Abstract
The variability of terminal restriction fragment polymorphism analysis applied to complex microbial communities was assessed statistically. Recent technological improvements were implemented in the successive steps of the procedure, resulting in a standardized procedure which provided a high level of reproducibility.
Terminal restriction fragment length polymorphism (T-RFLP) analysis is a robust, high-resolution, high-throughput, rapid, and cost-effective method for studying the structures of microbial communities (3, 10). T-RFLP analysis is based on group-specific variations in the restriction patterns of molecular markers essential to all life forms (i.e., rRNA genes) or unique to a particular physiological group (e.g., ammonia-oxidizing and sulfate-reducing bacteria) which generate specific and characteristic terminal restriction fragment (T-RF) patterns from mixed fluorescently labeled amplicon pools of environmental nucleic acid extracts. This analysis has developed recently into one of the favorite techniques for the rapid assessment of the structures of bacterial communities. Refinements of the technique and data analysis have been introduced (5, 8, 11, 14, 20-22). Improvements have been made to the sampling procedure (16), to the DNA extraction and amplification steps (17, 19, 26), and to enzymatic restriction digestion (2, 6). Statistical analysis has also been improved in the treatment of the raw data and the selection of logical binning and clustering algorithms resulting, for instance, in the alignment of replicate profiles into a single consensus profile (1, 13). Finally, recent developments have been proposed for the statistical analysis of the profiles using multivariate techniques from numerical ecology (4, 7, 9, 23-25, 27).
Both the resolution and reproducibility of T-RFLP analysis have already been assessed using artificially created bacterial communities (12) comprising up to 30 different clones or bacterial species. However, to the best knowledge of the authors, so far no study has been conducted to assess statistically the dissimilarities obtained in the electropherogram profiles when more complex bacterial communities from natural samples have been analyzed. The main purpose of this report is then to assess statistically the resolution and reproducibility of a standardized T-RFLP protocol, as applied to the analysis of 16S rRNA gene pools from complex communities. The statistical analysis was carried out at successive steps of the procedure, from the initial PCR amplification to the sizing of the obtained T-RFs.
The samples used for this study were taken from a sequencing batch bubble column reactor inoculated with activated sludge from a municipal wastewater treatment plant and operated in such a way as to produce aerobic granular sludge able to remove carbon, nitrogen, and phosphate from an artificial wastewater sample containing acetate, ammonium, and phosphate. Samples were taken at different steps of operation of the reactor systems. The standardized protocol used in the present report is presented in detail in the supplemental material. Note that the methodology implied in the extraction of the total bacterial DNA is not discussed in the context of this work. The T-RFLP protocol was conceived on the basis of recent developments made in the protocol at various stages of the T-RFLP analysis and was implemented with optimized procedures allowing us to minimize potential biases and to ensure a high degree of reproducibility. Whenever possible, technological advances in instrumentation were included, as for instance with the application of optimized electrophoresis conditions and the use of more complex sizing standards and brighter fluorochromes. The use of relatively large and precise amounts of digested PCR fragments (200 ng per replica) also contributed to a drastic reduction of the background noise, which was usually observed to be equal to only about 10 relative fluorescence units (RFU).
Numerical treatment and analysis of the data were carried out with R (R Development Core Team) and the Vegan library (18). We used asymmetric dissimilarity indices to compare T-RFLP profiles using the Jaccard formula, so that the double absence of a T-RF was not considered a resemblance between two profiles (15). The Jaccard dissimilarity was applied to binary data, i.e., the presence/absence of T-RFs. Moreover, to take into account the relative intensity of T-RF areas within each profile in the comparison, we used Ruzicka dissimilarity, which is the Jaccard index applied to quantitative data. Both dissimilarity measures range from 0 (identical profiles) to 1 (different profiles with no T-RF in common). Numerical treatment of the data was also carried out on the modified results, so as to reduce potential biases induced by the inconsistent presence of T-RFs showing very small amounts of fluorescence. T-RF signals just above the detection threshold (low signal-to-noise ratio) can be a cause of suboptimal fingerprinting reproducibility. For this reason, small-area T-RFs (<300 RFU) were suppressed when they were not present in all replicate profiles of a sample.
PCR amplification.
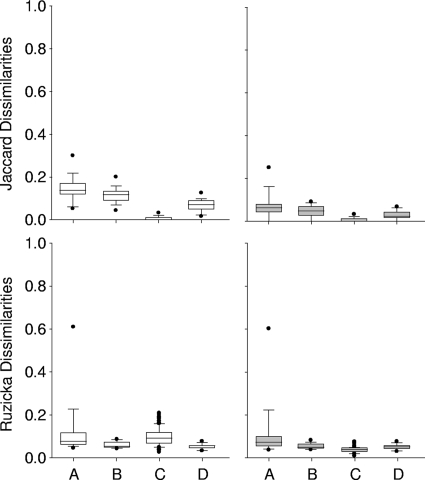
The variability induced by the PCR amplification step was assessed using PCR products from 15 DNA samples, which were all amplified in triplicate (Fig. 1). The PCR products were purified and digested individually, before being analyzed in three consecutive runs using the same capillary per sample. The average Jaccard dissimilarities computed on the three profiles were 0.147 ± 0.061 when all T-RFs were considered and 0.075 ± 0.065 when small-area T-RFs were removed from the profiles, indicating the strong influence from the contributions of inconsistent minor T-RF areas, although the total amount of fluorescence which was removed was only 0.32% ± 0.37%. On the other hand, the average Ruzicka dissimilarities stayed constant and were 0.129 ± 0.142 and 0.120 ± 0.143, respectively. The data analysis of the corresponding electropherograms showed that in addition to the variability induced by small-area T-RFs, a significant difference was found in the total sum of fluorescence obtained for each profile, as well as notable discrepancies in the amounts of fluorescence calculated for major T-RF areas. A very high Ruzicka dissimilarity was observed in one sample only, with a value of 0.602, indicating a possible selective amplification phenomenon. Neither the quality of the raw DNA (measured in terms of the A260/A280 ratios) nor the relative complexity of the involved community (expressed in terms of total T-RF numbers) could explain the anomalous results obtained in the analysis at this stage. Nevertheless, these deviating results were obtained only infrequently, i.e., for 3 out of 15 samples. When these three outlier samples were withdrawn from the calculation, the Jaccard dissimilarities decreased slightly to 0.134 ± 0.045 when all T-RFs were considered and to 0.047 ± 0.027 when small-area T-RFs were removed. The Ruzicka dissimilarities decreased quite significantly to 0.085 ± 0.036 and 0.0669 ± 0.019, respectively.
FIG. 1.
Whisker plots showing the Ruzicka and Jaccard dissimilarities computed for three steps of the T-RFLP analysis. Shown are dissimilarity values obtained when we tested the impacts of PCR amplification (A), enzymatic restriction (B), the electrophoresis conditions and fragment size (intercapillary variability) (C), and the run-to-run variability using 15 different samples in three successive runs (D). White bars, raw data sets; gray bars, data sets without inconsistent small-area T-RFs (<300 RFU in peak area); •, outliers.
Restriction enzyme digestion.
Fifteen DNA samples were amplified once, and the amplification products were digested in triplicate with HaeIII. The Jaccard and Ruzicka dissimilarities were then calculated between three T-RFLP profiles per sample. The Ruzicka dissimilarities were 0.061 ± 0.015 when all T-RFs were considered and 0.055 ± 0.014 when small-area T-RFs (>300 RFU) were removed from the profiles. As was to be expected, the removal of these T-RFs had a more marked effect on the Jaccard dissimilarities, with a significant change from 0.115 ± 0.038 to 0.046 ± 0.028, although they accounted for only 0.32% ± 0.37% of the total sum of fluorescence.
Electrophoresis analysis.
A single DNA sample was PCR amplified once using a large reaction volume and was digested in a single reaction. Forty-five aliquots of the digested PCR product were processed in three series of successive analyses using 15 capillaries of the electrophoresis device (plus one capillary as a control) so as to assess the intercapillary variability (Fig. 1C). The dissimilarities between each couple of samples were computed per run and averaged. The total sum of fluorescence (surface area of all T-RFs) was about 106 RFU. On average, 118 ± 5 T-RFs (peak height, >50 RFU) were taken into account for further analysis. This number was reduced to 94 ± 0.4 T-RFs when inconsistent small-peak areas were removed (peak areas of <300 RFU). The latter correspond to 0.58% ± 0.14% of the total sum of fluorescence values. When only the presence of the T-RFs was taken into account, the average intercapillary Jaccard dissimilarities were 0.003 ± 0.006. When T-RFs of small areas were removed, the Jaccard dissimilarities stayed almost constant at 0.0027 ± 0.006. In other words, the three replicates obtained per capillary showed a very strong resemblance, since more than 99% of all T-RFs could be found in all profiles, independently of their respective fluorescence contribution. When the contribution of the surface area of each T-RF was taken into account, the dissimilarities according to the Ruzicka index were 0.099 ± 0.041 when all T-RFs were considered and 0.040 ± 0.015 when small-area T-RFs were removed. When replicates obtained by different capillaries were compared, slightly larger dissimilarities were observed. The calculated Ruzicka dissimilarities were occasionally higher, with a maximum at 0.228, whereas the Jaccard dissimilarities remained low. The careful observation of the results showed that not all capillaries behaved in the same way in the assessment of the fluorescence of large-area T-RFs (>50,000 RFU), even though their respective sizing was correct.
Run-to-run variability was assessed once using 15 PCR-amplified samples and a large reaction volume that was digested with HaeIII in one single reaction. Three aliquots were analyzed from three consecutive runs using the same capillary per sample (Fig. 1D). The dissimilarities between the three T-RFLP profiles were comparable to those already described above. The average dissimilarities obtained between the replicates were 0.068 ± 0.030 and 0.054 ± 0.013 for the Jaccard and Ruzicka dissimilarities, respectively. The higher values obtained when the same weight was given to all T-RFs on a presence/absence basis (Jaccard) were induced mainly by a certain inconsistency in the analysis of T-RFs with small peak areas, which represented on average only 0.19% ± 0.19% of the totality of the fluorescence. When these T-RFs were withdrawn from the profiles, the Jaccard dissimilarities calculated between the three profiles decreased to 0.028 ± 0.021. In contrast, the Ruzicka dissimilarities remained stable, at 0.052 ± 0.013.
Conclusions.
The proposed protocol allowed us to assign peak areas down to 300 RFU to T-RFs with a high degree of confidence in the majority of cases. However, T-RFs with very small areas (<300 RFU) exhibited a high volatility, which could result in important dissimilarities between replicates, although they accounted for only 0.05% to 0.70% of the total sum of fluorescence. The withdrawal of peak areas smaller than 300 RFU allowed for a substantial decrease of both Ruzicka and Jaccard dissimilarities computed between replicates. These dissimilarities could generally be reduced to values below 0.1, corresponding to an excellent replicate reproducibility. It is, however, difficult to suggest a specific threshold for the dissimilarity values in order to accept or refuse the results of a fragment analysis when other structures of bacterial communities are involved. In general, an increase in the number of T-RFs showing small areas would be translated into increasing dissimilarities, as a consequence of their volatility. Hypothetically, the volatility could be related to the targeted gene, to the degree of oligotrophy, or to the distribution of species and their inherent relative contributions within the communities. Dissimilarities between T-RFLP profiles are thus probably not exclusively influenced by technological biases but also by the intrinsic nature of the bacterial communities.
Supplementary Material
Acknowledgments
This research was supported by the project BIOTOOL (GOCE-003998) of the European Commission under the Sixth Framework Programme.
We thank Sébastien Gabus for providing us with DNA samples from the sequencing batch bubble column reactors, Om Prakash (Department of Zoology, University of Delhi, India) for the literature survey, Arvind Shah (IMT, Neuchâtel, Switzerland), and Noam Shani (EPFL-LBE, Lausanne, Switzerland) for their relevant remarks and suggestions on the manuscript, as well as three anonymous reviewers for their constructive comments.
Footnotes
Published ahead of print on 11 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdo, Z., U. M. E. Schuette, S. J. Bent, C. J. Williams, L. J. Forney, and P. Joyce. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929-938. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood, C. B., T. Marsh, S. H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clement, B. G., L. E. Kehl, K. L. DeBord, and C. L. Kitts. 1998. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J. Microbiol. Methods 31:135-142. [Google Scholar]
- 4.Culman, S. W., H. G. Gauch, C. B. Blackwood, and J. E. Thies. 2008. Analysis of T-RFLP data using analysis of variance and ordination methods: a comparative study. J. Microbiol. Methods 75:55-63. [DOI] [PubMed] [Google Scholar]
- 5.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 69:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engebretson, J. J., and C. L. Moyer. 2003. Fidelity of select restriction endonucleases in determining microbial diversity by terminal-restriction fragment length polymorphism. Appl. Environ. Microbiol. 69:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzjohn, R. G., and I. A. Dickie. 2007. TRAMPR: an R package for analysis and matching of terminal-restriction fragment length polymorphism (TRFLP) profiles. Mol. Ecol. Notes 7:583-587. [Google Scholar]
- 8.Frey, J. C., E. R. Angert, and A. N. Pell. 2006. Assessment of biases associated with profiling simple, model communities using terminal-restriction fragment length polymorphism-based analyses. J. Microbiol. Methods 67:9-19. [DOI] [PubMed] [Google Scholar]
- 9.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 10.Grant, A., and L. A. Ogilvie. 2003. Terminal restriction fragment length polymorphism data analysis. Appl. Environ. Microbiol. 69:6342-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann, M., J. Enkerli, and F. Widmer. 2007. Residual polymerase activity-induced bias in terminal restriction fragment length polymorphism analysis. Environ. Microbiol. 9:555-559. [DOI] [PubMed] [Google Scholar]
- 12.Hartmann, M., and F. Widmer. 2008. Reliability for detecting composition and changes of microbial communities by T-RFLP genetic profiling. FEMS Microbiol. Ecol. 63:249-260. [DOI] [PubMed] [Google Scholar]
- 13.Hewson, I., and J. A. Fuhrman. 2006. Improved strategy for comparing microbial assemblage fingerprints. Microb. Ecol. 51:147-153. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan, C. W., and C. L. Kitts. 2003. Variation between observed and true terminal restriction fragment length is dependent on true TRF length and purine content. J. Microbiol. Methods 54:121-125. [DOI] [PubMed] [Google Scholar]
- 15.Legendre, P., and L. Legendre. 1998. Numerical ecology, 2nd ed. Elsevier, Amsterdam, The Netherlands.
- 16.Lehman, R. M. 2007. Understanding of aquifer microbiology is tightly linked to sampling approaches. Geomicrobiol. J. 24:331-341. [Google Scholar]
- 17.Luna, G. M., A. Dell'Anno, and R. Danovaro. 2006. DNA extraction procedure: a critical issue for bacterial diversity assessment in marine sediments. Environ. Microbiol. 8:308-320. [DOI] [PubMed] [Google Scholar]
- 18.Oksanen, J., R. Kindt, P. Legendre, B. O'Hara, G. L. Simpson, and M. H. H. Stevens. 2008. vegan: community ecology package. R package version 1.11-0. http://cran.r-project.org/; http://vegan.r-forge.r-project.org/.
- 19.Osborne, C. A., M. Galic, P. Sangwan, and P. H. Janssen. 2005. PCR-generated artefact from 16S rRNA gene-specific primers. FEMS Microbiol. Lett. 248:183-187. [DOI] [PubMed] [Google Scholar]
- 20.Osborne, C. A., G. N. Rees, Y. Bernstein, and P. H. Janssen. 2006. New threshold and confidence estimates for terminal restriction fragment length polymorphism analysis of complex bacterial communities. Appl. Environ. Microbiol. 72:1270-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey, J., K. Ganesan, and R. K. Jain. 2007. Variations in T-RFLP profiles with differing chemistries of fluorescent dyes used for labeling the PCR primers. J. Microbiol. Methods 68:633-638. [DOI] [PubMed] [Google Scholar]
- 22.Qiu, X. Y., L. Y. Wu, H. S. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Z. Zhou. 2001. Evaluation of PCR-generated chimeras: mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramette, A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudi, K., M. Zimonja, P. Trosvik, and T. Naes. 2007. Use of multivariate statistics for 16S rRNA gene analysis of microbial communities. Int. J. Food Microbiol. 120:95-99. [DOI] [PubMed] [Google Scholar]
- 25.Schutte, U. M. E., Z. Abdo, S. J. Bent, C. Shyu, C. J. Williams, J. D. Pierson, and L. J. Forney. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80:365-380. [DOI] [PubMed] [Google Scholar]
- 26.Sipos, R., A. J. Szekely, M. Palatinszky, S. Revesz, K. Marialigeti, and M. Nikolausz. 2007. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targeting bacterial community analysis. FEMS Microbiol. Ecol. 60:341-350. [DOI] [PubMed] [Google Scholar]
- 27.Trosvik, P., B. Skanseng, K. S. Jakobsen, N. C. Stenseth, T. Naes, and K. Rudi. 2007. Multivariate analysis of complex DNA sequence electropherograms for high-throughput quantitative analysis of mixed microbial populations. Appl. Environ. Microbiol. 73:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.