mu. 2007. Phylogenetic diversity of the archaeal community in a continental high-temperature, water-flooded petroleum reservoir. Curr. Microbiol. 55:382-388. [DOI] [PubMed] [Google Scholar]
- 31.Li, H., S.-Z. Yang, B.-Z. Mu, Z.-F. Rong, and J. Zhang. 2006. Molecular analysis of the bacterial community in a continental high-temperature and water-flooded petroleum reservoir. FEMS Microbiol. Lett. 257:92-98. [DOI] [PubMed] [Google Scholar]
- 32.Li, H., S.-Z. Yang, B.-Z. Mu, Z.-F. Rong, and J. Zhang. 2007. Molecular phylogenetic diversity of the microbial community associated with a high-temperature petroleum reservoir at an offshore oilfield. FEMS Microbiol. Ecol. 60:74-84. [DOI] [PubMed] [Google Scholar]
- 33.Lipp, J. S., Y. Morono, F. Inagaki, and K.-U. Hinrichs. 2008. Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454:991-994. [DOI] [PubMed] [Google Scholar]
- 34.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda-Tello, E., M.-L. Fardeau, C. Joulian, M. Magot, P. Thomas, J.-L. Tholozan, and B. Ollivier. 2007. Petrotoga halophila sp. nov., a thermophilic, moderately halophilic, fermentative bacterium isolated from an offshore oil well in Congo. Int. J. Syst. Evol. Microbiol. 57:40-44. [DOI] [PubMed] [Google Scholar]
- 37.Miroshnichenko, M. L., A. I. Slobodkin, N. A. Kostrikina, S. L'Haridon, O. Nercessian, S. Spring, E. Stackebrandt, E. A. Bonch-Osmolovskaya, and C. Jeanthon. 2003. Deferribacter abyssi sp. nov., an anaerobic thermophile from deep-sea hydrothermal vents of the Mid-Atlantic Ridge. Int. J. Syst. Evol. Microbiol. 53:1637-1641. [DOI] [PubMed] [Google Scholar]
- 38.Myhr, S., B.-L. P. Lillebø, E. Sunde, J. Beeder, and T. Torsvik. 2002. Inhibition of microbial H2S production in an oil reservoir model column by nitrate injection. Appl. Microbiol. Biotechnol. 58:400-408. [DOI] [PubMed] [Google Scholar]
- 39.Nemati, M., T. J. Mazutinec, G. E. Jenneman, and G. Voordouw. 2001. Control of biogenic H2S production with nitrite and molybdate. J. Ind. Microbiol. Biotechnol. 26:350-355. [DOI] [PubMed] [Google Scholar]
- 40.Orphan, V. J., L. T. Taylor, D. Hafenbradl, and E. F. Delong. 2000. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 66:700-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruesse, E., C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies, and F. O. Gloeckner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raskin, L., J. M. Stromley, B. E. Rittmann, and D. A. Stahl. 1994. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60:1232-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rees, G. N., G. S. Grassia, A. J. Sheehy, P. P. Dwivedi, and B. K. C. Patel. 1995. Desulfacinum infernum gen. nov., sp. nov., a thermophilic sulfate-reducing bacterium from a petroleum reservoir. Int. J. Syst. Bacteriol. 45:85-89. [Google Scholar]
- 44.Rosnes, J. T., T. Torsvik, and T. Lien. 1991. Spore-forming thermophilic sulfate-reducing bacteria isolated from North Sea oil field waters. Appl. Environ. Microbiol. 57:2302-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scholle, P. A., T. Albrechtsen, and H. Tirsgaard. 1998. Formation and diagenesis of bedding cycles in uppermost Cretaceous chalks of the Dan Field, Danish North Sea. Sedimentology 45:223-243. [Google Scholar]
- 47.Schwermer, C. U., G. Lavik, R. M. M. Abed, B. Dunsmore, T. G. Ferdelman, P. Stoodley, A. Gieseke, and D. de Beer. 2008. Impact of nitrate on the structure and function of bacterial biofilm communities in pipelines used for injection of seawater into oil fields. Appl. Environ. Microbiol. 74:2841-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slobodkin, A. I., C. Jeanthon, S. L'Haridon, T. Nazina, M. Miroshnichenko, and E. Bonchosmolovskaya. 1999. Dissimilatory reduction of Fe(III) by thermophilic bacteria and archaea in deep subsurface petroleum reservoirs of Western Siberia. Curr. Microbiol. 39:99-102. [DOI] [PubMed] [Google Scholar]
- 49.Stetter, K. O., and R. Huber. 1998. The role of hyperthermophilic prokaryotes in oil fields, p. 369-375. In C. R. Bell, M. Brylinsky, and P. Johnston-Green (ed.), Microbial biosystems: new frontiers. Proceedings of the 8th International Symposium on Microbial Ecology (ISME-8), Halifax, Nova Scotia, Canada.
- 50.Stetter, K. O., R. Huber, E. Blochl, M. Kurr, R. D. Eden, M. Fielder, H. Cash, and I. Vance. 1993. Hyperthermophilic archaea are thriving in deep North Sea and Alaskan oil reservoirs. Nature 365:743-745. [Google Scholar]
- 51.Sunde, E., and T. Torsvik. 2005. Microbial control of hydrogen sulfide production in oil reservoirs, p. 201-213. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington, DC.
- 52.Takai, K., H. Kobayashi, K. H. Nealson, and K. Horikoshi. 2003. Deferribacter desulfuricans sp. nov., a novel sulfur-, nitrate- and arsenate-reducing thermophile isolated from a deep-sea hydrothermal vent. Int. J. Syst. Evol. Microbiol. 53:839-846. [DOI] [PubMed] [Google Scholar]
- 53.Telang, A. J., S. Ebert, J. M. Foght, D. W. S. Westlake, G. E. Jenneman, D. Gevertz, and G. Voordouw. 1997. Effect of nitrate injection on the microbial community in an oil field as monitored by reverse sample genome probing. Appl. Environ. Microbiol. 63:1785-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vance, I., and D. R. Thrasher. 2005. Reservoir souring: mechanisms and prevention, p. 123-142. In B. Ollivier and M. Magot (ed.), Petroleum microbiology. ASM Press, Washington, DC.
- 55.Wagner, M., A. J. Roger, J. L. Flax, G. A. Brusseau, and D. A. Stahl. 1998. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 180:2975-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.