Abstract
Cyanuric acid, a metabolic intermediate in the degradation of many s-triazine compounds, is further metabolized by cyanuric acid hydrolase. Cyanuric acid also accumulates in swimming pools due to the breakdown of the sanitizing agents di- and trichloroisocyanuric acid. Structurally stable cyanuric acid hydrolases are being considered for usage in pool water remediation. In this study, cyanuric acid hydrolase from the thermophile Moorella thermoacetica ATCC 39073 was cloned, expressed in Escherichia coli, and purified to homogeneity. The recombinant enzyme was found to have a broader temperature range and greater stability, at both elevated and low temperatures, than previously described cyanuric acid hydrolases. The enzyme had a narrow substrate specificity, acting only on cyanuric acid and N-methylisocyanuric acid. The M. thermoacetica enzyme did not require metals or other discernible cofactors for activity. Cyanuric acid hydrolase from M. thermoacetica is the most promising enzyme to use for cyanuric acid remediation applications.
s-Triazine compounds have diverse applications as herbicides, resins, and disinfectants. The s-triazine herbicides, such as atrazine, help to promote high-yield, sustainable agriculture. Melamine, or triamino-s-triazine, is a high-volume industrial chemical. Melamine-based polymers have outstanding thermosetting properties, ideal for their use in kitchen utensils and plates, as high-pressure laminates such as Formica, and as whiteboards. Di- and tri-chloroisocyanuric acids find widespread application as disinfectants, algicides, and bactericides. The chlorinated isocyanuric acids are used in wastewater treatment, in the textile industry as bleaching compounds, and in preventing and curing diseases in husbandry and fisheries. A major use of these compounds is for swimming pool chlorination. They have outstanding performance for maintaining a high, stable chlorine content by dissolving slowly in water, allowing a continuous metered dosing of chlorine.
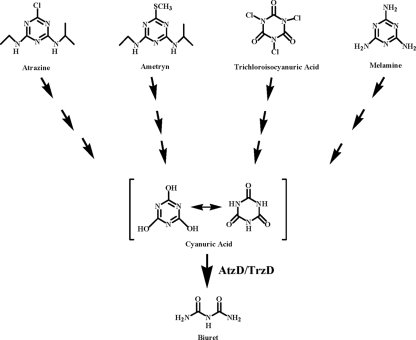
Degradation of these and other s-triazine compounds results in the production of cyanuric acid (Fig. 1). Cyanuric acid has come under increased scrutiny because of its potential involvement in comediating toxicity resulting from the ingestion of melamine (10). Recently, melamine has been found in adulterated pet food and baby formula. Melamine and its metabolite cyanuric acid cocrystallize at low concentrations and are implicated in acute renal failure in cats that have consumed adulterated food products (10). Cyanuric acid degradation is also of interest from the perspective of environmental remediation. The use of di- or trichloroisocyanuric acid in pool water results in spontaneous chemical dechlorination that disinfects the water but also produces, as a by-product, large amounts of cyanuric acid. High levels of cyanuric acid perturb the equilibrium, thus preventing dechlorination by additional chlorinated isocyanuric acid, such that disinfection is not achieved. As a result, swimming pools must be emptied and refilled, using water and causing discharge issues. It would be desirable to remediate pool water in situ, conserving water, saving money, and extending pool water use. In this context, there is a need to better understand cyanuric acid degradation and to identify highly stable biological catalysts to use for this purpose.
FIG. 1.
Atrazine, ametryn, trichloroisocyanuric acid, and melamine are all metabolized via cyanuric acid that is transformed to biuret by the action of cyanuric acid hydrolases.
Microbial enzymatic degradation of cyanuric acid has been studied previously (3, 4, 8, 18). Two distinct but homologous enzymes, AtzD from Pseudomonas sp. strain ADP (8) and TrzD from Pseudomonas sp. strain NRRLB-12227 (now called Acidovorax avenae subsp. citrulli) (11), have been studied in detail. These enzymes, known as cyanuric acid hydrolases, catalyze the conversion of cyanuric acid to biuret (Fig. 1). Biuret is not considered toxic to humans and degrades more readily than cyanuric acid.
Barbiturase is the only protein known to be homologous to cyanuric acid hydrolase that has a defined and different physiological function. Barbiturase catalyzes the conversion of barbituric acid to ureidomalonic acid in organisms that catabolize pyrimidines by the oxidative pathway. Barbiturase is unstable at 4°C in the absence of ethylene glycol and dithiothreitol (DTT). Furthermore, activity is completely lost when the protein is maintained at 55°C for 30 min (20). AtzD and TrzD are relatively stable at 4°C, but they lose activity when frozen (our unpublished data). Moreover, the thermostability properties of AtzD and TrzD are not well studied, but these enzymes are derived from mesophilic bacteria. In this context, we initiated a search to identify a stable cyanuric acid hydrolase. Enzymes that are more stable in response to temperature changes are more stable in response to many environmental factors. Thus, a thermostable enzyme would be most applicable to pool water and other remediation efforts.
We employed bioinformatic techniques that identified a cyanuric acid hydrolase homolog in Moorella thermoacetica ATCC 39073, an anaerobic, acetogenic bacterium that is able to grow at 65°C. The gene was cloned into E. coli, the protein was expressed at high levels, the recombinant E. coli strain degraded cyanuric acid, and the enzyme was obtained in homogeneous form by a convenient one-step purification. The enzyme's function as a cyanuric acid hydrolase was confirmed, and it was shown to be significantly more stable than other known members of the cyanuric acid protein family.
MATERIALS AND METHODS
Chemicals.
5-Nitrobarbituric acid, 5-methylbarbituric acid, 2-methylamino-4,6-dihydroxy-s-triazine, 1,3,5-trimethoxy-s-triazine, methyl isocyanurate, 2-(3-methoxypropylamino)-4,6-dihydroxy-s-triazine, triphenoxy-s-triazine, 2-secbutyl-4,6-dihydroxy-s-triazine, N-phenyl ammelide, 2-ethylamino-4,6-dihydroxy-s-triazine, tri-isopropoxy-s-triazine, trimethyl isocyanurate, and 1,3-dimethyl isocyanurate were prepared as previously described (19). Hexazinone, 4,6-dihydroxy-2-mercaptopyrimidine, and 4,6-dihydroxy-5-nitro-pyrimidine were purchased from Sigma-Aldrich.
Bacterial strains and growth conditions.
M. thermoacetica ATCC 39073, formerly known as Clostridium thermaceticum (6), was obtained from the ATCC and grown anaerobically at 55°C in serum bottles (125 ml) or in rubber-stoppered, screw-cap infusion flasks (1,200 ml; Muller-Krempel, Bulach, Switzerland). The bacterium was grown in an anaerobic medium containing the following per liter: 5 g yeast extract, 5 g tryptone, 6.4 g Na2HPO4, 6.1 g NaH2PO4, 0.4 g NH4Cl, 0.3 g MgCl2·6H2O, 0.05 g CaCl2·2H2O, 0.04 g Fe(NH4)(SO4)2·6H2O, and 10 ml of a mineral mix which was previously described (12). After autoclaving, 18 g glucose and 0.2 mg each of biotin, cyanocobalamin, flavin mononucleotide, folic acid, nicotinic acid, p-aminobenzoic acid, and thiamine pyrophosphate were added individually. Immediately before inoculation, 10 ml of a reducing solution, containing 0.36 g of Na2S·9H2O and 0.36 g of cysteine HCl, was added. The pH of all media was adjusted to 6.6 prior to the addition of 2 g/liter NaHCO3. The medium was flushed with oxygen-free N2-CO2 (80:20 [vol/vol]) for 30 min prior to sealing of tubes with butyl rubber stoppers.
Protein identification and comparisons.
The BLAST algorithm was used to identify homologs of AtzD, TrzD, and barbiturase. Sequence alignments were done with Clustal W (21), and protein trees were constructed with the maximum likelihood algorithm in PHYLIP (5). The protein sequences (GenBank accession numbers) used are as follows: Pseudomonas sp. strain ADP AtzD (AAK50331), Pseudomonas sp. strain NRRLB-12227 (now called Acidovorax avenae subsp. citrulli) TrzD (AAC61577), Rhodococcus erythropolis barbiturase (CAC86669), Chelatobacter heintzii (AAK52819), Moorella thermoacetica ATCC 39073 (YP_430955), Bradyrhizobium japonicum USDA 110 (BAC52546), and Arthrobacter sp. strain AD25 (ABK41866).
Cloning and expression.
Total genomic DNA was extracted from cell pellets of M. thermoacetica ATCC 39073, Pseudomonas sp. strain NRRLB-12227, and Pseudomonas sp. strain ADP, as previously described (13, 22). Open reading frame (ORF) Moth_2120 from M. thermoacetica ATCC 39073 was PCR amplified, using primers 5′-GCGAATTCCATATGCAAAAAGTTGAAGTCTT-3′ and 5′-GCCAAGCTTCTACACCCTGGCAATAACAG-3′ (NdeI and HindIII restriction enzyme sites, respectively, are underlined). The gene was cloned into a pET28b+ vector (Novagene, Madison, WI). The resulting vector, pET28b+::Moth_2120, was transformed into E. coli BRL21(DE3)pLysS and grown at 37°C in Luria-Bertani medium (13) containing 50 μg kanamycin and 25 μg chloramphenicol per ml. When the culture reached an optical density at 600 nm of 0.5, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the induced cells were grown overnight at 30°C. A similar cloning, expression, and purification system was used to clone and His tag the trzD gene from Pseudomonas sp. strain NRRLB-12227 and atzD from Pseudomonas sp. strain ADP to obtain the encoded enzymes for comparison. Primers for cloning atzD were atzdf-14ndeI (5′-CGCGGGAATTCGACATATGTATCACATCG-3′) and atzdr1101hindIII (5′-CAGAGACCGAAAGCTTGGCAGGTC-3′). Primers for trzD were trzdf-13ndeI (5′-CAAGGAGTCCCATATGCAAGCGC-3′) and trzdr1108hindIII (5′-GACATAGGTAAGCTTCCTCTATTAAGC-3′).
Enzyme purification.
For enzyme purification, 2 liters of E. coli BRL21(DE3)(pET28b+::Moth_2120) cells were grown as described above. The culture was centrifuged at 10,000 × g for 20 min at 4°C, and the pellet was resuspended (1 ml per gram of cells) in 100 mM potassium phosphate buffer, pH 7.0. The cell suspension was passed three times through a chilled Amicon French pressure cell, operated at 140 MPa, and the crude cell lysate was centrifuged at 18,000 × g for 90 min at 4°C. The enzyme was purified using a 5-ml HiTrap chelating HP column (Amersham Pharmacia Biotech AB, Uppsala, Sweden) and a Pharmacia FPLC LKB system (Amersham Pharmacia Biotech AB, Uppsala, Sweden). The column was prepared per the manufacturer's instructions and equilibrated with 60 ml of 100 mM potassium phosphate buffer, pH 7.0. After loading of the protein, the column was washed with a series of step gradients, consisting of 0.05 M, 0.1 M, 0.25 M, and 0.5 M imidazole in 100 mM potassium phosphate buffer (pH 7.0). Purified protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis for purity, with subunit molecular weight determination by comparison to broad-range standards (Bio-Rad Laboratories, Hercules, CA). The purified protein was dialyzed at 4°C against 100 mM potassium phosphate buffer (pH 7.0) to remove imidazole.
Enzyme assay and kinetic constant determinations.
Enzyme activity was monitored on a Beckman DU 640 spectrophotometer (Beckman Coulter, Fullerton, CA). Cyanuric acid and barbituric acid concentrations were measured at 214 nm (extinction coefficient = 9,200 cm−1 M−1) and 256 nm, respectively. Reactions were carried out in 0.5 ml 25 mM Tris-HCl buffer, pH 8.0, with 100 μM cyanuric acid or barbituric acid at 30°C. Reactions were initiated by the addition of the enzyme. Kinetic parameters were determined by obtaining rates at cyanuric acid concentrations ranging from 10 to 110 μM. The data were plotted using Lineweaver-Burk plots.
Conversion of cyanuric acid to biuret was confirmed using high-pressure liquid chromatography (HPLC). Control samples, standards, and enzymatic reactions were set up in 5 mM phosphate buffer (pH 7.0). Samples were analyzed by HPLC, using a Hewlett-Packard HP 1100 system equipped with a photodiode array detector interfaced to an HP Chemstation. A mixed-mode C8/anion 7μ column (250 by 4.6 mm) (Alltech, Deerfield, IL) was used with an isocratic mobile phase consisting of 95% methanol and 5% 5 mM phosphate buffer (pH 7.0).
Substrate specificity analysis.
The following 17 compounds which are structurally analogous to cyanuric acid were tested as substrates: 5-nitrobarbituric acid, 5-methylbarbituric acid, 2-methylamino-4,6-dihydroxy-s-triazine, 1,3,5-trimethoxy-s-triazine, 2-(3-methoxy propylamino)-4,6-dihydroxy-s-triazine, triphenoxy-s-triazine, 2-secbutyl-4,6-dihydroxy-s-triazine, N-phenyl ammelide, 2-ethylamino-4,6-dihydroxy-s-triazine, tri-isopropoxy-s-triazine, trimethyl isocyanurate, 1,3-dimethyl isocyanurate, hexazinone, 4,6-dihydroxy-2-mercaptopyrimidine, 4,6-dihydroxy-5-nitro-pyrimidine, barbituric acid, and methyl isocyanurate. The compounds (100 μM) were incubated with 50 μg of purified enzyme in 25 mM Tris-HCl buffer (pH 8.0) for 30 min. Changes in the UV spectra were recorded. In cases where changes in the spectra were detected, an appropriate wavelength with absorbance differences between the substrate and product was chosen for further kinetic study. For N-methylisocyanurate, absorbance was measured at 214 nm (extinction coefficient = 9,500 cm−1 M−1).
Effects of pH and temperature on enzymatic activity.
The optimum pH of the enzyme was determined at 30°C with the following buffers: 25 mM NaHCO3/Na2CO3, pH 5 to 7; 25 mM Tris-HCl, pH 7 to 9; NaHCO3/Na2CO3, pH 9.5 to 10.5. The temperature optimum was determined by assaying the enzyme activity in 25 mM Tris-HCl, pH 8.0, at temperatures ranging from 25°C to 70°C.
Metal chelator effects.
Purified protein was incubated at room temperature with 5 mM 1,10-phenanthroline, 8-hydroxyquinoline-5-sulfonic acid, or EDTA for 24 h. PD-10 desalting columns (GE Healthcare) were used, per the manufacturer's instructions, to remove the chelator from the enzyme. Enzymatic activity was measured with cyanuric acid as the substrate, as described above.
Effects of metals.
The enzyme was incubated with a 0.2 mM concentration of each metal salt for 60 min at 4°C. For each treatment, specific activity was determined as described above. The final metal concentration in the assay buffer was 0.1 mM. The following salts were also tested at the final concentrations indicated: 1 mM CaCl2 and 2 mM MgCl2.
Metal analysis.
The enzyme was hydrolyzed with 6 M hydrochloric acid at 110°C for 24 h under vacuum. Metal content and protein concentrations were determined as previously described (2).
RESULTS
Identification of ORF Moth_2120.
The BLAST algorithm was used to search GenBank for amino acid sequences homologous to AtzD, TrzD, and barbiturase. A subgroup of the sequences identified were found to cluster most closely to the cyanuric acid hydrolases (Fig. 2). The protein from Chelatobacter heintzii was identical to TrzD, and the protein from Arthrobacter AD25 was 99% identical to AtzD. The protein from Bradyrhizobium japonicum USDA 110 was 56%, 66%, and 44% identical to TrzD, AtzD, and barbiturase, respectively. A predicted protein sequence derived from the genome sequence of Moorella thermoacetica ATCC 39073 (gi83590946; locus tag Moth_2120) was 64%, 57%, and 48% identical to TrzD, AtzD, and barbiturase, respectively. This revealed two divergent homologs to known cyanuric acid hydrolases, but the known ability of M. thermoacetica to grow at elevated temperatures made this protein more attractive for further study.
FIG. 2.
Phylogenetic tree showing relatedness of cyanuric acid hydrolases known or identified in different bacteria. Branch lengths are proportional to the number of amino acid substitutions, as indicated by the scale bar below the tree.
Protein characterization and determination of catalytic activity.
The putative cyanuric acid hydrolase homolog encoded by ORF Moth_2120 from M. thermoacetica ATCC 39073 was cloned and expressed in E. coli as described above. The resultant E. coli strain, unlike the wild-type strain, showed clearing zones on agar plates containing a suspension of 130 mM cyanuric acid. The recombinant protein was expressed with an N-terminal His tag that allowed its purification to homogeneity in a single step. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed a single protein band corresponding to 40 kDa. This agrees with the calculated molecular mass of 38.9 kDa for a polypeptide encoded by this ORF.
The purified enzyme in vitro hydrolyzed cyanuric acid with a specific activity of 15.7 μmol/min/mg but did not show detectable activity with barbituric acid, even with the use of 50 μg of protein in a single assay. The product of the reaction was confirmed to be biuret via coelution with standards in HPLC. These results indicate that ORF Moth_2120 is a cyanuric acid hydrolase. Consistent with this view, the enzyme showed a physiologically reasonable Km value of 110 μM with cyanuric acid as the substrate. The kcat was determined to be 10.6 s−1, with a kcat/Km value of 1.0 × 105 M−1 s−1. These can be compared to the published values for Km and kcat of 57 μM and 6.8 s−1 for AtzD (8) and of 50 μM and 250 s−1 for TrzD (11).
Substrate specificity.
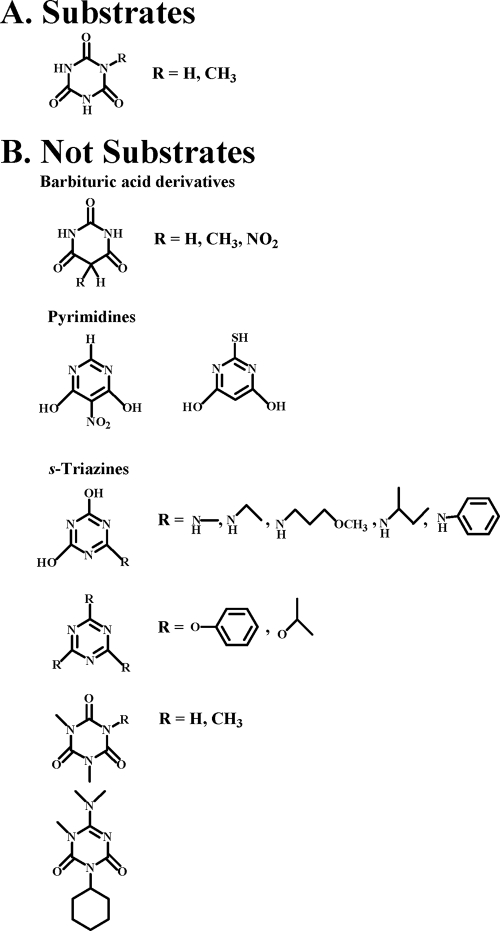
Seventeen compounds that are structurally analogous to cyanuric acid were tested as substrates for hydrolysis by the Moorella cyanuric acid hydrolase (Fig. 3). Of these, only methyl isocyanurate was a substrate, with a rate of 0.13 μmol/min/mg, slightly less than 1% of the specific activity of the preferred substrate cyanuric acid. Based on these analyses, the substrates for this enzyme appear to require three nitrogen atoms in a six-member ring, as is typical of s-triazine compounds, three carbonyl oxygen atoms on the carbons of the ring, and at least two of the ring nitrogens having hydrogens, while the other nitrogen atom can be bonded to a hydrogen or a methyl group.
FIG. 3.
Substrate specificity of cyanuric acid hydrolase from M. thermoacetica ATCC 39073. (A) Compounds that are substrates. (B) Compounds that are not substrates.
Optimum pH and temperature of the enzyme.
The activity of cyanuric acid hydrolase from M. thermoacetica ATCC 39073 was determined at pH values ranging from 5.5 to 10.5. Maximum activity was achieved at pH 8.0. This pH optimum agrees with that of the other characterized cyanuric acid hydrolases (8, 11). The effects of temperature on enzyme activity were also determined in the range of 25 to 70°C. The activity of the enzyme steadily increased with temperature, reaching a maximum, 24 μmol per min per mg protein, at 70°C. Temperatures greater than 70°C could not be assayed due to the limitations of our equipment. The optimum temperature was determined to be 70°C, which is much higher than the 45 to 50°C range reported for TrzD (11).
Temperature stability of the enzyme.
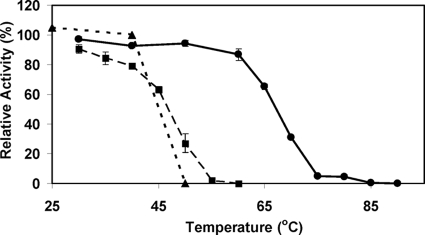
The thermostability of the enzyme at pH 8.0 was also tested by incubating the enzyme for 30 min prior to determining the activity (Fig. 4). At 50°C, 95% of the initial activity remained with the Moorella enzyme, while at 70°C, 30% of the initial activity remained. This contrasts with the case for other members of this family of proteins. AtzD had no loss of activity at 40°C, but all activity was lost when it was incubated at 50°C. Likewise, TrzD lost most of its activity when the temperature was increased to 50°C (Fig. 4). Barbiturase is also thermally sensitive and was reported to have no residual activity after a 30-min incubation at 55°C (20). These results suggest that the Moorella cyanuric acid hydrolase is structurally and catalytically more stable than other members of this family of enzymes.
FIG. 4.
Stability of enzyme activity versus temperature at pH 8.0 for purified cyanuric acid hydrolase from M. thermoacetica ATCC 39073 (•) compared with TrzD (▪) and AtzD (▴). Enzymes were maintained at the temperature indicated for 30 min prior to assay under standard conditions.
The storage stability of the enzyme at 4°C and −80°C in the presence and absence of various additives was also examined (Table 1). Among the additives were 10% ethylene glycol, 0.2 mM DTT, 25% glycerol, and 25% polyethylene glycol 4000 (PEG 4000). The enzymatic activity was monitored every 2 weeks for a total of 8 weeks. The enzyme without additives showed no loss of activity after 8 weeks at both storage temperatures. This suggested that the enzyme was stable at low temperatures. Though the enzymatic activity was unchanged in the frozen sample after 8 weeks, the specific activity of the sample stored at 4°C increased to 142% and 136% of initial specific activity compared to those in the samples without additives and with 0.2 mM DTT, respectively. The cause for this increase is unknown at this time, but similar observations have been seen with other enzymes (9, 10). The addition of 25% PEG 4000 had a stimulatory effect upon activity prior to storage, increasing the activity by 270%. However, the activity of the PEG-stimulated enzyme subsequently decreased to around 1.4 to 1.6 times the initial activity without additives over the 8-week time course at both temperatures. All other additives, including 10% ethylene glycol and 25% glycerol, had little or no positive effect on sustaining activity.
TABLE 1.
Storage stability of purified cyanuric acid hydrolase from M. thermoacetica ATCC 39073 at pH 8.0 as a function of time and additivesa
| Temp (°C) | Additive | Activity (μmol per min per mg protein)b |
||||
|---|---|---|---|---|---|---|
| 0 wk | 2 wk | 4 wk | 6 wk | 8 wk | ||
| 4 | Enzyme only | 11.9 ± 0.2 | 12.5 ± 0.2 | 15.9 ± 0.7 | 16.6 ± 0.9 | 16.9 ± 1.0 |
| 10% Ethylene glycol | 12.3 ± 1.2 | 13.8 ± 0.3 | 11.3 ± 0.3 | 10.5 ± 0.9 | 11.4 ± 0.6 | |
| 0.2 mM DTT | 13.4 ± 0.9 | 15.3 ± 0.5 | 17.7 ± 0.7 | 14.8 ± 0.3 | 18.3 ± 0.6 | |
| 25% Glycerol | 14.3 ± 1.9 | 13.4 ± 0.1 | 12.2 ± 0.5 | 9.8 ± 0.5 | 12.0 ± 1.3 | |
| 25% PEG 4000 | 31.8 ± 1.8 | 20.0 ± 0.2 | 31.0 ± 0.5 | 22.2 ± 0.6 | 18.7 ± 0.1 | |
| −80 | Enzyme only | 11.9 ± 0.2 | 14.9 ± 0.1 | 14.3 ± 0.3 | 12.3 ± 0.4 | 11.7 ± 1.1 |
| 10% Ethylene glycol | 12.3 ± 1.2 | 13.3 ± 0.3 | 14.1 ± 0.3 | 13.6 ± 0.8 | 13.0 ± 0.2 | |
| 0.2 mM DTT | 13.4 ± 0.9 | 14.7 ± 0.2 | 14.3 ± 0.3 | 12.2 ± 0.3 | 11.8 ± 0.3 | |
| 25% Glycerol | 14.3 ± 1.9 | 14.0 ± 0.2 | 14.0 ± 0.9 | 10.7 ± 1.0 | 11.0 ± 1.2 | |
| 25% PEG 4000 | 31.8 ± 1.8 | 21.4 ± 0.7 | 22.0 ± 0.4 | 21.0 ± 0.3 | 17.1 ± 0.4 | |
Values are means ± standard errors for three replicates.
Activity was measured at 25°C by using a standard assay.
Effects of metal ions and chelators.
The effects of divalent metals upon native enzyme activity were monitored (Table 2). Zn(II) had the greatest inhibitory effect, with slight decreases in activity observed in the presence of Cu(II) and Ni(II). At higher concentrations, Ca(II) and Mg(II) caused slight increases, to 121% and 119% of native activity, respectively. Chelators, including EDTA, o-phenanthroline, and 8-hydroxyquinoline-5-sulfonic acid, failed to alter activity, even with 24-h incubations. These data suggested that either a metal could not be removed or metals were not necessary for catalytic activity.
TABLE 2.
Effects of metal ions on activity of purified cyanuric acid hydrolase from M. thermoaceticaa
| Metal | Concentration (mM) | Relative activity |
|---|---|---|
| CoCl2 | 0.1 | 92 ± 5 |
| MnSO4 | 0.1 | 96 ± 3 |
| ZnSO4 | 0.1 | 51 ± 4 |
| CuCl2 | 0.1 | 83 ± 3 |
| FeCl3 | 0.1 | 105 ± 4 |
| NiCl2 | 0.1 | 79 ± 3 |
| CaCl2 | 1.0 | 121 ± 7 |
| MgCl2 | 2.0 | 119 ± 2 |
Values are means ± standard errors for three replicates.
Quantitation of bound metal.
The metal content of the enzyme was analyzed by inductively coupled plasma emission spectroscopy. The only metals detected above background were zinc and nickel, which were present at 0.08 and 0.05 M metal-to-subunit stoichiometries, respectively. These results indicated that no catalytically significant metals were present in an enzyme preparation with a significant catalytic rate having a kcat/Km value of 1.0 × 105 M−1 s−1. Similar results were obtained with AtzD (8).
DISCUSSION
The cyanuric acid hydrolase enzymes AtzD from Pseudomonas sp. strain ADP (8) and TrzD from Pseudomonas sp. strain NRRLB-12227 (11) have been purified and characterized. These enzymes were identified in organisms isolated for their ability to degrade atrazine and melamine, respectively. Both organisms mineralize the compounds via metabolic pathways that proceed through cyanuric acid as an intermediate. Cyanuric acid also forms abiotically via the spontaneous decomposition of chlorinated isocyanuric acids (1). The breakdown is an intended process, as it serves to disinfect pools by slowly generating hypochlorite. After a time, however, cyanuric acid accumulates, rendering further chlorination ineffective. Various methods to remove cyanuric acid chemically have been tried, but none have proven commercially successful to date. To deal with this issue, enzymatic treatment of cyanuric acid in pool water has been considered. One impediment to this application has been the relative instability of the cyanuric acid hydrolases known to date.
The known cyanuric acid hydrolases, AtzD and TrzD, have been observed to lose activity during freezing (unpublished data), a typical method used to maintain enzyme stability and to deter microbial and proteolytic breakdown of proteins. The goal of this study was to identify a more stable cyanuric acid hydrolase enzyme for use in bioremediation applications. In this context, we identified an AtzD/TrzD homolog from M. thermoacetica ATCC 39073. This is the first cyanuric acid hydrolase purified from an organism not previously shown to degrade s-triazine compounds. Genome context gives little indication of a native function for this gene. Upstream of Moth_2120 are genes that encode a CdaR transcriptional regulator, two hypothetical proteins, an FdrA protein implicated in regulating diverse cellular processes through FtsH, and another hypothetical protein. Downstream there are genes encoding the following proteins: a hypothetical protein, uracil-xanthine permease, carbamate kinase, and methyltransferase.
The AtzD gene homolog from M. thermoacetica was cloned, the recombinant enzyme was purified, and the characteristics of the enzyme were studied with respect to catalysis and stability. Table 3 compares the properties of this new enzyme with those of AtzD and TrzD. The cyanuric acid hydrolase from M. thermoacetica ATCC 39073 was shown to have cyanuric acid hydrolase activity, with Km and kcat values for cyanuric acid of 110 μM and 10.6 s−1, respectively. The enzyme displayed a high degree of substrate discrimination, only catalyzing reactions with cyanuric acid and its close structural analog N-methylisocyanuric acid, but with no other analogous structure. This mirrors the restricted reactivity found with AtzD (8) and indicates that although the sequences of the cyanuric acid hydrolases are only 57 to 64% identical, the proteins share a similar substrate range. The key difference between the M. thermoacetica cyanuric acid hydrolase and AtzD/TrzD is in the greater stability of the former enzyme. The new enzyme had its highest catalytic activity at 70°C (the highest temperature that our assays were able to maintain), was stable when stored for 30 min at 50°C, and was able to be stored frozen for long periods. In combination, these properties make this a superior enzyme for cyanuric acid remediation.
TABLE 3.
Comparison of cyanuric acid hydrolases
| Property | Value or description (reference) |
||
|---|---|---|---|
| Moorella enzyme | TrzD | AtzD | |
| Molecular mass (calculated) (kDa) | 38.9 | 39.4 | 38.2 |
| pH optimum | 8.0 | 8.0-8.5 (11) | 8.2 (8) |
| Temp optimum (°C) | 70 | 45-50 (11) | 30 (7) |
| Thermostabilitya (°C) | 20-65 | 20-45 | 20-40 |
| kcat (s−1) | 10.6 | 250 (11) | 6.8 ± 0.7 (8) |
| Km (μM) | 110 | 50 (11) | 57 ± 10 (8) |
| kcat/Km (s−1 M−1) | 1.0 × 105 | 5 × 106 | 1.2 × 105 |
| Known substrates | Cyanuric acid, N-methylisocyanuric acid | Cyanuric acid (11) | Cyanuric acid, N-methylisocyanuric acid (8) |
| Metal content | None | NDb | None (8) |
| Metal effects on activity | Cu(II), Zn(I), and Ni(II), inhibitory at 0.2 mM; Ca(II) and Mg(II), slight increase at 1.0 and 2.0 mM, respectively | Mg(II) and Mn(II), no effect at 1 mM; Co(II), Cu(II), and Fe(II), slightly inhibitory at 1 mM; Zn(II), greatly inhibitory at 1 mM (11) | No stimulatory effects with Zn(II), Cu(II), Fe(II), Co(II), or Ni(II) (8) |
| Influence of chelators on activity | No effect of EDTA, o-phenanthroline, or 8-hydroxyquinoline-5-sulfonic acid | No effect of EDTA (11) | No effect of EDTA or o-phenanthroline |
Temperature where activity was above 50% after a 30-min incubation.
ND, not determined.
Barbiturase, a cyanuric acid homolog, has been proposed to be a member of the amidohydrolase superfamily (20). In general, amidohydrolase superfamily members contain one or two metal atoms per subunit, with zinc being the most commonly identified metal. The enzymes AtzA, AtzB, AtzC, and TrzN, which are in the pathway of atrazine degradation, are all metalloenzymes (14-17) and are members of this superfamily. However, sequence analysis of the AtzD/TrzD/barbiturase family of proteins revealed that they are not members of this common superfamily. Instead, these enzymes are very distant from any other proteins of known function and cluster as an isolated island in sequence space. Because of the lack of sequence and evolutionary links to other well-characterized amidase enzymes, this family must be considered independently. Consistent with this, a metal was not necessary for catalysis by the M. thermoacetica cyanuric acid hydrolase. Previously, active AtzD was also found not to contain significant levels of metals (8).
In conclusion, the cyanuric acid hydrolase from M. thermoacetica ATCC 39073 is not a metalloenzyme. Furthermore, it is an outstanding thermophilic enzyme that is stable at high or low temperatures.
Acknowledgments
This research was funded, in part, by a grant from the University of Minnesota BioTechnology Institute Biocatalysis Initiative.
Footnotes
Published ahead of print on 18 September 2009.
REFERENCES
- 1.Cantú, R., O. Evans, F. K. Kawahara, J. A. Shoemaker, and A. P. Dufour. 2000. An HPLC method with UV detection, pH control, and reductive ascorbic acid for cyanuric acid analysis in water. Anal. Chem. 72:5820-5828. [DOI] [PubMed] [Google Scholar]
- 2.de Souza, M. L., M. J. Sadowsky, and L. P. Wackett. 1996. Atrazine chlorohydrolase from Pseudomonas sp. ADP: gene sequence, enzyme purification and protein characterization. J. Bacteriol. 178:4894-4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devers, M., N. El Azhari, N. U. Kolic, and F. Martin-Laurent. 2007. Detection and organization of atrazine-degrading genetic potential of seventeen bacterial isolates belonging to divergent taxa indicate a recent common origin of their catabolic functions. FEMS Microbiol. Lett. 273:78-86. [DOI] [PubMed] [Google Scholar]
- 4.Eaton, R. W., and J. S. Karns. 1991. Cloning and comparison of the DNA encoding ammelide aminohydrolase and cyanuric acid amidohydrolase from three s-triazine-degrading bacterial strains. J. Bacteriol. 173:1363-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 6.Fontaine, F. E., W. H. Peterson, E. McCoy, M. J. Johnson, and G. J. Ritter. 1942. A new type of glucose fermentation by Clostridium thermoaceticum. J. Bacteriol. 43:701-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fruchey, I. 2001. Purification and characterization of AtzD: a novel cyanuric acid amidohydrolase from Pseudomonas sp. strain ADP. M.S. thesis. University of Minnesota, St. Paul.
- 8.Fruchey, I., N. Shapir, M. J. Sadowsky, and L. P. Wackett. 2003. On the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP. Appl. Environ. Microbiol. 69:3653-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein, L. D., P. H. Jennings, and H. V. Marsh, Jr. 1971. A preliminary investigation of l-phenylalanine ammonia lyase activity in asparagus: distribution and response to storage, excision and incubation. Plant Cell Physiol. 12:657-661. [Google Scholar]
- 10.Gudnason, G. V., and W. F. Shipe. 1962. Factors affecting the apparent activity and heat sensitivity of xanthine oxidase in milk. J. Dairy Sci. 40:187. [Google Scholar]
- 11.Karns, J. S. 1999. Gene sequence and properties of an s-triazine ring-cleavage enzyme from Pseudomonas sp. strain NRRLB-12227. Appl. Environ. Microbiol. 65:3512-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsili, E., J. B. Rollefson, D. B. Baron, R. M. Hozalski, and D. R. Bond. 2008. Microbial biofilm voltammetry: direct electrochemical characterization of catalytic electrode-attached biofilms. Appl. Environ. Microbiol. 74:7329-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 14.Seffernick, J. L., A. Aleem, J. P. Osborne, G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2007. Hydroxyatrazine N-ethylaminohydrolase (AtzB): an amidohydrolase superfamily enzyme catalyzing deamination and dechlorination. J. Bacteriol. 189:6989-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seffernick, J. L., H. McTavish, J. P. Osborne, M. L. de Souza, M. J. Sadowsky, and L. P. Wackett. 2002. Atrazine chlorohydrolase from Pseudomonas sp. strain ADP is a metalloenzyme. Biochemistry 41:14430-14437. [DOI] [PubMed] [Google Scholar]
- 16.Shapir, N., J. P. Osborne, G. Johnson, M. J. Sadowsky, and L. P. Wackett. 2002. Purification, substrate range, and metal center of AtzC: the N-isopropylammelide aminohydrolase involved in bacterial atrazine metabolism. J. Bacteriol. 184:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapir, N., C. Pedersen, O. Gil, L. Strong, J. Seffernick, M. J. Sadowsky, and L. P. Wackett. 2006. TrzN from Arthrobacter aurescens TC1 is a zinc amidohydrolase. J. Bacteriol. 188:5859-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith, D., S. Alvey, and D. E. Crowley. 2005. Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol. Ecol. 53:265-273. [DOI] [PubMed] [Google Scholar]
- 19.Smolin, E. M., and L. Rapoport. 1959. s-Triazines and derivatives, p. 269-308. In A. Weissberger (ed.), The chemistry of heterocyclic compounds. Interscience Publishers Inc., New York, NY.
- 20.Soong, C. L., J. Ogawa, E. Sakuradani, and S. Shimizu. 2002. Barbiturase, a novel zinc-containing amidohydrolase involved in oxidative pyrimidine metabolism. J. Biol. Chem. 277:7051-7058. [DOI] [PubMed] [Google Scholar]
- 21.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Maarel, M. J., M. Jansen, R. Haanstra, W. G. Meijer, and T. A. Hansen. 1996. Demethylation of dimethylsulfoniopropionate to 3-S-methylmercaptopropionate by marine sulfate-reducing bacteria. Appl. Environ. Microbiol. 62:3978-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]