Abstract
Factors potentially contributing to the lower incidence of Lyme borreliosis (LB) in the far-western than in the northeastern United States include tick host-seeking behavior resulting in fewer human tick encounters, lower densities of Borrelia burgdorferi-infected vector ticks in peridomestic environments, and genetic variation among B. burgdorferi spirochetes to which humans are exposed. We determined the population structure of B. burgdorferi in over 200 infected nymphs of the primary bridging vector to humans, Ixodes pacificus, collected in Mendocino County, CA. This was accomplished by sequence typing the spirochete lipoprotein ospC and the 16S-23S rRNA intergenic spacer (IGS). Thirteen ospC alleles belonging to 12 genotypes were found in California, and the two most abundant, ospC genotypes H3 and E3, have not been detected in ticks in the Northeast. The most prevalent ospC and IGS biallelic profile in the population, found in about 22% of ticks, was a new B. burgdorferi strain defined by ospC genotype H3. Eight of the most common ospC genotypes in the northeastern United States, including genotypes I and K that are associated with disseminated human infections, were absent in Mendocino County nymphs. ospC H3 was associated with hardwood-dominated habitats where western gray squirrels, the reservoir host, are commonly infected with LB spirochetes. The differences in B. burgdorferi population structure in California ticks compared to the Northeast emphasize the need for a greater understanding of the genetic diversity of spirochetes infecting California LB patients.
In the United States, Lyme borreliosis (LB) is the most commonly reported vector-borne illness and is caused by infection with the spirochete Borrelia burgdorferi (3, 9, 52). The signs and symptoms of LB can include a rash, erythema migrans, fever, fatigue, arthritis, carditis, and neurological manifestations (50, 51). The black-legged tick, Ixodes scapularis, and the western black-legged tick, Ixodes pacificus, are the primary vectors of B. burgdorferi to humans in the United States, with the former in the northeastern and north-central parts of the country and the latter in the Far West (9, 10). These ticks perpetuate enzootic transmission cycles together with a vertebrate reservoir host such as the white-footed mouse, Peromyscus leucopus, in the Northeast and Midwest (24, 35), or the western gray squirrel, Sciurus griseus, in California (31, 46).
B. burgdorferi is a spirochete species with a largely clonal population structure (14, 16) comprising several different strains or lineages (8). The polymorphic ospC gene of B. burgdorferi encodes a surface lipoprotein that increases expression within the tick during blood feeding (47) and is required for initial infection of mammalian hosts (25, 55). To date, approximately 20 North American ospC genotypes have been described (40, 45, 49, 56). At least four, and possibly up to nine, of these genotypes are associated with B. burgdorferi invasiveness in humans (1, 15, 17, 49, 57). Restriction fragment length polymorphism (RFLP) and, subsequently, sequence analysis of the 16S-23S rRNA intergenic spacer (IGS) are used as molecular typing tools to investigate genotypic variation in B. burgdorferi (2, 36, 38, 44, 44, 57). The locus maintains a high level of variation between related species, and this variation reflects the heterogeneity found at the genomic level of the organism (37). The IGS and ospC loci appear to be linked (2, 8, 26, 45, 57), but the studies to date have not been representative of the full range of diversity of B. burgdorferi in North America.
Previous studies in the northeastern and midwestern United States have utilized IGS and ospC genotyping to elucidate B. burgdorferi evolution, host strain specificity, vector-reservoir associations, and disease risk to humans. In California, only six ospC and five IGS genotypes have been described heretofore in samples from LB patients or I. pacificus ticks (40, 49, 56) compared to approximately 20 ospC and IGS genotypes identified in ticks, vertebrate hosts, or humans from the Northeast and Midwest (8, 40, 45, 49, 56). Here, we employ sequence analysis of both the ospC gene and IGS region to describe the population structure of B. burgdorferi in more than 200 infected I. pacificus nymphs from Mendocino County, CA, where the incidence of LB is among the highest in the state (11). Further, we compare the Mendocino County spirochete population to populations found in the Northeast.
MATERIALS AND METHODS
Tick collection, spirochete culture, and DNA extraction.
Questing I. pacificus nymphs were collected from 78 dense woodlands in Mendocino County, CA, in 2004 during their peak activity period (late April to early June), as previously described (19). Up to 100 ticks were tested for the presence of B. burgdorferi at each site, and this target number of ticks was reached in 51 (65%) sites. Total DNA was extracted from individual ticks using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol for animal tissues. DNA was extracted similarly from scrapings of frozen B. burgdorferi cultures derived from questing I. pacificus nymphs and adults collected between 1987 and 1999 from Marin, Sonoma, and Mendocino Counties in California. These isolates are part of the Borrelia collection housed at the University of California, Berkeley. B. burgdorferi isolate CA8, passage 7, derived from an I. pacificus adult collected in Sonoma County, CA, in 1987 (32), was cultivated in Barbour-Stoenner-Kelly II medium (4). The DNA was extracted from passage 9 as previously described (8). Data presented throughout this report refer to I. pacificus nymphs collected in 2004 unless otherwise noted.
Sequence analysis. (i) 5S-23S rRNA spacer region, IGS, and ospC.
Amplification of the 5S-23S rRNA spacer region of B. burgdorferi by PCR was performed as described earlier (34) with minor modifications. Cycling conditions included a 4-min denaturation step at 95°C, followed by 40 cycles consisting of 95°C for 40 s, 52°C for 40 s for outer primers (58°C for internal primers), and 72°C for 1 min, followed by a 10-min extension at 72°C. Only DNA from ticks determined by sequence analysis of the 5S-23S region to be infected with B. burgdorferi (n = 263) was subjected to further PCR amplification using primers for IGS and ospC as previously described (8) with minor modifications. IGS and ospC PCRs were conducted using Phusion DNA polymerase (New England Biolabs, Ipswich, MA) or Amplitaq DNA polymerase (Applied Biosystems, Foster City, CA). The ospC PCR was performed using 35 amplification cycles for both first and second reactions and an annealing temperature of 50°C. Products from positive tick samples were purified using either a QIAquick PCR Purification Kit (Qiagen) for 5S-23S PCR products or a ZR-96 DNA Clean and Concentrator (Zymo Research, Orange, CA) for IGS and ospC products.
Both strands of purified 5S-23S DNA were sequenced by the University of California, Berkeley, CA, DNA Sequencing Facility using internal PCR primers (34). Purified IGS and ospC PCR products were sequenced by Polymorphic DNA Technologies, Alameda, CA. Primers used for IGS sequencing included a forward internal (nested) PCR primer (8) and another, overlapping forward primer, 5′-TTT CGC TAA AGT GCA AGG A-3′ (B. Travinsky et al., unpublished data). ospC sequencing was carried out using internal PCR primers (8).
The 5S-23S, IGS, and ospC sequences were assembled and manually edited using Sequencher 4.6 (Gene Codes Corp, Ann Arbor, MI). 5S-23S contigs were aligned with sequences selected from the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/index.html) using Clustal X (version 1.83.1) (54) and edited to 158 bp using Mesquite (version 2.6; http://mesquiteproject.org). B. burgdorferi assignment was based on phylogenetic analysis using the neighbor-joining method implemented in PAUP* (version 4.0 beta; Sinauer, Sunderland, MA) (uncorrected P distances).
IGS contigs were manually edited in Sequencher, resulting in 805- to 812-bp sequences which were subsequently aligned using Clustal X. The alignment was manually edited for minor errors using MacClade (version 4; Sinauer, Sunderland, MA). IGS sequences were assigned to genotype based on both neighbor-joining distance analysis in PAUP* and direct sequence comparison to IGS sequences available in the GenBank. IGS genotypes, designated by their GenBank accession number, are defined by as little as a 1-nucleotide (nt) difference. Position 1 of the IGS alignment corresponds to position 2424 of B. burgdorferi B31, GenBank accession number U03396, and position 444532 of the B31 genome sequence NC_001318. ospC contigs were edited similarly in Sequencher and MacClade, resulting in a 549-bp alignment. ospC sequences were compared to those available in GenBank, and genotype assignments were based on direct sequence comparison. Position 1 of the ospC alignment corresponds to position 396 of B. burgdorferi B31 ospC gene sequence U01894 and position 16993 of the B31 cp26 genome sequence NC_001903.
Nomenclature for ospC genotypes follows the groups system described by Seinost et al. (49) and Wang et al. (56). Sequences were designated new ospC genotypes when two or more tick samples contained a nucleotide sequence that differed from other genotypes by >8% (56). When ospC sequences were <8% different in sequence identity, alleles were assigned a name including the genotype and polymorphic nucleotide position number, e.g., ospC A (nt 123). The prevalence of ospC alleles in I. pacificus nymphs was statistically compared to the prevalence in I. scapularis nymphs in a study by Wormser et al. (57) using Fisher's exact test (two-tailed).
(ii) 16S rRNA and flagellin.
To amplify regions of the 16S rRNA and flagellin (flaB) genes of California B. burgdorferi isolate CA8, PCR was performed as previously described (5) with some modification. Cycling parameters included an initial incubation for 3 min at 95°C and a final extension step of 75°C for 7 min using Phusion DNA polymerase. Primers used for flaB DNA amplification produced 641-bp products whose sequences were aligned using Clustal X, version 1.83, with other B. burgdorferi species (Fig. 1). 16S rRNA PCR produced 1,336-bp products whose sequences were aligned with other Borrelia species (Fig. 1). Phylogenetic analysis of alignments using neighbor-joining and maximum-likelihood criteria were performed with Phylo_win, version 2.0 (http://pbil.univ-lyon1.fr/software/phylowin.html) (23).
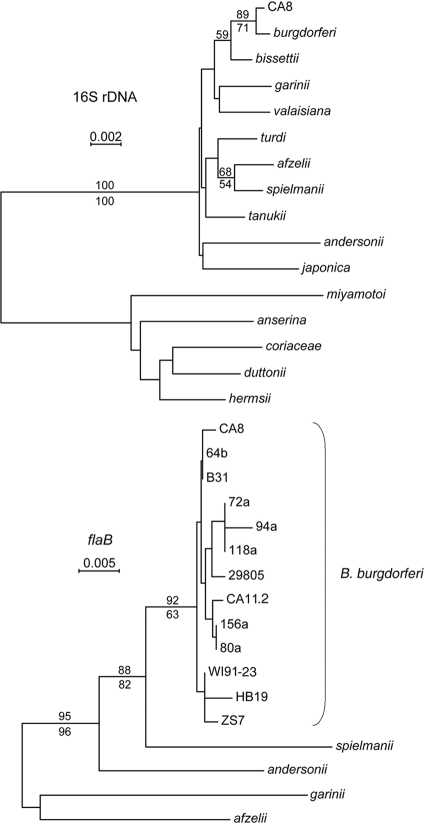
FIG. 1.
Neighbor-joining distance phylograms for partial 16S rRNA gene (observed differences) and flaB (Tajima-Nei method) sequences of selected Borrelia species and B. burgdorferi strains. The upper panel includes relapsing-fever species as the outgroup. Nodes with bootstrap support (1,000 iterations) of ≥50% by distance criteria (above the line) and by maximum-likelihood criteria (below the line) are shown. CA8 is the representative strain expressing ospC H3. All B. burgdorferi strains, with the exception of CA8 and HB19, were subjected to whole-genome sequencing, and the sequences were obtained from the GenBank.
Habitat analysis of genotypes.
Associations between habitat type and ospC genotypes were examined using Fisher's exact test (two-tailed). Collection sites were previously classified into seven habitat types based on field assessments (19), and we lumped six of these into two new broader categories: (i) hardwood-fir, composed of habitats that are suitable for LB reservoir western gray squirrels (46), and (ii) redwood-pine-tanoak, where western gray squirrels are not typically observed (31). The hardwood-fir category combined two habitat types: (i) hardwood with ≥90% Quercus sp. oaks, Pacific madrone (Arbutus menziesii), and California bay (Umbellularia californica) and (ii) mixed hardwood/conifer with ≥80% Quercus spp. oaks, Pacific madrone, California bay, Douglas fir (Pseudotsuga menziesii), and ponderosa pine (Pinus ponderosa). Tanoak (Lithocarpus densiflorus) represented 0 to 16% of trees at sites within these habitat types. The redwood-pine-tanoak category combined four habitat types: (i) redwood with ≥65% redwood (Sequoia sempervirens); (ii) coastal pine with ≥90% bishop pine (Pinus muricata), redwood, and Douglas fir; (iii) inland pine with ≥65% ponderosa pine and Douglas fir; and (iv) tanoak with >60% tanoak, redwood, and California bay. Quercus oaks were absent from or were rare in these habitat types. Four sites from a mixed habitat type previously referred to as tanoak-madrone-conifer (19) were excluded from the habitat analysis, meaning that 12 ticks infected with B. burgdorferi ospC genotype H3 (n = 3), ospC E3 (n = 3), ospC G (n = 4), ospC H (n = 1), and ospC M (nt 348) (n = 1) were eliminated.
To visualize the distributions of the three unique ospC genotypes in relation to habitat type (hardwood-fir versus redwood-pine-tanoak) throughout Mendocino County, site locations were coded by habitat type and displayed in map format using ArcGIS, version 9.3 (ESRI, Redlands, CA). The relative ospC genotype frequencies were calculated by using the number of ticks with a particular genotype at a site, divided by the total number of ticks containing that genotype across all sites (n = 209 ticks across 74 sites).
Nucleotide sequence accession numbers.
DNA sequences are available in GenBank for ospC (FJ932732 to FJ932736) and IGS (EU886969 to EU886976 and FJ932731) loci of B. burgdorferi derived from I. pacificus and for the 16S rRNA (GQ247740) and flaB (GQ247741) loci of B. burgdorferi isolate CA8. Accession numbers for taxa used in the 16S rRNA gene-based phylogenetic analysis are U42298, AJ009753, AJ224141, D67024, NR_025861, NR_025983, M88329, L46696, U42292, U42284, L46701, NR_025874, EU135595, AM182229, and CP000395. With the exception of B. burgdorferi HB19 (X75200), B. garinii (L42885), B. andersonii (D83763), and B. afzelii (X75202), sequences used in the flaB gene phylogenetic analysis were taken from whole genomes (complete or in progress) available in GenBank (AE000783, NZ_ABKA00000000, NZ_ABGJ00000000, NZABGK00000000, NZ_ABGI00000000, NZ_ABJX00000000, NZ_ABJY00000000, NZ_ABCV00000000, NZ_ABJU00000000, NZ_ABJW00000000, CP001205, and NZ_ABKB02000003).
RESULTS
Frequency distribution of ospC genotypes.
ospC DNA was successfully amplified in 227 (86.3%) of 263 B. burgdorferi-infected ticks collected in Mendocino County in 2004. Twelve ospC alleles, or unique sequences, belonging to 11 ospC genotypes were detected (Table 1). Exact matches to previously published sequences were found for eight of the ospC genotypes: A, B, D, E3, F, G, H, and T. We identified two new ospC genotypes, H3 and I3, and two new ospC alleles (sequence variants of genotypes): B (nt 59), which belongs to genotype B, and M (nt 348), which belongs to genotype M (Table 1). Because the ospC H3 nucleotide sequence was >8% different from ospC genotypes submitted to GenBank to date, we designated it a new ospC genotype (56). It was most similar (84%) to ospC genotype E (GenBank accession no. AY275221). Compared to AY275221, the H3 sequence had 90 nt changes throughout the 549-bp alignment, conferring 42 amino acid changes, as well as a deletion at positions 367 to 369 (amino acid 123) and a 3-bp insertion at positions 424 to 426. I3 is a hybrid of two previously described genotypes, F and A. I3 is nearly an exact match to genotype F (L42896) (39) from nucleotides 1 to 385, with two exceptions: a nucleotide change from A → G at position 184, conferring an N → D change in amino acids at position 62, and a nucleotide change from G → A at position 277, conferring a D → N amino acid change at position 93. I3 positions 386 to 549 are an exact match to sequences from genotype A (AY275213). Although ospC I3 is divergent from both F and A by less than 8%, its unique hybrid status and moderately high frequency in ticks (see below) led us to designate it a new genotype.
TABLE 1.
ospC alleles identified in B. burgdorferi-infected I.pacificus ticks collected in 2004 and from 1987 to 1999 in Mendocino County
| ospC genotype or allele | GenBank accession no. | Sequence length (nt; no gaps) | GenBank comparison data |
Position of nucleotide changeb | Nucleotide change | Amino acid change | |||
|---|---|---|---|---|---|---|---|---|---|
| Best match accession no. | Strain identifier | % Similarity | Reference | ||||||
| A | 528 | AY275213 | B31 | 100 | 8 | None | |||
| B | 531 | AY275215 | 1-24 | 100 | 8 | None | |||
| B (nt 59)a | FJ932735 | 531 | AY275215 | 1-24 | 99 | 8 | 59 | C to T | T to M |
| D | 531 | L25413 | CA-11.2A | 100 | 56 | None | |||
| E3 | FJ932732 | 531 | EF592545 | 18.74/cp26 | 100 | Unpublished | None | ||
| F | 525 | L42896 | 27579 | 100 | 39 | None | |||
| Ic | 528 | EU377752 | CA92-1096 | 100d | 40 | None | |||
| I3a | FJ932734 | 525 | L42896 | 27579 | 94 | 39 | |||
| G | 528 | AY275223 | 2-43 | 100 | 8 | None | |||
| H3a | FJ932733 | 525 | AY275221 | NP40 (ospC E) | 84 | 8 | |||
| H | 528 | EF053519 | LDS79 | 100 | 17 | None | |||
| M (nt 348)a | FJ932736 | 534 | AY275218 | Bve | 99 | 17 | 348 | T to C | None |
| T | 537 | AY275222 | 10-33 | 100 | 17 | None | |||
Novel ospC genotype or allele.
Based on 549-nt alignment (including gaps) unless otherwise indicated. Position number 1 of the alignment corresponds to position 396 of GenBank no. U01894 and position 16993 of the B31 cp26 genome sequence (GenBank no. NC_001903). For I3 and H3, see the text.
Identified only in an I. pacificus male collected in 1992 (Table 3).
Based on alignment of nt positions 4 to 487.
ospC sequences could not be fully typed in 22 (9.7%) of 227 amplicons, which presumably represented mixed B. burgdorferi infections. In 17 of these ticks, at least one ospC genotype was decipherable, and these genotypes were included in the overall ospC allele frequency given in Table 2. B. burgdorferi genotype H3 was the most frequently encountered (24%) ospC genotype detected in spirochete-infected I. pacificus nymphs (Table 2). The second-most-common genotype found was E3 (17.2%), followed by H (12.2%), A (10.9%), F (9.5%), D and G (8.1% each), B (4.5%), M (nt 348) (2.7%), I3 (2.3%), and B (nt 59) and T (0.5%) (Table 2).
TABLE 2.
ospC allele frequency in B.burgdorferi DNA amplified from I. pacificus nymphs collected in Mendocino County in 2004
| ospC allele | No. of ticks with allele |
ospC allele frequency in tick population (%) | |
|---|---|---|---|
| −MIa | +MIb | ||
| H3 | 51 | 53 | 24.0 |
| E3 | 32 | 38 | 17.2 |
| H | 24 | 27 | 12.2 |
| A | 23 | 24 | 10.9 |
| F | 21 | 21 | 9.5 |
| D | 17 | 18 | 8.1 |
| G | 17 | 18 | 8.1 |
| B | 8 | 10 | 4.5 |
| M (nt 348) | 5 | 6 | 2.7 |
| I3 | 5 | 5 | 2.3 |
| B (nt 59) | 1 | 1 | 0.5 |
| T | 1 | 1 | 0.5 |
| Total | 205 | 222 | 100.0 |
−MI, not including mixed infections.
+MI, including mixed infections.
We also determined the ospC sequence type for 35 B. burgdorferi frozen cultures derived from I. pacificus nymphs or adults collected in three northern Californian counties (Marin, Sonoma, and Mendocino) between 1987 and 1999 (Table 3). We then compared the genotype frequencies in Mendocino County nymphs collected in 2004 to the genotype frequencies in cultures derived solely from Mendocino County nymphs (n = 14). ospC A was detected in 50% of cultured nymphs and 10.9% of nymphs collected in 2004 (Fisher's exact test, P < 0.01). ospC H was detected in 28.6% of cultured nymphs compared to 12.2% of nymphs collected in 2004 (P = 0.09). ospC F had a similar prevalence in Mendocino County nymph cultures (14.3%) compared to DNA from nymphs collected in 2004 (9.5%; P = 0.63). We detected ospC I3 in 7.1% of cultures compared to 2.3% in ticks collected in 2004 (P = 0.31). A single example of ospC I was identified in culture isolate CA337 made from a male I. pacificus in Mendocino County in 1992, which matched 481 nt (positions 4 to 487) of isolate CA92-1096 made from a human skin biopsy in Sonoma County in 1992 (J. Piesman, personal communication) (40) (Table 1). ospC genotype I was not detected in ticks collected in 2004. Only one example of the predominant ospC genotype detected in the nymphs collected in 2004, H3, was found in culture isolate CA8 obtained in 1987 from an I. pacificus female in Sonoma County.
TABLE 3.
ospC allele frequencies in California B.burgdorferi culture isolates from nymphal or adult I. pacificus ticks, 1987 to 1999
| ospC genotype | No. of cultures positive for the allele (frequency [%]) | California isolate no. | 5S-23S RFLP typea | Tick life stage and sexb | County | Year of isolation |
|---|---|---|---|---|---|---|
| A | 16 (45.7) | 17 | A | M | Mendocino | 1990 |
| 18 | A | M | Mendocino | 1990 | ||
| 19 | A | F | Mendocino | 1990 | ||
| 382 | A | Unknown | Mendocino | 1993 | ||
| 535 | A | N | Mendocino | 1998 | ||
| 536 | A | N | Mendocino | 1998 | ||
| 537 | A | N | Mendocino | 1999 | ||
| 4 | A | M | Sonoma | 1987 | ||
| 5 | A | F | Sonoma | 1987 | ||
| 6 | A | M | Sonoma | 1987 | ||
| 538 | A | N | Mendocino | 1999 | ||
| 543 | A | N | Mendocino | 1998 | ||
| 544 | A | N | Mendocino | 1998 | ||
| 3 | B | F | Marin | 1986 | ||
| 9 | Not typed | F | Marin | 1988 | ||
| 540 | Not typed | N | Mendocino | 1999 | ||
| D | 3 (8.6) | 15 | B | F | Mendocino | 1990 |
| 172 | B | Unknown | Mendocino | Unknown | ||
| 338 | B | M | Mendocino | 1992 | ||
| F | 3 (8.6) | 7 | A | F | Sonoma | 1987 |
| 358 | B | N | Mendocino | 1992 | ||
| 542 | B | N | Mendocino | 1998 | ||
| I3 | 4 (11.4) | 10 | B | F | Mendocino | 1988 |
| 11 | B | F | Mendocino | 1989 | ||
| 12 | B | Unknown | Mendocino | 1989 | ||
| 360 | B | N | Mendocino | 1992 | ||
| G | 1 (2.9) | 566 | B | M | Sonoma | 1999 |
| H3 | 1 (2.9) | 8 | Not typed | F | Sonoma | 1987 |
| H | 6 (17.1) | 533 | A | N | Mendocino | 1998 |
| 534 | A | N | Mendocino | 1998 | ||
| 541 | A | N | Mendocino | 1998 | ||
| 567 | A | M | Sonoma | Unknown | ||
| 336 | Not typed | M | Mendocino | 1992 | ||
| 569 | Not typed | N | Mendocino | 1999 | ||
| I | 1 (2.9) | 337 | B | M | Mendocino | 1992 |
Reference 7.
M, adult male; F, adult female; N, nymph.
We also evaluated the relationship between ospC genotype and RFLP sequence type of the 5S-23S rRNA IGS (7) (Table 3). Linkage between RFLP pattern and ospC genotype was observed in 28 out of 30 (93%) samples typed by both methods. With the exception of isolate CA3, all ospC genotype A and H cultures (n = 16 and 6, respectively) were 5S-23S RFLP pattern A. All other ospC genotypes (D, F, I3, G, G3, and I) were RFLP pattern B except for CA7 (RFLP type A).
Frequency distribution of IGS genotypes.
Using an 812-bp alignment, 13 IGS alleles defined by as little as a 1-nt difference were identified in I. pacificus nymphs, nine of which have not been previously described (Table 4). Exact matches to sequences formerly named IGS genotype 5 (GenBank accession no. AY275201), subtype 3A (DQ437500), and subtype 6A (AY275202) (8) were found in addition to an exact match to GenBank sequence EF649786, which is most similar to genotype 4 described by Bunikis et al. (8) (Table 5). The most divergent Californian IGS allele (GenBank accession no. EU886974) had seven single nucleotide changes compared to the IGS sequence AY275201 and a 5-bp deletion at positions 794 to 798 (Table 4). Based on its nucleotide sequence divergence and its strict association with novel ospC genotype H3 (Table 5), we propose that this sequence represents a new B. burgdorferi IGS genotype.
TABLE 4.
Signature polymorphic nucleotides of 5S-23S IGS sequence types identified in B.burgdorferi-infected I. pacificus nymphs collected in Mendocino County, 2004
| GenBank accession no. | Former IGS genotype or subtype nameb | Residue at polymorphic positionc |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 226 | 239 | 274 | 289 | 314 | 419 | 435 | 489 | 516 | 538 | 684 | 706 | 713 | 725 | 729 | Δd | ||
| AY275189a | 1A | T | G | T | G | A | C | C | G | G | A | C | A | C | G | G | No |
| EU886973 | T | G | T | G | A | C | C | G | G | A | T | A | C | G | G | No | |
| AY275194a | 2D | T | G | T | G | A | T | C | G | G | A | T | A | T | G | G | No |
| EU886969 | T | G | T | G | A | T | C | G | G | A | T | A | C | G | G | No | |
| AY275201 | 5 | C | G | T | G | A | T | C | G | G | A | T | A | C | G | G | No |
| EU886970 | C | A | T | G | A | T | C | G | G | A | T | A | C | G | G | No | |
| FJ932731 | C | G | T | G | A | T | C | G | G | G | T | A | C | G | G | No | |
| EU886976 | C | G | T | G | A | T | C | G | A | A | T | A | C | G | G | No | |
| EU886975 | C | G | T | G | A | T | C | G | G | A | T | A | C | A | G | No | |
| EU886971 | C | G | C | G | A | T | C | G | G | G | T | A | C | G | G | No | |
| EU886972 | C | G | C | G | A | T | T | G | G | G | T | A | C | G | G | No | |
| EU886974 | NG | T | G | T | A | T | C | C | A | G | A | T | G | C | G | T | Yes |
TABLE 5.
ospC/IGS biallelic profiles found in B.burgdorferi-infected I.pacificus nymphs in Mendocino County in 2004a
| ospC genotype or allele | IGS allele (accession no.)b | Former IGS genotype or subtype namec | No. of ticks positive for the allele (% of total)d |
|---|---|---|---|
| H3 | AY275201 | 5 | 1 (0.5) |
| EU886971 | 5 | 1 (0.5) | |
| EU886974 | NG | 42 (21.9) | |
| AY275202 | 6A | 1 (0.5) | |
| E3 | AY275201 | 5 | 11 (5.7) |
| EU886976 | 5 | 3 (1.6) | |
| EU886975 | 5 | 17 (8.9) | |
| H | EU886969 | 2 | 22 (11.5) |
| A | EU886973 | 1 | 17 (8.9) |
| AY275201 | 5 | 1 (0.5) | |
| EU886971 | 5 | 1 (0.5) | |
| AY275202 | 6A | 2 (1.0) | |
| F | EF649786 | 4 | 21 (10.9) |
| D | EU886970 | 5 | 17 (8.9) |
| G | EU886971 | 5 | 8 (4.2) |
| AY275201 | 5 | 1 (0.5) | |
| EU886972 | 5 | 7 (3.6) | |
| B | DQ437500 | 3A | 6 (3.1) |
| AY275201 | 5 | 1 (0.5) | |
| I3 | EF649786 | 4 | 5 (2.6) |
| M (nt 348) | AY275202 | 6A | 5 (2.6) |
| B (nt 59) | AY275201 | 5 | 1 (0.5) |
| T | EU886975 | 5 | 1 (0.5) |
| Total | 192 (100.0) |
Dominant biallelic profiles for each ospC genotype are in bold.
GenBank accession numbers correspond either to newly submitted sequences (Table 4) or to sequences existing in the database.
Based on an exact match or similarity to previously described sequences (8). NG, new genotype.
Results from mixed infections are not included.
ospC and IGS biallelic profiles.
Because regions of both IGS and ospC of B. burgdorferi were sequenced in individual ticks, we were able to investigate linkage between the two loci. IGS and ospC biallelic profiles could be analyzed in 192 ticks after mixtures and samples that were PCR negative for one of the two loci were eliminated (Table 5). ospC alleles D, H, F, I3, and M (nt 348) had strict associations with IGS alleles. ospC alleles H3, A, and B/B (nt 59) were associated with more than one IGS allele although a dominant allele linkage was evident (Table 5). The linked alleles H3 and EU886974 represented 21.9% of all ticks analyzed (Table 5). IGS allele EU886974 was not found associated with another ospC allele. The second-most-common biallelic profile was ospC H/IGS EU886969 (11.5%), followed by F/EF649786 (10.9%). ospC biallelic profiles E3/EU886975, A/EU886973, and D/EU886970 were all found at the same frequency (8.9%). The next most common profile was E3/AY275201 (5.7%), followed by G/EU886971 (4.2%) and G/EU886972 (3.6%), B/DQ437500 (3.1%), I3/4C and M (nt 348)/AY275202 (2.6%), and T/EU886975 (0.5%) (Table 5). With the exception of ospC alleles G and T, all IGS linkages in I. pacificus ticks were similar to those reported from the northeastern and midwestern United States (8, 26, 40). Comparison of ospC E3 linkage to IGS alleles in other areas of B. burgdorferi transmission will be reported elsewhere (Travinsky et al., unpublished data).
B. burgdorferi population structure of Mendocino County versus the northeastern United States.
We compared the frequency distribution of ospC alleles found in I. pacificus nymphs collected in 2004 with that found in I. scapularis ticks collected in the northeastern United States in three studies (45, 56, 57) (Table 6). Eight ospC genotypes, C, E, I, J, K, N, O, and U, identified in I. scapularis were absent from I. pacificus nymphs collected in 2004. ospC K made up the majority of alleles found in I. scapularis in two out of three of the studies used for comparison (15.7% [45] and 19.5% [57]). In this comparison, three ospC genotypes, E3, H3, and I3, were found only in California.
TABLE 6.
Comparison of ospC allele frequency distribution in I.pacificus nymphs from Mendocino County (2004) versus in I.scapularis nymphs or adults from three studies conducted in the northeastern United States
| ospC genotype | Presence of the genotype in ticks of the indicated region |
Statistical comparison (P)e | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Shelter Island, NY (n = 40)a |
Long Island and the coast from MA to SC (n = 203)b |
Dutchess County, NY (n = 451)c |
Mendocino County, CA (n = 205)d |
||||||
| No. positive | Frequency (% of total) | No. positive | Frequency (% of total) | No. positive | Frequency (% of total) | No. positive | Frequency (% of total) | ||
| A | 12 | 16.2 | 40 | 8.2 | 59 | 13.1 | 24 | 10.9 | 0.46 |
| B | 12 | 16.2 | 51 | 10.5 | 44 | 9.8 | 11 | 5.0 | 0.036 |
| C | 11 | 14.9 | NA | 0 | 0.0 | 0 | 0.0 | ||
| D | 9 | 12.2 | 34 | 7.0 | 40 | 8.9 | 18 | 8.1 | 0.88 |
| E | 4 | 5.4 | 34 | 7.0 | 30 | 6.7 | 0 | 0.0 | <0.001 |
| F | 6 | 8.1 | 38 | 7.8 | 31 | 6.9 | 20 | 9.0 | 0.35 |
| G | 5 | 6.8 | 40 | 8.2 | 25 | 5.5 | 18 | 8.1 | 0.24 |
| H | 7 | 9.5 | 39 | 8.0 | 24 | 5.3 | 27 | 12.2 | 0.003 |
| I | 1 | 1.4 | NA | 15 | 3.3 | 0 | 0.0 | 0.004 | |
| J | 2 | 2.7 | 14 | 2.9 | 9 | 2.0 | 0 | 0.0 | 0.034 |
| K | 5 | 6.8 | 76 | 15.7 | 88 | 19.5 | 0 | 0.0 | <0.001 |
| L | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | |
| M | 0 | 0.0 | 44 | 9.1 | 39 | 8.6 | 6 | 2.7 | 0.003 |
| N | 0 | 0.0 | 40 | 8.2 | 9 | 2.0 | 0 | 0.0 | 0.034 |
| O | 0 | 0.0 | 0 | 0.0 | 1 | 0.2 | 0 | 0.0 | 1 |
| T | 0 | 0.0 | 19 | 3.9 | 23 | 5.1 | 1 | 0.5 | 0.001 |
| U | 0 | 0.0 | 16 | 3.3 | 14 | 3.1 | 0 | 0.0 | 0.007 |
| E3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 38 | 17.2 | <0.001 |
| I3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 5 | 2.3 | 0.004 |
| H3 | 0 | 0.0 | 0 | 0.0 | 0 | 0.0 | 53 | 24.0 | <0.001 |
| Total | 74 | 100.0 | 485 | 100.0 | 451 | 100.0 | 221 | 100.0 | |
Of the ospC alleles that the northeastern and Californian studies shared, A, D, F, and G occurred at similarly high frequencies in the two locations (10.9%, 8.1%, 9.0%, and 8.1%, respectively, for Mendocino County versus a range of 8.2 to 16.2%, 7 to 12.1%, 6.9 to 8.1%, and 5.5 to 8.2%, respectively, for northeastern studies) (Table 6). The frequency (5.0%) of ospC B in Mendocino County nymphs was about one-third to one-half that in ticks from the Northeast (range of 9.8 to 16.2%). ospC H, the third-most-frequent allele in California ticks (12.2%), was slightly less common in I. scapularis (range of 5.3 to 9.5%). ospC allele M was more common in I. scapularis (range of 0 to 9.1%) than in I. pacificus (2.7%). ospC T was less common in Mendocino County (0.5% versus a range of 0 to 5.1%) although only a single tick with this sequence type was identified during our study.
Because Wang et al. (56) and Qiu et al. (45) genotyped B. burgdorferi in adult ticks, we could directly compare ospC allele frequencies in our study only to those found in Dutchess County, NY, I. scapularis nymphs by Wormser et al. (57). This decision was made based on the assumption that allele frequencies may differ in nymphs and adults due to differences in host preference (12) and because adult ticks are likely infected with multiple alleles (45). The frequencies of ospC alleles A, D, F, and G were similar between studies (Table 6). All other shared alleles were in dissimilar proportions, including B, E3, H, H3, I3, M, and T. Only alleles H, E3, H3, and I3 were more common in Mendocino County than in Dutchess County.
Genetic characterization of Californian B. burgdorferi strain CA8.
The genetic divergence and abundance of ospC H3 in Mendocino County nymphs prompted further investigation into the relationship between isolates containing the allele and other B. burgdorferi and Borrelia sp. strains. We analyzed partial sequences of the B. burgdorferi 16S rRNA and flaB genes (Fig. 1) using DNA extracted from B. burgdorferi culture isolate CA8 whose ospC sequence belonged to genotype H3 (Table 3). The neighbor-joining distance phylograms comparing CA8 to select Borrelia species (16S rRNA locus) and to select B. burgdorferi strains (flaB locus) confirmed both its status as B. burgdorferi and its genetic divergence from previously characterized B. burgdorferi isolates.
Habitat associations of novel and abundant California ospC alleles.
We analyzed the geographic distributions and relative frequencies of three ospC genotypes, H3, E3 and I3, which are unique and abundant in California (Fig. 2). The relative frequencies of ospC H3 and E3 were highest in the central and southeastern regions of the county, which overlapped previously reported areas of high predicted acarological risk of exposure to I. pacificus nymphs (22).
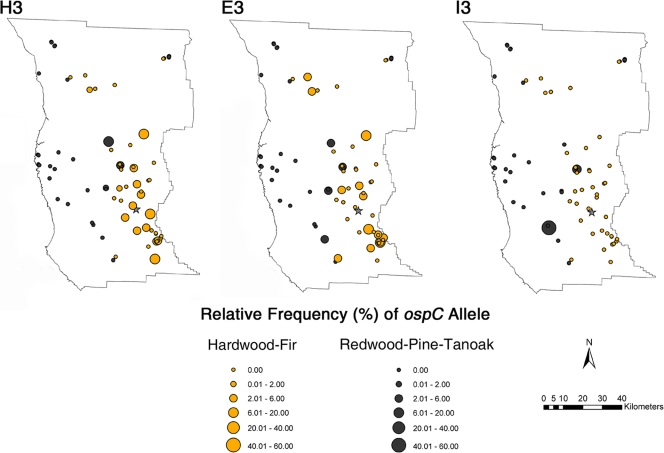
FIG. 2.
Relative frequency distribution of B. burgdorferi ospC alleles H3, E3, and I3 found in I. pacificus nymphs across 74 woodland collection sites in Mendocino County. Circle diameters correspond to relative frequency values (displayed as percentages). The largest city in the county, Ukiah, is indicated by a star.
We hypothesized that ospC H3 and E3 would be significantly associated with hardwood-fir habitats because the primary LB reservoir in Mendocino County, the western gray squirrel, is commonly infected with B. burgdorferi in these habitats (20, 31, 46). In our analysis, tick collection sites that contained ospC H3 were more likely to be hardwood-fir habitats than redwood-pine-tanoak habitats (Fisher's exact test, P < 0.05). The same association was not found for ospC E3 (P = 0.238). Of the 164 B. burgdorferi-infected ticks in hardwood-fir habitats, 23.2% carried ospC H3, 17.1% were infected with E3, and none carried I3. Among the 45 B. burgdorferi-infected ticks found in redwood-pine-tanoak habitats, 26.7% carried ospC H3, 13.3% were infected with E3, and 11.1% carried I3. Redwood-pine-tanoak sites containing a high percentage of ospC H3 were located near hardwood-fir habitats in inland Mendocino County (Fig. 2). Redwood-pine-tanoak sites with little or no ospC H3 were located in the western, coastal region of Mendocino County.
DISCUSSION
California B. burgdorferi population structure and LB.
We conducted the first large-scale, systematic survey of B. burgdorferi ospC and IGS sequence types in western North America. Our findings highlight differences between B. burgdorferi genotypes in I. pacificus ticks and those found in the eastern LB vector, I. scapularis. For example, the most abundant B. burgdorferi ospC allele in Mendocino County ticks, H3, had not been described in the Northeast, and E3 had been identified only in ticks from the north-central United States (Travinsky et al., unpublished). We identified seven B. burgdorferi strains defined by ospC alleles B, D, G, T, H3, I3, and E3, California representatives of which are lacking from a recent study of B. burgdorferi evolution and geographic population structure in North America and Europe (40). Inclusion of these samples in future studies using methods suitable for inferring spirochete evolution, such as multilocus sequence typing based on housekeeping genes, may clarify the relationship between far-western B. burgdorferi populations and those found in the other parts of the United States and Europe (27, 40).
Sequence variation in the ospC locus of B. burgdorferi has been linked to variation in the probability of disseminated spirochete infection and in the clinical manifestations of LB (15, 17, 28, 49, 57). The lack of previous reports of ospC H3 in clinical studies enables us only to speculate about its pathogenic potential in humans. Although E3 has been identified only in ticks in the north-central United States, a sequence similar to the E3 allele (GenBank accession number EU482056) was recently described in a clinical isolate from the Northeast (26). Given the high frequency of H3 and E3 genotypes in I. pacificus and their location in areas of high acarological risk to humans (22), it is possible that humans in Mendocino County regularly are being exposed to these genotypes during a tick bite. However, further classification of tick collection sites by human usage is required to assess the likelihood of human exposure to particular genotypes.
ospC allele frequency distributions in tick populations do not always correspond to frequency distributions found in LB patient skin lesions or secondary sites of infection (49). This may be related to differences in the spirochete detection methods used for clinical samples (i.e., cultivation) versus ticks (i.e., PCR on extracted DNA), resulting in culture bias for particular genotypes. A limited amount of information exists regarding the relationship between ospC genotype and the pathogenesis of LB in California. To our knowledge, B. burgdorferi isolates derived from blood or cerebrospinal fluid of California patients have been ospC genotype A (n = 3 isolates), while this genotype and others including H, I, and M (n = 1 isolate each) have been found in the skin (40, 49). Thorough analysis of ospC sequence types in California LB patients is sorely needed and is essential to understanding the relationship between ospC allele frequency in ticks and human infections in the region.
I. pacificus nymphs lacked two ospC genotypes, I and K, which are considered to be highly invasive (15) or associated with disseminated human infection in the Northeast (49, 57). Genotype K is abundant in I. scapularis as it has been found in 6.8% (49, 56), 15.7% (45), and 19.5% of ticks (57) and has been isolated in about 30 to 40% of human skin and blood cultures in northeastern studies (49, 57). Although ospC genotype I was identified in a single tick from Mendocino County in 1992 (CA337), its absence, as well as the absence of genotype K, in the B. burgdorferi-positive ticks genotyped throughout the county in 2004 is notable. It is possible that the lack of genotypes I and K in I. pacificus is related to the lower incidence of human cases in the Far West, but extensive testing of California B. burgdorferi human isolates is needed to draw any conclusions regarding spirochete genetics and regional differences in disease.
Vertebrate-vector-spirochete interactions in California.
California and the Northeast differ in climate and topography, as well as in reservoir and vector host species involved in enzootic LB transmission cycles. These transmission cycles are also geographically separated by about 3,000 miles. Therefore, it is not surprising that we detected major differences in B. burgdorferi population structures between the two regions. Approximately half of the ospC genotypes previously found in the Northeast were not detected in California ticks, meaning that the genetic diversity of B. burgdorferi is comparatively lower in the Far West. One possible explanation is that B. burgdorferi-refractory lizards (29, 30, 33, 59), the primary larval and nymphal hosts for I. pacificus in Mendocino County (18), are reducing the overall diversity of spirochetes. If so, only genotypes that can replicate well, or persist in, nonlizard tick hosts might be selected for.
Western gray squirrels, who account for the majority of infectious feeds to I. pacificus larvae in oak woodland habitats (20), appear to be particularly susceptible to ospC genotypes H3 and E3. In preliminary studies, over 50% of western gray squirrels collected in Mendocino County were infected with B. burgdorferi ospC genotypes H3 and E3 (S. Leonhard, Y. A. Girard, D. J. Salkeld and R. S. Lane, unpublished data). This prevalence is similar to the combined prevalence of ospC H3 and E3 in I. pacificus nymphs (41%). The positive association of ospC H3 with hardwood-fir habitats further supports the notion that these squirrels are members of an ospC H3 niche in Mendocino County (6).
Genotypic diversity of Californian B. burgdorferi.
In 2007, three adjacent counties in north-coastal California, Trinity, Humboldt, and Mendocino, and one inland county, Amador, reported the highest incidence of LB cases in the state (≥5 per 100,000 person-years) (11). B. burgdorferi in other counties in California where Lyme disease is endemic may have different population structures than in Mendocino County, particularly where vertebrate reservoirs other than the western gray squirrel may maintain spirochetes. Variability in both the size (molecular mass) and the amount of ospC protein expressed by B. burgdorferi isolates from 26 counties throughout California (48) suggests that statewide spirochete diversity also may exist at a nucleotide level.
Strong linkage between the IGS and ospC loci indicate that B. burgdorferi is clonal (8, 26). When linkages between ospC and IGS were analyzed in our study, identification of nondominant biallelic profiles (Table 5) suggested the presence of mixed infections in which the IGS primers amplified the gene of one strain while the ospC primers amplified the gene of a different strain. This is a potential limitation of using PCR and DNA sequencing alone. Another explanation is that multiple IGS alleles are truly associated with a single ospC allele. Because our methods possibly underestimated the multiplicity of infection in I. pacificus, other methods such as reverse line blot or PCR-single-strand conformation polymorphism (45, 56) may be needed to further examine multiple infections in I. pacificus ticks. However, because the spirochete infection rate in I. pacificus in California was generally lower than in I. scapularis in the Northeast (53), we would expect to find a lower degree of multiplicity in I. pacificus (45).
Our report of B. burgdorferi genotypes in I. pacificus nymphs represents a snapshot of the spirochete population in Mendocino County vectors. Although we cannot directly compare the frequency distribution of alleles in B. burgdorferi culture isolates to the 2004 tick samples for a number of reasons (e.g., different life stages, multiple counties, multiple years of collection, and nonsystematic sampling), it is remarkable that, with the exception of CA8 from adjacent Sonoma County, the two predominant ospC genotypes in nymphs, H3 and E3, were absent in culture-derived DNA. It is possible that a temporal shift in allele frequency may have occurred in the spirochete population, as observed in areas of the Northeast (43, 45). More likely, differences in allelic frequencies between the sample sets are due to selection by culture, a phenomenon observed by others studying the allele frequencies of IGS, p66, and ospA (37, 41). Given the high frequency of ospC A in the culture isolates, this genotype may outgrow ospC H3 and E3 in a mixed infection.
PCR-RFLP typing using the 5S-23S or 16S-23S rRNA spacer regions has been used to sort Borrelia genospecies and genotypes within a genospecies (36, 42). Brown et al. (7), using PCR-RFLP to genotype northern Californian B. burgdorferi isolates based on MseI and DraI fragment patterns, defined only two genotypes, referred to as A and B. We improved upon this earlier effort by sequencing a portion of the ospC gene and were able to define seven different genotypes (A, D, F, I, I3, G, and H) within the same samples. Further characterization of northern California B. burgdorferi isolates by sequence analysis of other loci and through studies of animal pathogenesis and vector competence will continue to improve our understanding of spirochete diversity in the Far West.
Conclusions.
Variation in the spirochete sequence types and abundance in California compared to the Northeast may be related to a number of factors including selection for genotypes within different tick vector species; differences in climate, habitat, and topography that affect the host-seeking phenology of such ticks; and differences in the type and abundance of reservoir hosts. Efforts to define the genetic background of B. burgdorferi in ticks, vertebrate hosts, and humans are important for developing effective LB diagnostics, vaccines, and public health interventions. Genetic variation in B. burgdorferi may influence both the severity of LB and the sensitivity of serological assays used to help diagnose it (58). Given the striking differences in spirochete population structure in I. pacificus compared to I. scapularis, it is likely that humans in California are exposed to a different subset of B. burgdorferi lineages than humans in the Northeast. Research is currently under way that should elucidate the role of California vertebrate reservoirs in B. burgdorferi transmission. This information, in conjunction with our current knowledge of high-risk habitats (13, 21, 22) and human behaviors that elevate exposure to infected ticks (34), will facilitate the design of interventions that target the unique B. burgdorferi transmission cycle in the Far West.
Acknowledgments
This work was supported by the National Institutes of Health grants AI22501 (R.S.L.) and AI24424 (A.G.B.) and by patent royalties (A.G.B.).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We gratefully acknowledge the technical help of Jeomhee Mun, Beth Slikas, Sarah Leonhard, and Joyce E. Kleinjan.
Footnotes
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Alghaferi, M. Y., J. M. Anderson, J. Park, P. G. Auwaerter, J. N. Aucott, D. E. Norris, and J. S. Dumler. 2005. Borrelia burgdorferi ospC heterogeneity among human and murine isolates from a defined region of northern Maryland and southern Pennsylvania: lack of correlation with invasive and noninvasive genotypes. J. Clin. Microbiol. 43:1879-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Attie, O., J. F. Bruno, Y. Xu, D. Qiu, B. J. Luft, and W. G. Qiu. 2007. Co-evolution of the outer surface protein C gene (ospC) and intraspecific lineages of Borrelia burgdorferi sensu stricto in the northeastern United States. Infect. Genet. Evol. 7:1-12. [DOI] [PubMed] [Google Scholar]
- 3.Bacon, R. M., K. J. Kugeler, and P. S. Mead. 2008. Surveillance for Lyme disease—United States, 1992-2006. MMWR Surveill. Summ. 57:1-9. [PubMed] [Google Scholar]
- 4.Barbour, A. G. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521-525. [PMC free article] [PubMed] [Google Scholar]
- 5.Barbour, A. G., G. O. Maupin, G. J. Teltow, C. J. Carter, and J. Piesman. 1996. Identification of an uncultivable Borrelia species in the hard tick Amblyomma americanum: possible agent of a Lyme disease-like illness. J. Infect. Dis. 173:403-409. [DOI] [PubMed] [Google Scholar]
- 6.Brisson, D., and D. E. Dykhuizen. 2004. ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics 168:713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, R. N., M. A. Peot, and R. S. Lane. 2006. Sylvatic maintenance of Borrelia burgdorferi (Spirochaetales) in Northern California: untangling the web of transmission. J. Med. Entomol. 43:743-751. [DOI] [PubMed] [Google Scholar]
- 8.Bunikis, J., U. Garpmo, J. Tsao, J. Berglund, D. Fish, and A. G. Barbour. 2004. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150:1741-1755. [DOI] [PubMed] [Google Scholar]
- 9.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 10.Burgdorfer, W., R. S. Lane, A. G. Barbour, R. A. Gresbrink, and J. R. Anderson. 1985. The western black-legged tick, Ixodes pacificus: a vector of Borrelia burgdorferi. Am. J. Trop. Med. Hyg. 34:925-930. [DOI] [PubMed] [Google Scholar]
- 11.California Department of Public Health. 2007. Vector-borne diseases survey annual report. http://www.cdph.ca.gov/programs/vbds/Pages/VBDSAnnualReports.aspx.
- 12.Castro, M. B., and S. A. Wright. 2007. Vertebrate hosts of Ixodes pacificus (Acari: Ixodidae) in California. J. Vector Ecol. 32:140-149. [DOI] [PubMed] [Google Scholar]
- 13.Clover, J. R., and R. S. Lane. 1995. Evidence implicating nymphal Ixodes pacificus (Acari: Ixodidae) in the epidemiology of Lyme disease in California. Am. J. Trop. Med. Hyg. 53:237-240. [DOI] [PubMed] [Google Scholar]
- 14.Dykhuizen, D. E., and G. Baranton. 2001. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 15.Dykhuizen, D. E., D. Brisson, S. Sandigursky, G. P. Wormser, J. Nowakowski, R. B. Nadelman, and I. Schwartz. 2008. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am. J. Trop. Med. Hyg. 78:806-810. [PMC free article] [PubMed] [Google Scholar]
- 16.Dykhuizen, D. E., D. S. Polin, J. J. Dunn, B. Wilske, V. Preac-Mursic, R. J. Dattwyler, and B. J. Luft. 1993. Borrelia burgdorferi is clonal: implications for taxonomy and vaccine development. Proc. Natl. Acad. Sci. USA 90:10163-10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earnhart, C. G., E. L. Buckles, J. S. Dumler, and R. T. Marconi. 2005. Demonstration of OspC type diversity in invasive human Lyme disease isolates and identification of previously uncharacterized epitopes that define the specificity of the OspC murine antibody response. Infect. Immun. 73:7869-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen, L., R. J. Eisen, and R. S. Lane. 2004. The roles of birds, lizards, and rodents as hosts for the western black-legged tick Ixodes pacificus. J. Vector Ecol. 29:295-308. [PubMed] [Google Scholar]
- 19.Eisen, L., R. J. Eisen, and R. S. Lane. 2006. Geographical distribution patterns and habitat suitability models for presence of host-seeking ixodid ticks in dense woodlands of Mendocino County, California. J. Med. Entomol. 43:415-427. [DOI] [PubMed] [Google Scholar]
- 20.Eisen, L., R. J. Eisen, J. Mun, D. J. Salkeld, and R. S. Lane. 2009. Transmission cycles of Borrelia burgdorferi and B. bissettii in relation to habitat type in northwestern California. J. Vector Ecol. 34:81-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisen, R. J., L. Eisen, and R. S. Lane. 2006. Predicting density of Ixodes pacificus nymphs in dense woodlands in Mendocino County, California, based on geographic information systems and remote sensing versus field-derived data. Am. J. Trop. Med. Hyg. 74:632-640. [PubMed] [Google Scholar]
- 22.Eisen, R. J., R. S. Lane, C. L. Fritz, and L. Eisen. 2006. Spatial patterns of Lyme disease risk in California based on disease incidence data and modeling of vector-tick exposure. Am. J. Trop. Med. Hyg. 75:669-676. [PubMed] [Google Scholar]
- 23.Galtier, N., M. Gouy, and C. Gautier. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543-548. [DOI] [PubMed] [Google Scholar]
- 24.Godsey, M. S., Jr., T. E. Amundson, E. C. Burgess, W. Schell, J. P. Davis, R. Kaslow, and R. Edelman. 1987. Lyme disease ecology in Wisconsin: distribution and host preferences of Ixodes dammini, and prevalence of antibody to Borrelia burgdorferi in small mammals. Am. J. Trop. Med. Hyg. 37:180-187. [DOI] [PubMed] [Google Scholar]
- 25.Grimm, D., K. Tilly, R. Byram, P. E. Stewart, J. G. Krum, D. M. Bueschel, T. G. Schwan, P. F. Policastro, A. F. Elias, and P. A. Rosa. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. USA 101:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanincova, K., D. Liveris, S. Sandigursky, G. P. Wormser, and I. Schwartz. 2008. Borrelia burgdorferi sensu stricto is clonal in patients with early Lyme borreliosis. Appl. Environ. Microbiol. 74:5008-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoen, A. G., G. Margos, S. J. Bent, M. Diuk-Wasser, A. G. Barbour, K. Kurtenbach, and D. Fish. 14 August 2009, posting date. Phylogeography of Borrelia burgdorferi in the eastern United States reflects multiple independent Lyme disease emergence events. Proc. Natl. Acad. Sci. USA. doi: 10.1073/pnas.0903810106. [DOI] [PMC free article] [PubMed]
- 28.Lagal, V., D. Postic, E. Ruzic-Sabljic, and G. Baranton. 2003. Genetic diversity among Borrelia strains determined by single-strand conformation polymorphism analysis of the ospC gene and its association with invasiveness. J. Clin. Microbiol. 41:5059-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane, R. S. 1990. Susceptibility of the western fence lizard (Sceloporus occidentalis) to the Lyme borreliosis spirochete (Borrelia burgdorferi). Am. J. Trop. Med. Hyg. 42:75-82. [DOI] [PubMed] [Google Scholar]
- 30.Lane, R. S., and J. E. Loye. 1989. Lyme disease in California: interrelationship of Ixodes pacificus (Acari: Ixodidae), the western fence lizard (Sceloporus occidentalis), and Borrelia burgdorferi. J. Med. Entomol. 26:272-278. [DOI] [PubMed] [Google Scholar]
- 31.Lane, R. S., J. Mun, R. J. Eisen, and L. Eisen. 2005. Western gray squirrel (Rodentia: Sciuridae): a primary reservoir host of Borrelia burgdorferi in Californian oak woodlands? J. Med. Entomol. 42:388-396. [DOI] [PubMed] [Google Scholar]
- 32.Lane, R. S., and J. A. Pascocello. 1989. Antigenic characteristics of Borrelia burgdorferi isolates from ixodid ticks in California. J. Clin. Microbiol. 27:2344-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane, R. S., and G. B. Quistad. 1998. Borreliacidal factor in the blood of the western fence lizard (Sceloporus occidentalis). J. Parasitol. 84:29-34. [PubMed] [Google Scholar]
- 34.Lane, R. S., D. B. Steinlein, and J. Mun. 2004. Human behaviors elevating exposure to Ixodes pacificus (Acari: Ixodidae) nymphs and their associated bacterial zoonotic agents in a hardwood forest. J. Med. Entomol. 41:239-248. [DOI] [PubMed] [Google Scholar]
- 35.Levine, J. F., M. L. Wilson, and A. Spielman. 1985. Mice as reservoirs of the Lyme disease spirochete. Am. J. Trop. Med. Hyg. 34:355-360. [DOI] [PubMed] [Google Scholar]
- 36.Liveris, D., A. Gazumyan, and I. Schwartz. 1995. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 33:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liveris, D., S. Varde, R. Iyer, S. Koenig, S. Bittker, D. Cooper, D. McKenna, J. Nowakowski, R. B. Nadelman, G. P. Wormser, and I. Schwartz. 1999. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J. Clin. Microbiol. 37:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liveris, D., G. P. Wormser, J. Nowakowski, R. Nadelman, S. Bittker, D. Cooper, S. Varde, F. H. Moy, G. Forseter, C. S. Pavia, and I. Schwartz. 1996. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:1306-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livey, I., C. P. Gibbs, R. Schuster, and F. Dorner. 1995. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol. Microbiol. 18:257-269. [DOI] [PubMed] [Google Scholar]
- 40.Margos, G., A. G. Gatewood, D. M. Aanensen, K. Hanincova, D. Terekhova, S. A. Vollmer, M. Cornet, J. Piesman, M. Donaghy, A. Bormane, M. A. Hurn, E. J. Feil, D. Fish, S. Casjens, G. P. Wormser, I. Schwartz, and K. Kurtenbach. 2008. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 105:8730-8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norris, D. E., B. J. Johnson, J. Piesman, G. O. Maupin, J. L. Clark, and W. C. Black. 1997. Culturing selects for specific genotypes of Borrelia burgdorferi in an enzootic cycle in Colorado. J. Clin. Microbiol. 35:2359-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postic, D., M. V. Assous, P. A. Grimont, and G. Baranton. 1994. Diversity of Borrelia burgdorferi sensu lato evidenced by restriction fragment length polymorphism of rrf (5S)-rrl (23S) intergenic spacer amplicons. Int. J. Syst. Bacteriol. 44:743-752. [DOI] [PubMed] [Google Scholar]
- 43.Qiu, W. G., E. M. Bosler, J. R. Campbell, G. D. Ugine, I. N. Wang, B. J. Luft, and D. E. Dykhuizen. 1997. A population genetic study of Borrelia burgdorferi sensu stricto from eastern Long Island, New York, suggested frequency-dependent selection, gene flow and host adaptation. Hereditas 127:203-216. [DOI] [PubMed] [Google Scholar]
- 44.Qiu, W. G., J. F. Bruno, W. D. McCaig, Y. Xu, I. Livey, M. E. Schriefer, and B. J. Luft. 2008. Wide distribution of a high-virulence Borrelia burgdorferi clone in Europe and North America. Emerg. Infect. Dis. 14:1097-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu, W. G., D. E. Dykhuizen, M. S. Acosta, and B. J. Luft. 2002. Geographic uniformity of the Lyme disease spirochete (Borrelia burgdorferi) and its shared history with tick vector (Ixodes scapularis) in the Northeastern United States. Genetics 160:833-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salkeld, D. J., S. Leonhard, Y. A. Girard, N. Hahn, J. Mun, K. A. Padgett, and R. S. Lane. 2008. Identifying the reservoir hosts of the Lyme disease spirochete Borrelia burgdorferi in California: the role of the western gray squirrel (Sciurus griseus). Am. J. Trop. Med. Hyg. 79:535-540. [PMC free article] [PubMed] [Google Scholar]
- 47.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwan, T. G., M. E. Schrumpf, R. H. Karstens, J. R. Clover, J. Wong, M. Daugherty, M. Struthers, and P. A. Rosa. 1993. Distribution and molecular analysis of Lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J. Clin. Microbiol. 31:3096-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seinost, G., D. E. Dykhuizen, R. J. Dattwyler, W. T. Golde, J. J. Dunn, I. N. Wang, G. P. Wormser, M. E. Schriefer, and B. J. Luft. 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 67:3518-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steere, A. C. 1989. Lyme disease. N. Engl. J. Med. 321:586-596. [DOI] [PubMed] [Google Scholar]
- 51.Steere, A. C., J. Coburn, and L. Glickstein. 2004. The emergence of Lyme disease. J. Clin. Investig. 113:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 53.Talleklint-Eisen, L., and R. S. Lane. 1999. Variation in the density of questing Ixodes pacificus (Acari: Ixodidae) nymphs infected with Borrelia burgdorferi at different spatial scales in California. J. Parasitol. 85:824-831. [PubMed] [Google Scholar]
- 54.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tilly, K., J. G. Krum, A. Bestor, M. W. Jewett, D. Grimm, D. Bueschel, R. Byram, D. Dorward, M. J. Vanraden, P. Stewart, and P. Rosa. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74:3554-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, I. N., D. E. Dykhuizen, W. Qiu, J. J. Dunn, E. M. Bosler, and B. J. Luft. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wormser, G. P., D. Brisson, D. Liveris, K. Hanincova, S. Sandigursky, J. Nowakowski, R. B. Nadelman, S. Ludin, and I. Schwartz. 2008. Borrelia burgdorferi genotype predicts the capacity for hematogenous dissemination during early Lyme disease. J. Infect. Dis. 198:1358-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wormser, G. P., D. Liveris, K. Hanincova, D. Brisson, S. Ludin, V. J. Stracuzzi, M. E. Embers, M. T. Philipp, A. Levin, M. Aguero-Rosenfeld, and I. Schwartz. 2008. Effect of Borrelia burgdorferi genotype on the sensitivity of C6 and 2-tier testing in North American patients with culture-confirmed Lyme disease. Clin. Infect. Dis. 47:910-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright, S. A., R. S. Lane, and J. R. Clover. 1998. Infestation of the southern alligator lizard (Squamata: Anguidae) by Ixodes pacificus (Acari: Ixodidae) and its susceptibility to Borrelia burgdorferi. J. Med. Entomol. 35:1044-1049. [DOI] [PubMed] [Google Scholar]