Abstract
Dominant bacteria in the different habitats in the Kuytun 51 Glacier were investigated using a 16S rRNA gene clone library sequencing technique. Results showed diverse bacteria on the glacial surface, with the dominant phyla being Proteobacteria, Cyanobacteria, and Bacteroidetes. UniFrac data showed distinct community patterns between the Kuytun and Himalayan Rongbuk glaciers.
Comparisons of geographically distinct glaciers worldwide have shown a great variation in microbial biomass and community structure (6, 8, 15, 18, 22, 34, 36). The variability is largely controlled by climatic and environmental factors, including geographic location (4, 12, 19, 24), wind direction, wind speed, light intensity, and availability of nutrients and liquid water (5, 6, 9, 13, 19, 24). There is some limited evidence of biogeographic effects on the distribution of microorganisms in the geographically different glaciers (7, 15, 25, 34, 36). However, main factors driving the dynamics of microbial community in glacial systems remain unclear.
The Kuytun 51 Glacier is a typical cold-based subcontinental glacier (11). The microbial communities in subcontinental glaciers have not been studied in detail, as there have been only a few reports on continental and polar glaciers (7, 16, 22, 23). To initially investigate the biogeographic effects on the distribution of microorganisms in glacial ice, six 16S rRNA gene libraries were established from the Kuytun 51 Glacier. Bacterial-community comparison was conducted among the Kuytun 51, John Evans (Canada) (22), Alaska Bench (22), and Himalayan Rongbuk (16) glaciers.
The glacial samples were obtained with extreme caution from the different habitats, solid ice, firn, and snow, at the terminus, middle, and top of the Kuytun 51 Glacier (84°24′E, 43°43′N) in the Tianshan Mountains in 2005. Numbers of live cells and total biomass in the melt water were determined with a flow cytometer in combination with cell marker propidium iodide and carboxyfluorescein diacetate by following the method of Amor et al. (3), except for cell staining for 15 min at 25°C. DNA nucleic acids were extracted from a portion (400 ml) of the melt water filtered through a sterile 0.22-μm-pore-size filter unit (Millipore) by following the protocols described by Zhou et al. (38). A neighbor-joining (28) phylogeny for the ClustalX (30) aligned sequences was constructed using MEGA 4.0 (27) with the maximum composite likelihood method (see the detailed methods in the supplemental material).
The Kuytun 51 Glacier contains a relatively large biomass, ranging from 7.27 × 104 to 1.29 × 106 cells/ml and 1.14 × 103 to 2.69 × 104 cells/ml for the total biomass and live biomass, respectively (Table 1); these numbers are higher than in most glaciers worldwide (Table 2). This could be attributed to the fact that Kuytun 51 Glacier is located near (within about 10 km of) human settlements and surrounded by vast deserts: the Gobi, Muyun Kum, Kyzyl Kum, and Kara Kum deserts (14). The frequent dust storm outbreaks that emerge in and around the arid and semiarid regions possibly carry a large number of microbes associated with dust onto the glacier via prevalent westerly winds (10, 14, 33). Biomass is generally higher in the mountainous glaciers than in the polar glaciers, which are far away and little influenced by the activities of human beings (Table 2) (8, 34). However, there are some exceptions, including low biomass in the Malan Glacier (35) and high biomass in the “silty” ice from the GISP 2 ice core (18). This may be attributed to the specific climatic and environmental conditions and local habitats. More data are necessary before definitive conclusions can be drawn because of different methods used in the estimation of biomass.
TABLE 1.
Bacterial biomasses in the different habitats of the Kuytun 51 Glacier
| Zone | Altitude (m above sea level) | Description | Total no. of cells (104 cells ml−1) | No. of live cells (102 cells ml−1) | % of cells that were live |
|---|---|---|---|---|---|
| Dry snow | 3,725 | 16.96 | 11.45 | 0.68 | |
| Firn | 3,601 | Firn zonea | 105.40 ± 23.54 | 232.34 ± 36.37 | 2.20 ± 0.88 |
| White | 81.86 ± 20.36 | 195.97 ± 30.78 | 2.39 ± 1.20 | ||
| Brown | 128.93 ± 27.14 | 268.71 ± 40.32 | 2.08 ± 0.32 | ||
| Ablation | 3,505 | Ablation zoneb | 34.92 ± 24.59 | 77.72 ± 51.26 | 2.22 ± 0.33 |
| Ice, H2O, and particles | 43.18 | 94.9 | 2.2 | ||
| Ice | 54.32 | 118.18 | 2.18 | ||
| H2O | 7.27 | 20.07 | 2.76 |
The average value was calculated by the analysis of three white and three brown snow samples collected in the firn zone of the glacial surface.
The average value was calculated by the analysis of ice, ice-water particles, and water samples collected in the ablation zone.
TABLE 2.
Biomass comparison among ice samples from the Kuytun 51 Glacier and other glaciers worldwide
| Glacier | Location | Altitude (m above sea level) | Glacial type | Total biomass (104 cells/ml) | Live biomassa | Reference |
|---|---|---|---|---|---|---|
| Kuytun Tienshan Mt. | 84°24′E | 3,500-4,000 | Subcontinental | 7.3-129 | 11.4 × 102-269 × 102 cells/ml | This study |
| 43°43′N | ||||||
| Rongbuk Hymalaya | 28°01′N | 6,000-8,000 | Continental | 2.1-90.4 | 0.1-5 CFU/ml | 16, 34 |
| 86°57′E | ||||||
| Guoqu Geladandong | 33°34′N | 6,621 | Continental | 0.3-83 (ice core) | ND | 15 |
| 91°10′E | ||||||
| Palong | 29°14′N | 4,618-5,974 | Marine | 0.68-37.2 | ND | 15 |
| 96°55′E | ||||||
| Kongsvege | 78°56′N | 1-20 | 2 | |||
| 11°52′E | ||||||
| Muztag Ata | 38°17′N | 6,350 | Continental | 0.2-21.7 (ice core) | 0-1.04 CFU/ml | 32 |
| 75°04′E | ||||||
| Guliya | 35°21′N | 6,200 | Subcontinental | 1 (ice core) | 0.07-1.8 CFU/ml | 8 |
| 81°31′E | ||||||
| Malan | 35°50′N | 5,680 | Continent | 0.005-0.04 (ice core) | <1-0.85 CFU/ml | 35 |
| 90°40′E | ||||||
| Puruogangri | 33°44′N | 6,000 | Subcontinental | 2.56-25.5 (ice core) | 0-7.6 CFU/ml | 37 |
| 89°20′E | ||||||
| Bench (Alaska) | 61.03°N | 950-1,600 | Subglacial | 6.6-37 | ND | 22 |
| 145.7°W | ||||||
| John Evans (Arctic) | 79°40′N | 100-1,500 | Polythermal | ND | 1-10 CFU/ml | 22 |
| 74°00′W | ||||||
| Antarctica | 78°27′S | 3,500 | Polar | 0.003-0.83 | 1 | |
| 106°48′E | ||||||
| GISP2 | 72°35′N | 3,203 | Polar | 6,100-9,100 (base ice) | ND | 18 |
| Greenland | 37°38′W | |||||
| South Pole | 90°S | 2,835 | Polar | 0.02-0.5 | 6 | |
| Taylor Dome, Antarctica | 77°47′S | 2,365 | Polar | ∼10 CFU | 7 | |
| 158°43′E |
ND, no determination.
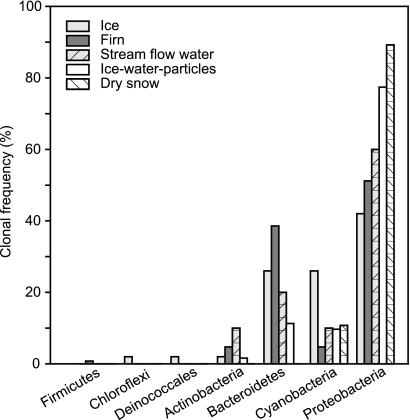
All 112 16S rRNA gene sequences obtained from the Kuytun 51 Glacier were >97% similar to known species from the different environments. They fell into 30 subphyla within 10 phyla: Alpha-, Beta-, Delta-, and Gammaproteobacteria, Bacteroidetes, Chloroflexi, Firmicutes, Actinobacteria, Deinococcales, and Cyanobacteria (see the supplemental material). There was an apparent difference in the phylogenetic distributions of bacteria in the glacier (Fig. 1). Proteobacteria were the most dominant bacteria, Bacteroidetes and Cyanobacteria were the second-most-common bacteria on the glacier surface, and Actinobacteria were present along the glacier surface, although they accounted for a small proportion of the total clones. Only a few clones belonging to the families Chloroflexi and Deinococcales and Firmicutes were found from the ice and firn niches.
FIG. 1.
Clonal frequency of the main phylogenetic groups based on the BLAST result of 16S rRNA gene clone sequencing in a single library.
The Kuytun Glacier bacteria represented two distinct metabolic types, including phototrophs and heterotrophs. Cyanobacteria were the dominant phototrophs across the glacial surface and closely related to the known species in four subgroups: Phormidium, Pseudanabaena, and Oscillatoria spp. and Stephanopyxidaceae. These phototrophic Cyanobacteria have frequently been reported from geographically different glaciers, such as the Alaska (26), Chile Tyndall (24), Svalbard (23), and Antarctic (7, 19, 29) glaciers. However, only one cyanobacterial clone has been reported from the Rongbuk Glacier (16). These results suggest that Cyanobacteria are very common in some glaciers but rare in other glaciers, which can be attributed to the possibly different sources of microbial loads on the glacier and the specific selection effect of local climatic and environmental conditions on microorganisms. The highly pigmented and structured biofilms may allow the Cyanobacteria to tolerate extreme environments and dominate in the glaciers (31).
Betaproteobacteria and Bacteroidetes are the most common phyla in the Kuytung 51 Glacier (Fig. 1), which is consistent with previous reports on the bacterial communities in glaciers (4, 15, 16, 23). This is possibly attributed to their tolerance to oligotrophic environments (21) and a wide spectrum of substrate range in low-nutrient media (20). Bacteroidetes are able to produce copious amounts of extracellular polymeric substances on surfaces and have high catabolic ability for complex and more recalcitrant organic matter (4).
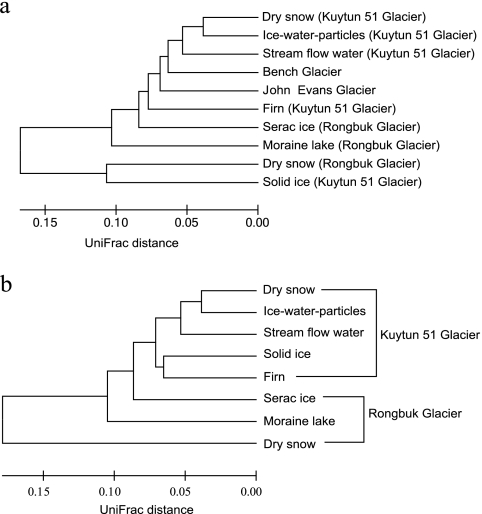
Also of significance are the apparent differences in dominant bacterial community compositions among the geographically distinct glaciers around the world (Fig. 2a and b). When subjected to UniFrac analysis, the dominant bacterial communities from the same glacier form distinct clusters from other glaciers (Fig. 2b). This promotes the idea of biogeography as an important factor with regard to microbial community structure.
FIG. 2.
Hierarchical clustering showing the overall phylogenetic distances between the bacterial communities on the Kuytun glacial surface (this study) and from John Evans Glacier, Canada (22), Alaska Bench Glacier (22), and East Rongbuk Glacier north of the Himalayas (16). (a) Comparison of communities among the Kuytun 51 Glacier, John Evans Glacier, Bench Glacier, and Rongbuk Glacier; (b) community comparison between the Kuytun 51 Glacier and Rongbuk Glacier (sequence KuyT-ice-3 with a later portion of the 16S rRNA gene was removed from the phylogenetic tree list). Distances were estimated with the weighted UniFrac algorithm (17). A sequence jackknifing technique was applied to each cluster to determine the sensitivity of the relationships to sample size.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the cloned sequences described in this study are EU263676 to EU263787.
Supplementary Material
Acknowledgments
We extend our special thanks to Lizhe An and Bingliang Xu for giving support for the completion of laboratory analysis for this study. We also thank Trista Vick very much for her kind help with the improvement of the English in this paper. We thank all of the members of the Kuytun Glacier 51 expedition for help with field sample collection.
This work was supported by the NSF Project of China (grants 40471025 and 40571038).
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abyzov, S. S., I. N. Mitskevich, and M. N. Poglazova. 1998. Microflora of the deep glacier horizons of central Antarctica. Microbiology 67:66-73. [Google Scholar]
- 2.Amato, P., R. Hennebelle, O. Magand, M. Sancelme, A. Delort, C. Barbante, C. Boutron, and C. Ferrari. 2007. Bacterial characterization of the snow cover at Spitzberg, Svalbard. FEMS Microbiol. Ecol. 59:255-264. [DOI] [PubMed] [Google Scholar]
- 3.Amor, K. B., P. Breeuwer, P. Verbaarschot, F. M. Rombouts, A. D. L. Akkermans, W. M. De Vos, and T. Abee. 2002. Multiparametric flow cytometry and cell sorting for the assessment of viable, injured, and dead bifidobacterium cells during bile salt stress. Appl. Environ. Microbiol. 68:5209-5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battin, T. J., A. Wille, B. Sattler, and R. Psenner. 2001. Phylogenetic and functional heterogeneity of sediment biofilms along environmental gradients in a glacial stream. Appl. Environ. Microbiol. 67:799-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatia, M., M. Sharp, and J. Foght. 2006. Distinct bacterial communities exist beneath a high Arctic polythermal glacier. Appl. Environ. Microbiol. 72:5838-5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpenter, E. J., S. Lin, and D. G. Capone. 2000. Bacterial activity in South Pole snow. Appl. Environ. Microbiol. 66:4514-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christner, B. C., B. H. Kvitko, and J. N. Reeve. 2003. Molecular identification of bacteria and eukarya inhabiting an Antarctic cryoconite hole. Extremophiles 7:177-183. [DOI] [PubMed] [Google Scholar]
- 8.Christner, B. C., E. Mosley-Thompson, L. G. Thompson, V. Zagorodnov, K. Sandman, and J. N. Reeve. 2000. Recovery and identification of viable bacteria immured in glacial ice. Icarus 144:479-485. [Google Scholar]
- 9.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microb. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 10.Husar, R. B., J. M. Prospero, and L. L. Stowe. 1997. Characterization of tropospheric aerosols over ocean with the NOAA/AVIIRR optical thickness operational product: passive remote sensing of tropospheric aerosol and atmospheric corrections from the new generation of satellite sensors. J. Geophys. Res. 102:16889-16909. [Google Scholar]
- 11.Jing, Z.-F., B.-S. Ye, K.-Q. Jiao, and H.-A. Yang. 2002. Surface velocity on the glacier no. 51 at Haxilegen of the Kuytun River, Tianshan Mountains. J. Glaciol. Geocryol. 24:563-566. [Google Scholar]
- 12.Kikuchi, Y. 1994. Glaciella, a new genus of freshwater Canthocampyidae (Copepoda Harpacticoida) from a glacier in Nepal, Himalayas. Hydrobiology 192:59-66. [Google Scholar]
- 13.Kohshima, S. 1994. Ecological characteristics of the glacier ecosystem. Jpn. J. Ecol. 44:93-98. [Google Scholar]
- 14.Li, X., D. Qin, G. Jiang, K. Duan, and H. Zhou. 2003. Atmospheric pollution of a remote area of Tianshan Mountain: ice core record. J. Geophys. Res. 108(D14):4406-4416. [Google Scholar]
- 15.Liu, Y., T. Yao, N. Jiao, S. Kang, B. Xu, Y. Zeng, S. Huang, and X. Liu. 2009. Bacteria diversity in the snow over Tibetan plateau glaciers. Extremophiles 13:411-423. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y., T. Yao, S. Kang, N. Jiao, Y. Zeng, S. Huang, and T. Luo. 2007. Microbial community structure in major habitats above 6000 m on Mount Everest. Chin. Sci. Bull. 52:2350-2357. [Google Scholar]
- 17.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miteva, V. I., P. P. Sheridan, and J. E. Brenchley. 2004. Phylogenetic and physiological diversity of microorganisms isolated from a deep Greenland glacier ice core. Appl. Environ. Microbiol. 70:202-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller, D. R., and W. H. Pollard. 2004. Gradient analysis of cryoconite ecosystems from two polar glaciers. Polar Biol. 27:66-74. [Google Scholar]
- 20.Noble, P. A., P. E. Dabinett, and J. Crow. 1990. A numerical taxonomic study of pelagic and benthic surface-layer bacteria in seasonally-cold coastal waters. Syst. Appl. Microbiol. 13:77-85. [Google Scholar]
- 21.Phung, N. T., J. Lee, K. H. Kang, I. S. Chang, G. M. Gadd, and B. H. Kim. 2004. Analysis of microbial diversity in oligotrophic microbial fuel cells using 16S rDNA sequences. FEMS Microbiol. Lett. 233:77-82. [DOI] [PubMed] [Google Scholar]
- 22.Skidmore, M., S. P. Anderson, M. Sharp, J. Foght, and B. D. Lanoil. 2005. Comparison of microbial community composition in two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl. Environ. Microbiol. 71:6986-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stibal, M., M. Sabacka, and K. Kastovska. 2006. Microbial communities on glacier surfaces in Svalbard: impact of physical and chemical properties on abundance and structure of cyanobacteria and algae. Microb. Ecol. 52:644-654. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi, N., and S. Koshima. 2004. A snow algal community on a Patagonian glacier, Tyndall glacier in the southern Patagonia Icefield. Arct. Antarct. Alpine Res. 36:91-98. [Google Scholar]
- 25.Takeuchi, N., J. Uetake, K. Fujita, V. Aizen, and S. Nikitin. 2006. A snow algal community on Akkem Glacier in the Russian Altai Mountains. Ann. Glaciol. 43:378-384. [Google Scholar]
- 26.Takeuchi, N. 2002. Surface albedo and characteristics of cryoconite on an Alaska glacier (Gulkana Glacier in the Alaska Range). Bull. Glaciol. Res. 19:63-70. [Google Scholar]
- 27.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 28.Tamura, K., M. Nei, and S. Kumar. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 101:11030-11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taton, A., S. Grubisic, E. Brambilla, R. De Wit, and A. Wilmotte. 2003. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl. Environ. Microbiol. 69:5157-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL-X Windows interface—flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vincent, W. F. 2000. Evolutionary origins of Antarctic microbiota: invasion, selection and endemism. Antarct. Sci. 12:374-385. [Google Scholar]
- 32.Xiang, S. R., T. D. Yao, G. J. Wu, Y. Chen, T. C. Shang, L. L. Pu, and L. Z. An. 2006. Deposition properties of bacterial populations in the Muztag Ata ice core. Quat. Sci. 26:185-191. [Google Scholar]
- 33.You, X. N., Q. Li, F. Wang, and Y. Zhu. 2006. Seasonal evolution of insoluble microparticles stratigraphy in glacier no. 1 percolation zone, Eastern Tianshan, China. Adv. Earth Sci. 21:1164-1170. [Google Scholar]
- 34.Zhang, S., S. Hou, X. Ma, D. Qin, and T. Chen. 2007. Culturable bacteria in Himalayan glacial ice in response to atmospheric circulation Biogeoscience 4:1-9. [Google Scholar]
- 35.Zhang, X., T. Yao, and X. Ma. 2002. Microorganism in a high altitude glacier ice in Tibet, China. Folia Microbiol. 47:241-245. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, X., X. Ma, N. Wang, and T. Yao. 2009. New subgroups of Bacteroidetes and diverse microorganisms in Tibetan plateau glacial ice provide a biological record of environmental conditions. FEMS Microbiol. Ecol. 67:21-29. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X. F., T. D. Yao, S. J. Xu, and L. Z. An. 2008. Phylogenetic and physiological diversity of microorganisms isolated from Puruogangri ice core. Microb. Ecol. 55:476-488. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.