Abstract
As polyphagous, holometabolous insects, tephritid fruit flies (Diptera: Tephritidae) provide a unique habitat for endosymbiotic bacteria, especially those microbes associated with the digestive system. Here we examine the endosymbiont of the olive fly [Bactrocera oleae (Rossi) (Diptera: Tephritidae)], a tephritid of great economic importance. “Candidatus Erwinia dacicola” was found in the digestive systems of all life stages of wild olive flies from the southwestern United States. PCR and microscopy demonstrated that “Ca. Erwinia dacicola” resided intracellularly in the gastric ceca of the larval midgut but extracellularly in the lumen of the foregut and ovipositor diverticulum of adult flies. “Ca. Erwinia dacicola” is one of the few nonpathogenic endosymbionts that transitions between intracellular and extracellular lifestyles during specific stages of the host's life cycle. Another unique feature of the olive fly endosymbiont is that unlike obligate endosymbionts of monophagous insects, “Ca. Erwinia dacicola” has a G+C nucleotide composition similar to those of closely related plant-pathogenic and free-living bacteria. These two characteristics of “Ca. Erwinia dacicola,” the ability to transition between intracellular and extracellular lifestyles and a G+C nucleotide composition similar to those of free-living relatives, may facilitate survival in a changing environment during the development of a polyphagous, holometabolous host. We propose that insect-bacterial symbioses should be classified based on the environment that the host provides to the endosymbiont (the endosymbiont environment).
Bacteria are diverse, abundant, and ubiquitous and have greatly influenced the evolution of eukaryotic life (38, 39). Bacterial symbionts of eukaryotes are generally studied in terms of benefiting or impairing their eukaryotic hosts (3, 7, 21, 26). The constraints that the host's life history and diet impose on their endosymbionts have yet to be considered in depth. Insect-bacterium symbioses, both mutualistic and pathogenic, are ideal for examining host constraints on endosymbionts. The diversity of insect lifestyles and diets provides a continuum of habitats for endosymbionts, exhibiting different selection pressures on insect-associated bacteria.
Tephritid fruit flies (Tephritidae: Diptera) provide a unique habitat for endosymbiotic bacteria, especially digestive-system microbes. First, most tephritids feed on fleshy fruit (such as olives) as larvae, and as adults they are optimal foragers on pollen, bird feces, phylloplane bacteria (15, 52), homopteran honeydew (44), and other food sources in the environment (15, 18). Thus, digestive-tract endosymbionts may be exposed to different nutrients during an insect's life time. Second, Diptera are holometabolous, undergoing complete metamorphosis. Vertically transmitted endosymbionts must survive the breakdown and rebuilding of the host tissues with which the bacteria associate (22). We predict that, in order to survive in such a changing environment, the endosymbiont would have a genome similar in size to those of free-living relatives, rather than a greatly reduced genome, as is seen in obligate nutritional mutualists, such as Buchnera aphidicola.
The interaction between the olive fly, Bactrocera oleae, and its digestive-system bacterial associates has been studied for more than a century (5, 9, 17, 28, 29, 32, 33, 36, 52, 55, 56), yet several important questions remain unanswered. Do all olive fly life stages have the same species of bacterial endosymbiont? With which organs of the digestive system do the bacteria associate? Where are the bacteria located in relation to host tissues (are they extracellular or intracellular)? How ubiquitous is the association between the olive fly and specific bacteria? Are the same species of bacteria found in olive flies from the Old World also in flies that have recently infested the United States? Is the nucleotide composition of the olive fly symbiont more similar to those of its free-living relatives or to those of obligate endosymbionts?
The purpose of this paper is to address the questions listed above and to characterize the olive fly-bacterial symbiosis more thoroughly by using microscopy and culture-dependent and -independent techniques to examine all life stages of wild olive flies from the United States. We provide a comprehensive description of the biology of the olive fly symbiosis throughout host development, and we highlight the distinctive nature of a highly variable endosymbiont environment provided by the host and its consequences for bacterial genome evolution. Better understanding of tephritid endosymbionts may provide insight into control methods for these significant agricultural pest insects.
MATERIALS AND METHODS
Collection and sampling.
In 2003, infected olives were collected, and a Tucson laboratory population (TLP) culture was established from emerged larvae. The TLP culture was maintained on surface-sterilized olives. Adult B. oleae flies were trapped in torula yeast-baited McPhail traps (as described in reference 8) in olive groves of the National Clonal Germplasm Repository at Wolfskill Experimental Orchard in Winters (Yolo County), CA, and in Tucson (Pima County), AZ, in 2005. Sixty adults (30 males and 30 females) were collected from the Winters site, and 40 adults (20 males and 20 females) were collected from Tucson (APHIS permit number 68318).
All olive fly life stages (eggs, third-instar larvae, and pupae) were collected from the TLP and California populations and were examined for bacterial microbiota. Life stages were surface sterilized by vortexing for 15 s in a 1% sodium hypochlorite (bleach)-0.1% Triton X mixture and then rinsing twice with distilled water. The rinse water was plated on Luria-Bertani medium (4) and used as a template for PCR with the universal prokaryote 16S rRNA primers 10F and 1507R (42). No colonies or PCR products were observed (data not shown). TLP females were presented with “agar olives,” a half-sphere of 2% agar wrapped in Parafilm (Pechiney Plastic Packaging, Chicago, IL) as an oviposition substrate. Eggs (24 h postoviposition) were aseptically removed from agar olives. Wandering third-instar larvae and pupae (48 h postpupation) were fixed in 95% ethanol.
Samples of different organs in larvae and 1-week-old TLP adults were also taken in order to determine in which tissues bacteria reside. Adult flies and third-instar larvae were surface sterilized as described above and dissected in a laminar flow hood. Larval and adult organs were preserved immediately in 95% ethanol for DNA extraction, and PCR was performed using 16S rRNA primers that amplified an Enterobacter sp. and “Candidatus Erwinia dacicola” as described below. Larval organs sampled included the mycetome (midgut gastric ceca) (n = 10) and salivary glands (n = 10). Adult organs analyzed were the esophageal bulb (evagination of the foregut) (n = 10), crop (n = 10), rectal sac (n = 8), ovaries (n = 5), ovipositor (n = 5), and testes (n = 8).
In addition, olives were screened in order to determine if “Candidatus Erwinia dacicola” and the Enterobacter sp. were present in drupe tissues. Bacterial presence or absence was determined across six different olive treatments: fresh olives lacking larvae (unripe and ripe), rotten olives lacking larvae (unripe and ripe), and rotten olives with larvae (unripe and ripe). Twenty olives were used per treatment. Samples of olive flesh were fixed in 95% ethanol for DNA extraction, followed by bacterium-specific amplification as described below.
Culture-dependent identification of bacterial associates.
Male and female olive flies from the TLP were surface sterilized as described above, homogenized in isolation buffer, and plated onto various media (4), including Luria broth, Kings medium B, nutrient broth agar, Erwinia-selective medium, Trypticase soy broth, and crushed olive mesocarp with agar. The rinse water was plated and checked for bacterial growth. All cultures were incubated at 28°C and checked daily, for 10 days, for colonies. Emergent colonies were picked and streaked for single colonies on the same type of medium from which they were collected. Bacteria from single colonies were grown overnight in liquid medium and were stored in 40% glycerol at −80°C.
Culture-independent identification of bacterial associates.
To identify bacteria present in the wild populations and within specific organs of olive flies in the lab population, surface-sterilized whole animals or fly tissues were snap-frozen in liquid nitrogen and homogenized using a disposable plastic pestle (BelArt Products, Pequannock, NJ) in a 1.5-ml microcentrifuge tube. DNA from fly tissues was extracted using the mouse tail protocol of the Qiagen (Valencia, CA) DNeasy kit. DNA from olive tissues was extracted using the DNeasy plant kit (Qiagen, Valencia, CA). Whole-fly DNA samples were amplified with the universal prokaryote 16S rRNA primers 10F and 1507R (42) to generate a nearly complete (∼1,489-bp) sequence. These PCR products were cloned using a TOPO-TA one-shot kit (Invitrogen, Carlsbad, CA) and were sequenced in both directions by the Genomic Analysis and Technology Core of the University of Arizona on an Applied Biosystems 3730XL sequencer. Sequences were manually edited (Sequencher; Gene Codes, Ann Arbor, MI). Edited sequences were subjected to BLAST analysis (1) against the GenBank database (http://www.ncbi.nlm.nih.gov/) to estimate taxonomic placement for initial primer design and subsequent phylogenetic analyses. Additional primers were designed from an alignment of several enteric bacteria to selectively amplify the 16S rRNA of “Ca. Erwinia dacicola” and the Enterobacter sp., amplified using universal primers. Primers EdF1 (5′-CTAATACCGCATAACGTCTTCG-3′) and EntF1 (5′-CTAATACCGCATAACGTCGCAA-3′) were paired with primer 1507R to generate a ∼1,300-bp sequence. EntF1 and EntR2 (5′-GAGTAATCCGATTAACGCTTG-3′) were used for subsequent screening of flies (∼400-bp sequence) for the Enterobacter sp. For additional resolution of bacterial identity, the ompA (outer membrane protein) primer set UF1ompA (5′-TTAACGGTGCGGCTGAGTTACAACG-3′)-UR2ompA (5′-ACACCTGGTACACTGGTGCTAAAC-3′) and the recA (recombinase A) primer set UF2RecA (5′-CCTGACCGATCTTGTCACC-3′)-UR2RecA (5′-GGTAAAGGCTCCATCATGCG-3′), producing a 400-bp fragment, were designed from an alignment of several enteric bacteria. The PCR program used was a 65-55 touchdown program: 94°C for 2 min, followed by 10 cycles of 94°C for 45 s, 65°C for 45 s, a decrease by 1°C to 55°C, and 72°C for 1.5 min. The 65-55 touchdown was followed by 21 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 1.5 min. Bacteria of each species were scored as present or absent. In samples for which no PCR product was generated, the universal prokaryote primers 10F and 1507R (42) were used to determine if any bacteria were present. The extracted DNA of a homogenized olive fly with the “Ca. Erwinia dacicola” symbiont was used as a positive-control template. Negative controls lacked DNA. PCR products were cloned, sequenced, and used for phylogenetic analysis. To ensure that the DNA extraction was successful, primers for the B. oleae adh gene (encoding alcohol dehydrogenase) (adhF2 [5′-GGCATACTCACCGATCCCAATGTAGA-3′] and adhR2 [5′-CATGAGTGGAATTGCCTCTAGCGT-3′]) were designed using an alignment of several insect adh genes. Analysis of variance was used to determine if different sexes or populations of olive flies had similar associated bacteria (StatView, version 5.0; SAS Institute, Cary, NC).
Identification of olive fly bacterial symbionts using phylogenetic analysis.
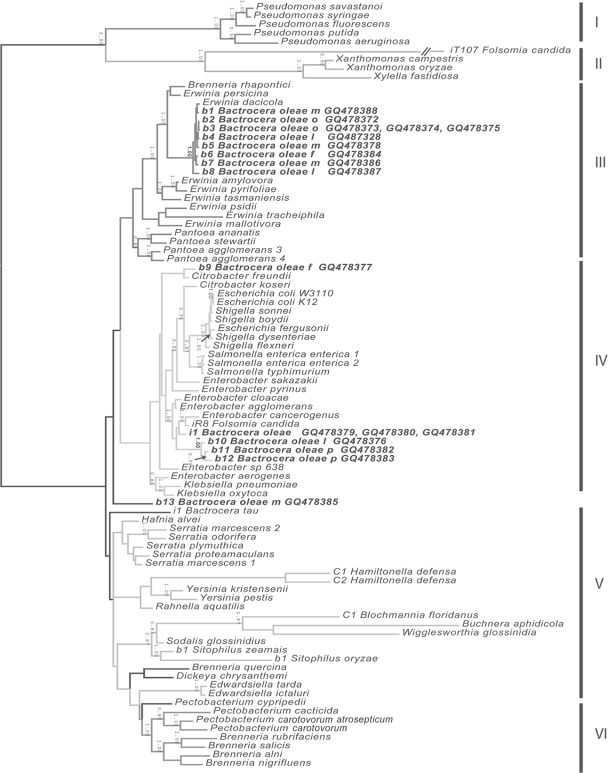
Nucleotide sequences from related bacteria, including free-living and insect endosymbiotic taxa, were obtained from NCBI for each locus (16S rRNA, ompA, recA) (see Fig. S1 in the supplemental material). All sequences generated during this study have been deposited in GenBank, and phylogenetic trees have been deposited in TreeBASE. Sequences were aligned separately for each locus and as a concatenated matrix using ClustalW (54) with default parameters. Codons of recA and ompA were translated to amino acids and converted back to nucleotides after amino acid alignment. All alignments were manually edited in Mesquite, version 2.01 (35). Nonalignable sections (i.e., regions of extensive small indels) were excluded from phylogenetic analysis (see Table S1 in the supplemental material). Only coding regions of recA and ompA were included for these loci. ModelTest (48) was used to compare models of evolution for individual loci (see Table S1 in the supplemental material), and the best model was chosen on the basis of the Akaike information criterion. A Bayesian analysis using MrBayes, version 3.1 (25), was performed to infer topologies, branch lengths, and support values for each locus and for the concatenated matrix. Table S1 in the supplemental material lists the partitioning and nucleotide evolution models as implemented by MrBayes. Two Metropolis-coupled Markov chain Monte Carlo (MCMC) analyses were run with four chains each for 5 million generations, with sampling every 1,000 generations. Burn-in periods (see Table S1 in the supplemental material) were estimated by graphing likelihood values and were defined by the generation when log-likelihoods appeared to converge to stationarity. All-compatible-partitions (allcompat) consensus trees were generated from post-burn-in MCMC samples. The taxa in clades I and II were selected as outgroups and were used to root the tree ( Fig. 1).
FIG. 1.
Phylogenetic identification of B. oleae symbionts. The results of Bayesian analysis of the concatenated sequences of the recA (∼870 bp), ompA (∼900 bp), and 16S rRNA (∼1,400 bp) genes are shown. Six groups (I to VI) that also occur in analyses of individual loci (see Table S2 in the supplemental material) are identified. Group V is an unresolved grade in all analyses. Posterior probabilities above 0.85 appear above and to the left of the corresponding nodes. The six clades have different grayscale values, and taxa incertae sedis are on black branches, as they change position from locus to locus. Designations beginning with “i” represent cultured isolates; those beginning with “b” represent bacteria amplified from the insect host named; and those beginning with “C” represent bacteria of Candidatus status. Samples of bacteria from Bactrocera oleae are in boldface, with the life stage (l, larva; p, pupa) or adult gender (m, male; f, female) or organ (o, ovipositor [female]) indicated.
Identification of bacteria by FISH.
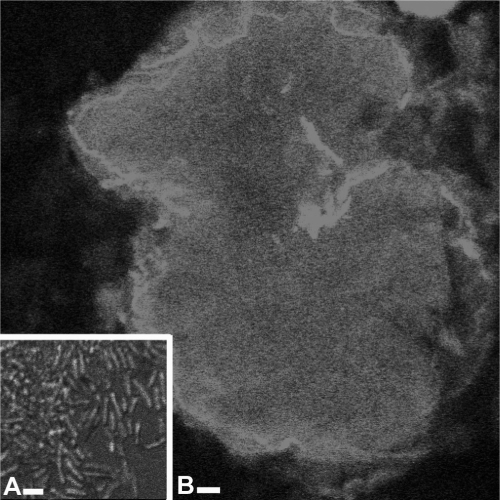
Fluorescent in-situ hybridization (FISH) was used to confirm the identity of the bacteria present in the esophageal bulbs of adults and the cultured bacterial isolates. Cultured Enterobacter sp. cells isolated from olive flies were grown overnight at 28°C, pelleted to remove medium, and washed twice in phosphate-buffered saline (PBS) at pH 7.0. Cells were fixed in 4% paraformaldehyde for 30 min at 22°C and were washed twice with PBS. Cells were dried on microscope slides (Fisherbrand Super Frost Plus; Fisher Scientific, Pittsburgh, PA) for FISH. Insect tissues fixed in paraformaldehyde had high levels of autofluorescence. Therefore, a different fixation protocol (19) was used. Flies were surface sterilized, and the esophageal bulb was dissected as described above. Esophageal bulbs were fixed in Carnoy's fixative for 4 days, rinsed twice for 5 min with 100% ethanol, and transferred to 6% H2O2 in ethanol for 4 days at 22°C. Bulbs were rinsed in 100% ethanol, followed by three washes, each 5 min long, in PBS plus 0.01% Tween. For hybridization, bulbs were rinsed in hybridization solution three times (for 5 min each time) and incubated overnight at 40°C in a mixture of the FISH probe, 20% formamide, and hybridization buffer. Samples were rinsed in 3 M sodium chloride and 0.3 M sodium citrate, followed by a PBS rinse. To protect the fluorescent samples from bleaching, a drop of Vectashield (Vector Labs, Burlingame, CA) was added. Samples were viewed under a Zeiss 510 Meta confocal microscope. Esophageal bulbs and cultured cells were both hybridized with a 6-carboxytetramethylrhodamine-labeled EUB338 (2) probe (data not shown), EdF1-Cy5, and EnF2-Cy3 (Fig. 2). To determine probe specificity, a range of formamide concentrations from 0 to 30% was used for both probes on both bacterial cells and esophageal bulbs. The optimal formamide concentration for both probes was 20%. Negative controls in PBS were also examined.
FIG. 2.
FISH of an adult esophageal bulb. (A) Whole mount of an adult esophageal bulb labeled with EdF1-Cy5, specific to “Ca. Erwinia dacicola.” Bar, 20 μm. (B) Higher magnification of “Ca. Erwinia dacicola” cells found within the esophageal bulb and labeled with EdF1-Cy5. Bar, 2 μm.
Bacterial localization within host tissues.
Larvae (third instar), 1-month-old fed adults, and recently eclosed (<24 h), unfed adults were collected from the TLP in order to determine where bacteria were located within host tissues by using transmission electron microscopy (TEM). Animals were surface sterilized, and the larval mycetomes and adult esophageal bulbs or ovipositors were collected. Samples were fixed in a mixture of 1.5% paraformaldehyde, 2% glutaraldehyde, 1.5% acrolein, and 1% dimethyl sulfoxide in 0.08 M cacodylate (pH 7.3) for 90 min at 20°C, rinsed for a total of 20 min in 0.08 M cacodylate (pH 7.3), postfixed in 2.0% OsO4 in 0.08 M cacodylate, and dehydrated with a standard ethanol series (30 to 100%) (modified from reference 37). Prior to embedding, samples were transferred into two exchanges of 100% propylene oxide for 10 min each and were embedded in Epon resin. Thick sections were cut on a Leica Ultramicrotome E and stained in 2% toluidine blue with 2% borax for basic histology. Thin sections (thickness, 60 to 90 nm) were placed on 100-μm-mesh Formvar-coated copper grids. Sections from two animals per organ type were viewed on a JEOL 100 CX II TEM. Microscopy was conducted at the University of Arizona's University Spectroscopy and Imaging Facilities. TEM negatives were scanned at 400 dpi using an Epson Perfection 2450 Photo scanner.
GC contents of representative loci.
The G+C nucleotide composition of olive fly endosymbionts was determined from recA, ompA, and 16S rRNA sequences. The total G+C content of each gene was determined by dividing the number of G+C nucleotides present by the total number of nucleotides in a given sequence. The G+C contents for the three loci were compared to the genome characteristics of the 32 sequenced genomes (free-living bacteria, plant pathogens, and obligate mutualists) used in our phylogenetic analyses. Regression analysis was used in each comparison to determine if the G+C contents of these loci were representative of the GC content of the whole genome (StatView, version 5.0; SAS Institute, Cary, NC). The mean and range of the G+C percentage for each locus were determined for the groups identified in the three-gene concatenated phylogeny (Table 1).
TABLE 1.
Percent G+C contents of recA, ompA, and 16S rRNA for species used in the phylogenetic analysisa
| Species | Mean (range) % G+C content |
||
|---|---|---|---|
| 16S rRNA | ompA | recA | |
| Clade I | 54.5 (53.8-55.9) (n = 6) | unb | 60 (58.2-63.9) (n = 4) |
| Clade II | 54.5 (53.8-54.96) (n = 4) | 65.5 (63-68) (n = 2) | 59.2 (52.1-63.3) (n = 3) |
| Clade III | 55.0 (54.2-55.6) (n = 12) | 52.7 (51.9-54.3) (n = 5) | 52.6 (49.8-55.4) (n = 11) |
| “Ca. Erwinia dacicola,” Italy | 54.7 (n = 1) | un | un |
| “Ca. Erwinia dacicola,” southwestern United States | 54.2 (n = 1) | 51.9 (51.8-52.2) (n = 3) | 53.6 (53.4-53.7) (n = 2) |
| Clade IV | 54.5 (48.9-55.7) (n = 21) | 54.6 (52.1-56.6) (n = 17) | 56.1 (51.9-61.2) (n = 16) |
| Enterobacter sp. | 56.1 (cultured), 54.7 (uncultured) (54.6-54.9) (n = 3) | 55.19 (n = 1) | 56.1 (n = 1) |
| Clade V | 52 (36-55) (n = 26) | 54.0 (14-57.2) (n = 16) | 47.9 (19-57.8) (n = 15) |
| Clade VI | 54.8 (53.8-55.8) (n = 7) | 52.1 (50.3-53.8) (n = 7) | 50.6 (48.4-54.0) (n = 7) |
The G+C composition was determined for each species in the clades identified in the three-gene concatenated phylogeny. Shown here are the mean, range, and number of species (n) for each clade. Also included are the statistics for the Enterobacter sp. and “Ca. Erwinia dacicola” sequenced from U.S. flies (this study) and for “Ca. Erwinia dacicola” from Italian flies (9).
un, unknown.
Nucleotide sequence accession numbers.
The sequences generated during this study have been deposited in GenBank under accession numbers GQ478372 to GQ478388 and GQ487328. Phylogenetic trees have been deposited in TreeBASE under study number S2481.
RESULTS
Specific bacteria were associated with olive fly populations.
Bacteria from surface-sterilized wild olive flies were identified by using sequences generated by PCR amplification of 16S rRNA, ompA, and recA. Phylogenetic analyses identified five clades of bacteria and one unresolved grade that appeared in allcompat consensus trees of the ompA (see Fig. S1B in the supplemental material), recA (see Fig. S1C in the supplemental material), and concatenated (Fig. 1; see also Table S1 in the supplemental material) analyses. Phylogenetic analyses of 16S rRNA alone (see Fig. S1A in the supplemental material) resulted in poorly resolved trees. Four bacterial species were recovered from olive flies (Fig. 1).
The most abundant bacterial species was identified from sequences obtained from whole animals (larvae, adults) and ovipositors (Fig. 1, samples b1 to b8). This abundant species had high sequence similarity to the entire 16S rRNA (99% BLAST identity) of “Ca. Erwinia dacicola” from Italian olive flies (9). Phylogenetic analyses of bacterial sequences amplified from adult and larval olive flies (Fig. 1, samples b1 to b8) and previously characterized “Ca. Erwinia dacicola” placed this bacterium within a subclade of clade III (Erwinia plus Pantoea) (posterior probability [PP] in concatenated analyses, 1.0). Phylogenetic analyses using ompA (see Fig. S1B in the supplemental material) and recA (see Fig. S1C in the supplemental material) revealed the monophyly of these samples and showed them to be allied to other Erwinia species. In concatenated analyses, the “Ca. Erwinia dacicola” clade was sister to a clade with plant pathogens Brenneria (Erwinia) rhapontici and Erwinia persicina (PP = 1.0) (Fig. 1). “Ca. Erwinia dacicola” is thus far unculturable (9; also data not shown).
The second most abundant bacterial species was identified from sequences obtained from larvae, pupae (Fig. 1, samples b10 to b12), and one cultured isolate (Fig. 1, sample i1) from the abdomen of a wild male olive fly. Several isolates from both larvae and male flies had a round, smooth colony morphology on Luria-Bertani (LB) and Erwinia-selective media. The cultured isolate and the 16S rRNA sequences amplified from olive fly larvae and pupae form clade IV (Enterobacteriaceae) (Fig. 1). The cultured isolate is a member of an unsupported sister clade that contains Enterobacter cancerogenus, Enterobacter agglomerans, Enterobacter cloacae (from the midgut of Leptinotarsa decemlineata), and a bacterium isolated from the collembolan Folsomia candida (PP in concatenated analyses, 1.0). Initial sequence similarity results (99% BLAST match) and phylogenetic analysis using 16S rRNA alone (see Fig. S1A in the supplemental material) suggested that the Enterobacter sp. isolate was closely related to Enterobacter cloacae or Enterobacter agglomerans. The recA analysis (see Fig. S1C in the supplemental material) groups the cultured bacterium with Enterobacter cancerogenus and Enterobacter cloacae (PP = 0.95). Further resolution was not obtained using ompA alone (see Fig. S1B in the supplemental material).
Two other bacterial species (Fig. 1, samples b9 and b13) were amplified only once in adult flies. Sample b9, from a wild female fly, was closely related to the plant pathogen Citrobacter freundii, although relationships were weak using ompA alone (see Fig. S1B in the supplemental material). Sample b13, from a wild California male fly, could not be identified using the taxon sampling of this phylogeny. BLAST results showed 99% similarity to Raoultella ornithinolytica.
“Ca. Erwinia dacicola” and the Enterobacter sp. were associated with all olive fly life stages.
PCR analysis revealed that “Ca. Erwinia dacicola” and the Enterobacter sp. were present in all life stages of wild olive flies surveyed, suggesting that they persist through metamorphosis (data not shown). Sequence analysis revealed that “Ca. Erwinia dacicola” was present in 94% of the females and 86% of the males. The sex of the flies did not influence the presence or absence of “Ca. Erwinia dacicola” (P = 0.2932; n = 113). In 9.8% of the adults, neither “Ca. Erwinia dacicola” nor the Enterobacter sp. was found. There were no flies with both bacteria. One male had only the Enterobacter sp. There was no difference in the frequency of “Ca. Erwinia dacicola” between individuals from California and those from Arizona (P = 0.7213; n = 113). “Ca. Erwinia dacicola” and the Enterobacter sp. were not present in either ripe or unripe olives without fly larvae. The rot tunnels that the larvae made while feeding on the olive always contained “Ca. Erwinia dacicola” in ripe (n = 20) and unripe (n = 20) olives (data not shown).
“Ca. Erwinia dacicola” and the Enterobacter sp. are located within larvae and adults.
“Ca. Erwinia dacicola” was consistently amplified from several structures of the B. oleae digestive tract. The endosymbiont was always present in the larval midgut (mycetome) (n = 10) and the evagination of the adult foregut (esophageal bulb) (n = 10). More than 70% of the 8 rectal sacs and 10 crops sampled had “Ca. Erwinia dacicola” (data not shown). None of the sampled larval salivary glands (n = 10) or adult testes (n = 8) or ovaries (n = 5) had either bacterium. Ovipositors of four of the five females sampled had “Ca. Erwinia dacicola.” The fifth ovipositor had only the Enterobacter sp.
Endosymbionts are both intracellular and extracellular.
Bacterial species present in adult esophageal bulbs were identified using FISH probes specific to “Ca. Erwinia dacicola” and Enterobacter 16S rRNA. In adults, all bacterial cells in the esophageal bulb hybridized with the general enteric bacterial probe EUB338 and the “Ca. Erwinia dacicola”-specific probe EdF1-Cy5 (Fig. 2A and B) but not with the Enterobacter-specific EnF2-Cy3 probe, demonstrating that these cells were “Ca. Erwinia dacicola.” The EnF2-Cy3 and EUB338 probes labeled the cultured Enterobacter sp. cells. Both of these results were confirmed using PCR. No labeling was seen in negative controls, and autofluorescence was very low (data not shown).
TEM revealed bacteria that were intracellular residents of larval cells (see Fig. 4A to D) (e.g., residing within the cellular membrane of epithelial cells of the digestive tissue) and extracellular residents of adult tissues (e.g., residing in the digestive system lumen but outside the cellular membrane of host cells) (Fig. 2A and B and 3A to D). In the adult, bacteria within the esophageal bulb (Fig. 3A to D) and ovipositor diverticula (Fig. 3E) had the classic rod-shaped morphology of an enteric bacterium (M1 type cells) and were surrounded by a matrix, suggesting biofilm development. These bacteria were longitudinal nearest the host epithelium, creating a very densely packed array of cells in cross section (Fig. 3B and E). In the esophageal bulb center, bacteria were more randomly oriented and may have included cells dispersing from the biofilm, since bacteria were observed in the stem connecting the esophageal bulb to the rest of the digestive system (data not shown). Other studies have noted bacteria dispersing from the esophageal bulb (9).
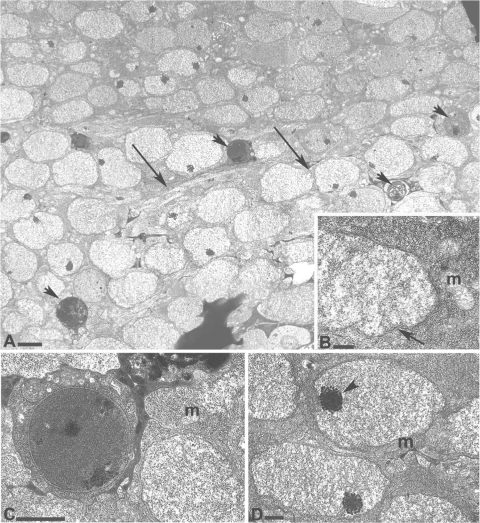
FIG. 4.
Intracellular bacteria of larval mycetome. (A) Larval midgut showing a field of bacteria with the amorphous morphology (M2). Four bacterial cells with the M1 morphology are indicated by arrowheads. Arrows point to a host cell membrane surrounding the bacteria. Bar, 5 μm. (B) Cell wall (arrow) of a gram-negative bacterium having the amorphous M2 morphology. Magnification, ×140,000. Bar, 4 μm. (C) High-magnification (×100,000) inset from panel A of a bacterial cell having the M1 morphology. Note the host cell membrane surrounding the bacterial cell. Bar, 2.5 μm. (D) M2 cells shown at ×72,000 magnification. Arrowhead points to electron-dense granules. Bar, 2 μm. Host mitochondria (m) are indicated in panels B, C, and D.
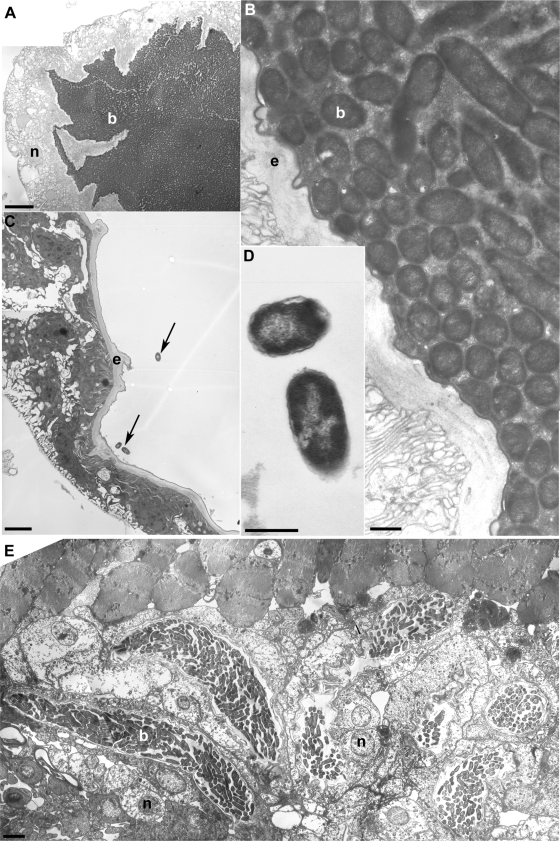
FIG. 3.
Extracellular bacteria in lumen of adult esophageal bulb and ovipositor. (A) Cross section through the esophageal bulb of a 1-month-old fed adult olive fly. A host nucleus (n) is identified in the epithelial tissue of the bulb that surrounds the bacteria. Extracellular bacteria (b) fill the esophageal bulb lumen. Bar, 80 μm. (B) Close-up of the esophageal bulb shown in panel A. Extracellular bacteria form a classic bacterial biofilm, with those closest to the host epithelia (e) seen in cross section. Bacteria farther away from the host epithelia are seen in longitudinal section, revealing a typical gram-negative rod shape. Endosymbionts are surrounded by an extracellular matrix. Bar, 5 μm. (C) Esophageal bulb of a recently eclosed, unfed adult olive fly. Arrows point to the three bacteria present in the esophageal bulb lumen. These bacteria are located near the host epithelium. Bar, 35 μm. (D) High (×190,000) magnification of the bacteria from panel C. Bar, 5 μm. (E) Cross section through the lumen of a female ovipositor showing seven diverticula filled with extracellular bacteria. Extracellular bacteria within one of the diverticula are indicated. The nuclei of two host cells next to the diverticulum are evident. These bacteria are similar in morphology to those seen in the adult esophageal bulb (panel B). Bar, 30 μm.
Although microscopy of the olive fly pupal stage was unsuccessful, recently eclosed (<24 h posteclosion) adults were examined to determine the number of bacteria present immediately after pupation. The esophageal bulbs of recently eclosed adults contained fewer than 10 bacterial cells closely associated with the bulb epithelium (Fig. 3C), and a few bacteria were loosely associated with membranes. In contrast, bulbs of older, fed adults were filled with bacteria (Fig. 3A and B).
In larvae, bacterial cells were present within host cells (intracellular) (Fig. 4A to D), instead of inside the digestive system lumen (extracellular), as in the adult esophageal bulb. Within host cells, the endosymbiont exhibited two bacterial morphologies. The majority of bacteria in host cells had an amorphous morphology (M2-type cells) (Fig. 4A, B, and D) similar to that of other insect intracellular endosymbionts, such as Buchnera (41), “Candidatus Carsonella” (53), and “Candidatus Ishikawaella” (24). The cytoplasm of the amorphous morphology was diffuse and granular, with electron-dense inclusion bodies clustered at one region of the cell (Fig. 4D). A second bacterial morphology, circular in cross section with a denser cytoplasm, typical of enteric bacteria and similar to what is seen in the adult stage (M1-type cells) (Fig. 4C), was rarely present. Although two distinct bacterial morphologies were present, PCR confirmed that “Ca. Erwinia dacicola” was the only bacterium found in the mycetome.
The G+C compositions of recA, ompA, and 16S rRNA from olive fly symbionts are similar to those of closely related, free-living species.
The G+C nucleotide contents of recA, ompA, and 16S rRNA were correlated with the overall genome G+C content (n = 32; R2 = 0.94, 0.97, and 0.70, respectively; P < 0.0001 in all cases), and the whole-genome G+C content is positively correlated with the genome size (n = 32; R2 = 0.90; P < 0.0001) in the sequenced bacterial genomes used in our phylogenetic analysis. The G+C contents of recA, ompA, and 16S rRNA were correlated with the genome size (n = 32; R2 = 0.76, 0.9, and 0.63, respectively; P < 0.0001 in all cases). The G+C compositions of recA, ompA, and 16S rRNA of both “Ca. Erwinia dacicola” and the Enterobacter sp. associated with olive flies are similar to those of closely related, free-living enteric bacteria (Table 1; see also Table S3 in the supplemental material). The genome sizes of the bacteria, estimated from the equation of the regression line of all three loci, were similar to the genome sizes of their free-living enteric bacterial relatives (Table 1; see also Table S3 in the supplemental material).
DISCUSSION
“Ca. Erwinia dacicola” is a tightly associated symbiont of the olive fly.
Culturable bacterial endosymbionts of the olive fly were first described by Petri in 1909 (47). A century later, Capuzzo et al. used molecular techniques to identify a specific uncultivable bacterium, “Candidatus Erwinia dacicola,” in Italian adult olive flies (9). Other studies have identified transient gammaproteobacterial species acquired during feeding (5, 52), as well as the alphaproteobacteria Acetobacter tropicalis (29) and Asaia spp. (52), within the olive fly.
Our study found two bacteria, a culturable Enterobacter sp. and the unculturable “Ca. Erwinia dacicola,” present in all life stages of wild olive flies, suggesting that these bacteria can be maintained through changes in diet and host metamorphosis. Although other low-abundance species that our universal 16S rRNA primers do not amplify may be present, we focused on symbionts consistently present at high density, not on all species within the olive fly digestive system.
“Candidatus Erwinia dacicola” is found both in the TLP and in 90% of wild flies surveyed. It is possible that the 10% of wild individuals testing negative for “Ca. Erwinia dacicola” were recently eclosed individuals with endosymbiont titers below the threshold detectable by PCR. qPCR of other recently eclosed flies showed extremely low densities of “Ca. Erwinia dacicola” (data not shown), and TEM revealed fewer than a dozen bacteria in the esophageal bulbs of recently eclosed flies (Fig. 3B), so PCR screens may underestimate endosymbiont presence. “Ca. Erwinia dacicola” has also been found within adult olive flies in Italy (9, 52) and Greece (29). The high frequency of association of “Ca. Erwinia dacicola” with the fly, its presence in different populations and in all life stages, its inability to be cultured on standard microbiological media, its vertical transmission to offspring, and its ability to reside within larval host cells suggest that this bacterium is a tightly associated endosymbiont of olive flies (46). To our knowledge, intracellular endosymbionts have not been reported in other tephritids (30), and other olive fly-associated bacteria, such as Acetobacter tropicalis, are exclusively extracellular (29, 52). An intracellular stage of “Ca. Erwinia dacicola” implies a more specific and longer-term interaction with the host (26).
The Enterobacter sp. is only occasionally found in the olive flies screened and is likely to be a nonspecific microbe. Enterobacter spp. are common in insects, including the other tephritids Rhagoletis completa (49), Rhagoletis pomonella (30), and Bactrocera tau (C. S. Prabhakar, P. Sood, B. A. Padder, S. S. Kanwar, V. Kapoor, P. K. Metha, and P. N. Sharma, unpublished data); cockroaches (12; also C. M. Gibson, unpublished data); fire ants (31); and cucumber beetles (D. R. Herndon and K. D. Spence, unpublished data). The Citrobacter sp. and sample b13 were thought to be phylloplane bacteria acquired during feeding (5, 14), since they were PCR amplified only once in wild-caught adults.
Our phylogenetic analyses demonstrate that “Ca. Erwinia dacicola” is closely related to several insect-vectored, plant-pathogenic Erwinia species, such as Erwinia amylovora, Erwinia pyrifoliae, Erwinia tracheiphila, and Pantoea (Erwinia) stewartii. These pathogens persist primarily in association with their plant hosts and insect vectors, with limited survival in the soil (50). These closely related plant pathogens differ from each other and from “Ca. Erwinia dacicola” in the duration and the intimacy of their association with their insect versus plant hosts. Erwinia amylovora and Erwinia pyrifoliae overwinter at canker margins and in plant host buds, causing wilt (34, 51). Insect vectors are attracted to these bacterium-filled exudates and transmit the pathogens to new infection sites (16, 23, 51). In contrast, Erwinia tracheiphila and Pantoea stewartii overwinter in beetle digestive systems, and disease incidence on host plants is correlated with insect survival (11, 20, 40). “Ca. Erwinia dacicola” is found (via PCR) only in the rot tunnels where larvae are present, presumably deposited in the olive through regurgitation or defecation. Whether the bacterium is metabolically active in rot or whether the host or symbiont is responsible for the rot is unknown.
“Ca. Erwinia dacicola” transitions between an intracellular and an extracellular existence.
During olive fly development, “Ca. Erwinia dacicola” resides intracellularly within larval midgut cells and extracellularly in the adult foregut. Other insect endosymbionts survive both intracellularly and extracellularly (10, 45); however, the ramifications of this transition for symbiosis and for endosymbiont evolution have not been discussed. The transition from an intracellular to an extracellular existence within the digestive system during insect development could be essential to endosymbiont survival in a polyphagous, holometabolous host. Prior to pupation, Diptera void their digestive tracts, reducing the number of extracellular microbes (22). During metamorphosis, larval digestive system tissues degrade, and adult tissues are assembled from larval midgut regenerative cells (27). Vertically transmitted endosymbionts must have mechanisms for surviving metamorphosis and reestablishing themselves in the adult. Although the mechanisms of endosymbiont survival during complete metamorphosis are unknown, we hypothesize that bacterial cells present in the regenerative cells recolonize the adult gut.
In adults, “Ca. Erwinia dacicola” resides in a bacterial biofilm in the digestive system lumen. Nutrient acquisition for opportunistic foragers, such as the adult olive fly, is often spatially and temporally patchy. The formation of bacterial biofilms may provide resistance to periods of low nutrients and other stresses (13). Olive fly endosymbionts could not be removed from adult flies fed bleach and antibiotics (rifampin [rifampicin], streptomycin, tetracycline, and ampicillin) (data not shown) once biofilms were established. The extracellular lifestyle could also be important for transmission to offspring. The presence of “Ca. Erwinia dacicola” in the ovipositor, but not in the ovaries or testes, suggests that olive fly endosymbionts (47) are vertically transmitted via smearing as the egg passes by the symbiont-rich ovipositor diverticulum (Fig. 3E) (52). Extracellular bacteria have more rigid and well-formed cell walls that may facilitate egg-smearing transmission.
The endosymbiont environment hypothesis and bacterial genome characteristics.
The olive fly, “Ca. Erwinia dacicola,” provides, to our knowledge, the first example of a tightly associated symbiosis in a polyphagous, holometabolous insect. This study expands our knowledge of the types of environments in which endosymbionts survive and leads us to propose that symbioses be discussed in terms of the endosymbiont environment the host provides. In insects, the endosymbiont environment ranges from constant to variable based on at least three axes: the host diet (monophagous versus polyphagous), the host life cycle (hemimetabolous versus holometabolous), and the location of the endosymbiont within host tissues (intracellular versus extracellular). The most constant environment for endosymbionts would be provided by monophagous, hemimetabolous insects, such as aphids and sharpshooters, where endosymbionts reside exclusively within bacteriomes and hosts feed exclusively on one food type. The most variable environment would be found in polyphagous, holometabolous insects, such as the olive fly. Endosymbionts of polyphagous, holometabolous insects must survive changes in insect diet and changes to tissues during complete metamorphosis. We suggest that the environment an insect endosymbiont experiences may greatly influence its genome composition, genome size, and evolution (A. M. Estes, unpublished data).
The nucleotide compositions and genome sizes of bacteria are directly related to the degree of environmental variability, since bacteria quickly accumulate mutations and lose genes that are not under selection (6, 43). Free-living bacteria experiencing a diversity of environmental conditions have more G+C-rich genes and larger genomes than obligately intracellular endosymbionts (3, 6, 21). Thus, we predicted that endosymbionts of polyphagous, holometabolous insects, such as the olive fly, would have genomes that are less A+T biased and larger than those of obligate endosymbionts of monophagous, holometabolous insects, such as aphids. Indeed, the nucleotide compositions of the three loci analyzed from “Ca. Erwinia dacicola” and the Enterobacter sp. were more similar to those for plant-pathogenic and free-living relatives than to those for obligately intracellular endosymbionts (Table 1; see also Table S3 in the supplemental material). The G+C nucleotide compositions of the recA, ompA, and 16S rRNA genes were shown to be a good proxy for genome size, and the genome sizes of both bacteria were estimated to be similar to those of free-living and plant-pathogenic relatives.
The data presented here suggest that polyphagous, holometabolous insects associate with specific microbes. This study of the olive fly endosymbiont, “Ca. Erwinia dacicola,” provides detailed microscopy to show that the bacterium associates with different host tissues and transitions between intracellular and extracellular stages during host development. This is one of the few examples of an insect endosymbiont that makes such a transition. Examination of digestive-system endosymbionts of other polyphagous, holometabolous insects will be essential to understanding the mechanisms that influence bacterial genome structure and evolution.
Supplementary Material
Acknowledgments
We thank Leland S. Pierson III for unlimited access to lab equipment and ancillary lab supplies; Elizabeth Arnold, Leland S. Pierson III, the Bronstein lab, Monica Alvarez, Antonio Belcari, and three anonymous reviewers for comments on the manuscript; Justin Clark for assisting with olive tissue samples; Hannah Burrack and the lab of Frank Zalom at the University of California, Davis, for California olive fly samples; David Bentley and Carl Boswell for microscopy assistance; Phat Tran for assistance with FISH; and the Pierson and Bronstein labs at the University of Arizona, especially Krishna Maddula, for helpful discussions.
This work was supported by a Doctoral Dissertation Improvement Grant, DEB-0608480 (to J.L.B. and A.M.E.); an Integrative Graduate Education and Research Traineeship, NSF-IGERT DGE 0114420, from the National Science Foundation (to A.M.E.); and a Grant-in-Aid from the Society for Integrative and Comparative Biology (to A.M.E.).
Footnotes
Published ahead of print on 18 September 2009.
Supplemental material for this article may be found at http://www.aem.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, S. G., C. Alsmark, B. Canbäck, W. Davids, C. Frank, O. Karlberg, L. Klasson, B. Antoine-Legault, A. Mira, and I. Tamas. 2002. Comparative genomics of microbial pathogens and symbionts. Bioinformatics 18(Suppl. 2):S17. [DOI] [PubMed] [Google Scholar]
- 4.Atlas, R. M., and L. C. Parks. 1993. Handbook of microbiological media. CRC Press, Boca Raton, FL.
- 5.Belcari, A., P. Sacchetti, G. Marchi, and G. Surico. 2003. La mosca delle olive e la simbiosi batterica. Infect. Fitopatol. 9:55-59. [Google Scholar]
- 6.Bentley, S. D., and J. Parkhill. 2004. Comparative genomic structure of prokaryotes. Annu. Rev. Genet. 38:771-791. [DOI] [PubMed] [Google Scholar]
- 7.Buchner, P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY.
- 8.Burrack, H. J., and F. G. Zalom. 2008. Olive fruit fly (Diptera: Tephritidae) ovipositional preference and larval performance in several commercially important olive varieties in California. J. Econ. Entomol. 101:750-758. [DOI] [PubMed] [Google Scholar]
- 9.Capuzzo, C., G. Firrao, L. Mazzon, A. Squartini, and V. Girolami. 2005. ‘Candidatus Erwinia dacicola’, a coevolved symbiotic bacterium of the olive fly Bactrocera oleae (Gmelin). Int. J. Syst. Evol. Microbiol. 55:1641-1647. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, Q., and S. Aksoy. 1999. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 8:125-132. [DOI] [PubMed] [Google Scholar]
- 11.Cook, K. A., R. A. Weinzierl, J. K. Pataky, P. D. Esker, and F. W. Nutter. 2005. Population densities of corn flea beetle (Coleoptera: Chrysomelidae) and incidence of Stewart's wilt in sweet corn. J. Econ. Entomol. 98:673-682. [DOI] [PubMed] [Google Scholar]
- 12.Cruden, D. L., and A. Markovetz. 1984. Microbial aspects of the cockroach hindgut. J. Arch. Microbiol. 138:131-139. [DOI] [PubMed] [Google Scholar]
- 13.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drew, R. A. I., and A. C. Lloyd. 1991. Bacteria in the life cycle of Tephritid fruit flies, p. 441-465. In P. Barbosa, V. A. Krischik, and C. G. Jones (ed.), Microbial mediation of plant-herbivore interactions. John Wiley and Sons, Inc., New York, NY.
- 15.Drew, R. A. I., and B. Yuval. 2000. The evolution of fruit fly feeding behavior, p. 731-749. In M. Aluja and A. L. Norrbom (ed.), Fruit flies (Tephritidae): phylogeny and evolution of behavior. CRC Press, Boca Raton, FL.
- 16.Emmett, B. J., and L. A. E. Baker. 1971. Insect transmission of fireblight. Plant Pathol. 20:41-45. [Google Scholar]
- 17.Estes, A. M., and E. A. Pierson. 2005. Understanding the interplay between the tephritid olive fly and its bacterial symbionts, abstr. 9. SICB Annu. Meet., San Diego, CA, 4 to 8 January 2005.
- 18.Fletcher, B. S. 1987. The biology of dacine fruit-flies. Annu. Rev. Entomol. 32:115-144. [Google Scholar]
- 19.Fukatsu, T., R. Koga, W. A. Smith, K. Tanaka, N. Nikoh, K. Sasaki-Fukatsu, K. Yoshizawa, C. Dale, and D. H. Clayton. 2007. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl. Environ. Microbiol. 73:6660-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Salazar, C., F. E. Gildow, S. J. Fleischer, D. Cox-Foster, and F. L. Lukezic. 2000. ELISA versus immunolocalization to determine the association of Erwinia tracheiphila in Acalymma vittatum (Coleoptera: Chrysomelidae). Environ. Entomol. 29:542-550. [Google Scholar]
- 21.Gil, R., A. Latorre, and A. Moya. 2004. Bacterial endosymbionts of insects: insights from comparative genomics. Environ. Microbiol. 6:1109-1122. [DOI] [PubMed] [Google Scholar]
- 22.Greenberg, B. 1959. Persistence of bacteria in the developmental stages of the housefly. III. Quantitative distribution in prepupae and pupae. Am. J. Trop. Med. Hyg. 8:613-617. [DOI] [PubMed] [Google Scholar]
- 23.Hildebrand, M., E. Dickler, and K. Geider. 2000. Occurrence of Erwinia amylovora on insects in a fire blight orchard. J. Phytopathol. 148:251-256. [Google Scholar]
- 24.Hosokawa, T., Y. Kikuchi, X. Meng, and T. Fukatsu. 2005. The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol. Ecol. 54:471-477. [DOI] [PubMed] [Google Scholar]
- 25.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa, H. 2003. Insect symbiosis: an introduction, p. 1-16. In K. Bourtzis and T. Miller (ed.), Insect symbiosis. CRC Press, Boca Raton, FL.
- 27.Jiang, C., E. H. Baehrecke, and C. S. Thummel. 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124:4673-4683. [DOI] [PubMed] [Google Scholar]
- 28.Konstantopoulou, M. A., D. G. Raptopoulos, N. G. Stavrakis, and B. E. Mazomenos. 2005. Microflora species and their volatile compounds affecting development of an alcohol dehydrogenase homozygous strain (Adh-I) of Bactrocera (Dacus) oleae (Diptera: Tephritidae). J. Econ. Entomol. 98:1943-1949. [DOI] [PubMed] [Google Scholar]
- 29.Kounatidis, I., E. Crotti, P. Sapountzis, L. Sacchi, A. Rizzi, B. Chouaia, C. Bandi, A. Alma, D. Daffonchio, P. Mavragani-Tsipidou, and K. Bourtzis. 2009. Acetobacter tropicalis is a major symbiont in the olive fruit fly Bactrocera oleae. Appl. Environ. Microbiol. 75:3281-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauzon, C. R. 2003. Symbiotic relationships of tephritids, p. 115-130. In K. Bourtzis and T. Miller (ed.), Insect symbiosis, vol. 1. CRC Press, Boca Raton, FL. [Google Scholar]
- 31.Lee, A. H., C. Husseneder, and L. Hooper-Bùi. 2008. Culture-independent identification of gut bacteria in fourth-instar red imported fire ant, Solenopsis invicta Buren, larvae. J. Invertebr. Pathol. 98:20-33. [DOI] [PubMed] [Google Scholar]
- 32.Lüthy, P., D. Studer, F. Jaquet, and C. Yamvrias. 1983. The bacterial symbiote of the olive fruit fly (Dacus oleae). Experientia 39:1436. [Google Scholar]
- 33.Lüthy, P., D. Studer, F. Jaquet, and C. Yamvrias. 1983. Morphology and in vitro cultivation of the bacterial symbiote of Dacus oleae. Mitt. Schweiz. Entomol. Ges. 56:67-72. [Google Scholar]
- 34.Maas Geesteranus, H. P., and P. M. de Vries. 1984. Survival of Erwinia amylovora bacteria on plant surfaces and their role in epidemiology. Acta Horticult. 151:55-61. [Google Scholar]
- 35.Maddison, W. P., and D. R. Maddison. 2007. Mesquite: a modular system for evolutionary analysis, version 2.0. http://mesquiteproject.org/mesquite/download/Mesquite2Manual.pdf.
- 36.Manousis, T., and D. Ellar. 1988. Dacus oleae microbial symbionts. Microbiol. Sci. 5:149-152. [PubMed] [Google Scholar]
- 37.Margaritis, L. H., F. C. Kafatos, and W. H. Petri. 1980. The eggshell of Drosophila melanogaster. I. Fine structure of the layers and regions of the wild type eggshell. J. Cell Sci. 43:1-35. [DOI] [PubMed] [Google Scholar]
- 38.Margulis, L., and R. Fester (ed.). 1991. Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. MIT Press, Cambridge, MA. [PubMed]
- 39.McFall-Ngai, M. J., B. Henderson, and E. G. Ruby (ed.). 2005. The influence of cooperative bacteria on animal host biology. Cambridge University Press, Cambridge, United Kingdom.
- 40.Menclas, B., C. C. Block, P. D. Esker, and F. W. Nutter. 2006. Quantifying the feeding periods required by corn flea beetles to acquire and transmit Pantoea stewartii. Plant Dis. 90:319-324. [DOI] [PubMed] [Google Scholar]
- 41.Moran, N. A., J. A. Russell, R. Koga, and T. Fukatsu. 2005. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71:3302-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munson, M. A., P. Baumann, M. A. Clark, L. Baumann, N. A. Moran, D. J. Voegtlin, and B. C. Campbell. 1991. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J. Bacteriol. 173:6321-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochman, H., and N. A. Moran. 2001. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292:1096-1099. [DOI] [PubMed] [Google Scholar]
- 44.Opp, S., M. Durbin, C. Patty, B. Mooney, and M. Johnson. 2007. Presence of honeydew reduces efficacy of GF-120 against olive fly and walnut husk fly, abstr. 54. 9th Annu. Exotic Fruit Fly Symp., Fresno, CA, 25 to 26 April 2007.
- 45.Pais, R., C. Lohs, Y. Wu, J. Wang, and S. Aksoy. 2008. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74:5965-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paracer, S., and V. Ahmadjian. 2000. Symbiosis: an introduction to biological associations, 2nd ed. Oxford University Press, Oxford, United Kingdom.
- 47.Petri, L. 1909. Ricerche sopra i batteri intestinali della mosca olearia, p. 1-129. Memorie della Regia Stazione di Patologia Vegetale di Roma, Rome, Italy.
- 48.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 49.Potter, S. E. 2001. Bacterial associations in the walnut husk fly, Rhagoletis completa Cresson. M.S. thesis. California State University, Hayward.
- 50.Purcell, A. H. 1982. Insect vector relationships with prokaryotic plant pathogens. Annu. Rev. Phytopathol. 20:397-417. [Google Scholar]
- 51.Rhim, S.-L., B. Völksch, L. Gardan, J.-P. Paulin, C. Landlotz, W.-S. Kim, and K. Geider. 1999. Erwinia pyrifoliae, an Erwinia species different from Erwinia amylovora, causes a necrotic disease of Asian pear trees. Plant Pathol. 48:514-520. [Google Scholar]
- 52.Sacchetti, P., A. Granchietti, S. Landini, C. Viti, L. Giovannetti, and A. Belcari. 2008. Relationships between the olive fly and bacteria. J. Appl. Entomol. 132:682-689. [Google Scholar]
- 53.Thao, M. L., N. A. Moran, P. Abbot, E. B. Brennan, D. H. Burckhardt, and P. Baumann. 2000. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl. Environ. Microbiol. 66:2898-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsiropoulos, G. J. 1983. Microflora associated with wild and laboratory reared adult olive fruit flies, Dacus oleae (Gmel.). Z. Ang. Entomol. 96:337-340. [Google Scholar]
- 56.Yamvrias, C., C. G. Panagopoulos, and P. G. Psallidas. 1970. Preliminary study of the internal bacterial flora of the olive fruit fly (Dacus oleae, Gmelin). Ann. Inst. Phytopathol. Benaki N.S. 9:201-206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.