Abstract
The gene encoding a novel modular xylanase from Cellulosimicrobium sp. strain HY-13 was identified and expressed in Escherichia coli, and its truncated gene product was characterized. The enzyme consisted of three distinct functional domains, an N-terminal catalytic GH10 domain, a fibronectin type 3 domain, and C-terminal carbohydrate-binding module 2.
Most known microbial xylanases, which decompose primarily β-1,4-xylosic polysaccharides in an endo fashion, are currently affiliated with the two glycoside hydrolase (GH) families 10 and 11. Compared to GH11 xylanases, GH10 xylanases generally have a molecular mass of >30 kDa and an acidic pI. In addition, GH10 xylanases are frequently found in nature as modular enzymes that consist of a catalytic GH10 domain with one or more substrate-binding domains, such as a cellulose-binding domain, carbohydrate-binding module (CBM), or xylan-binding domain (1, 5, 12). However, no modular xylanase with a fibronectin type 3 (Fn3) domain has been characterized to date, even though Fn3 modules are often found in bacterial carbohydrolases such as cellulases, amylases, pullulanases, polygalacturonidases, and chitinases. It is now believed that the Fn3 domains in bacterial carbohydrolases participate in promotion of the hydrolysis of carbohydrate substrates by modifying their surfaces (10, 17).
Gut microorganisms from invertebrates have recently attracted a great deal of attention as sources of novel fibrolytic enzymes with unique molecular structures and distinct substrate specificities (2-4, 7, 15). However, no study has been conducted to evaluate xylanolytic enzymes from the gut microorganisms of earthworms that may participate in the digestion of cellulosic or hemicellulosic foods taken up by the hosts. Here, we report a novel GH10 xylanase (XylK1) with an Fn3 domain from Cellulosimicrobium sp. strain HY-13 KCTC 11302BP (11), which was isolated from the digestive tract of the earthworm Eisenia fetida.
Amplification of a partial sequence of the Cellulosimicrobium sp. strain HY-13 xylanase gene from the genomic DNA was conducted using the degenerate primers designed on the basis of conserved regions (WDVVNE and ITELDI) in the GH10 xylanases. The upstream primer (KF) was 5′-TGGGACGTCSTCAACGAG-3′, and the downstream primer (KR) was 5′-GATGTCGAGCTCSGTGAT-3′, which produced a 342-bp DNA fragment. Cloning of the full xylK1 gene was performed by repeated genomic walking and nested-PCR methods using a DNA Walking SpeedUp premix kit (Seegene). To overproduce mature XylK1, its encoding gene was cloned into the NdeI/HindIII sites of a pET-28a(+) vector (Novagen). Likewise, a partial sequence containing the GH10 domain (Ala34 to Leu345) of XylK1 was amplified using the primers tKF (5′-CATATGGCCACCGAGCCGCTCG-3′) and tKR (5′-AAGCTTTCAGGACCTCGGCGATCGC-3′) and subsequently also cloned into the same expression vector. When overexpressed in recombinant Escherichia coli BL21 cells harboring pET-28a(+)/xylK1, most recombinant proteins (rXylK1) were produced as inactive inclusion bodies. Therefore, after solubilization of the isolated inclusion bodies, on-column refolding and purification of rXylK1 was conducted using a HisTrap HP (GE Healthcare, Sweden) (5-ml) column attached to a fast-performance liquid chromatography (LC) system (Amersham Pharmacia Biotech, Sweden) according to the manufacturer's instructions. The active rXylK1 proteins were then purified to electrophoretic homogeneity by gel permeation chromatography using a HiLoad 26/60 Superdex 200 prep-grade (Amersham Biosciences, Sweden) column, as previously described (11). Like rXylK1, the recombinant proteins (rXylK1ΔFn3) without both an Fn3 domain and CBM 2 were also purified by the method described above because they were produced as insoluble inclusion bodies. The relative molecular mass of the denatured rXylK1 was evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12% gel, and the protein concentrations were assayed by using the Bradford reagent (Bio-Rad). Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analysis was conducted using an Ultraflex III MALDI-TOF mass spectrometer (Bruker Daltonics, Germany) at the Korea Basic Science Institute (Daejeon, South Korea). The binding capacity of recombinant enzymes with/without the Fn3 domain to carbohydrate polymers was determined as described elsewhere (4). Xylanase activity was routinely assayed by measuring the amount of reducing sugars released from birch wood xylan by using the 3,5-dinitrosalicylic acid reagent. The standard assay mixture (0.5 ml) consisted of birch wood xylan (1.0%) or p-nitrophenyl (PNP)-sugar derivatives (5 mM) with suitably diluted enzyme solution (0.05 ml) in 50 mM sodium phosphate buffer (pH 6.0), and the catalytic reaction was performed at 55°C for 10 min. One international unit of xylanase activity for xylans or PNP-sugar derivatives was defined as the amount of enzyme required to produce 1 μmol of reducing sugar or PNP, respectively, per min under standard assay conditions. Enzymatic hydrolysis of birch wood xylan (10 mg) (Sigma Co.), xylooligosaccharides (1 mg each) (Megazyme International Ireland, Ireland), and cellooligosaccharides (1 mg each) (Seikagaku Biobusiness Co., Japan) was conducted using purified rXylK1 (2 μg) in 0.1 ml of 50 mM sodium phosphate buffer (pH 6.0) for 3 or 6 h at 37°C, during which time the enzyme remained fairly stable. The reaction mixture was then heated to 100°C for 5 min to stop the enzyme reaction. The hydrolysis products were identified by LC-MS, as previously described (11).
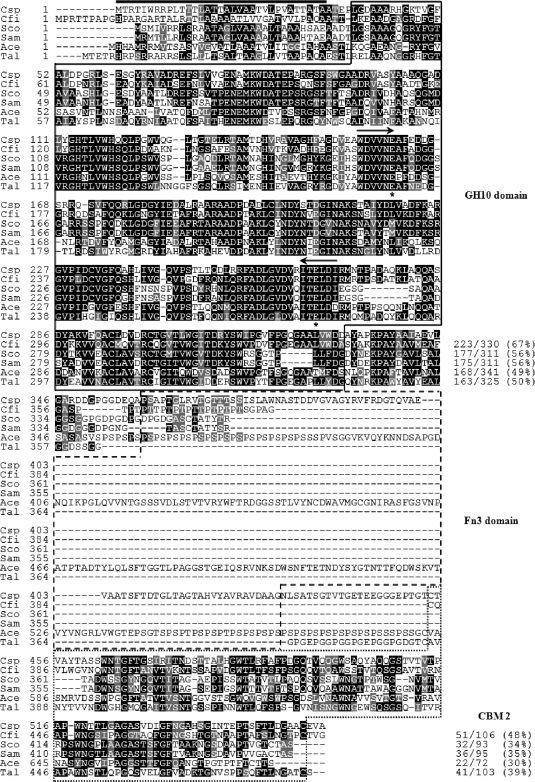
The isolated XylK1 gene (GenBank accession no. FJ859907) contained a 1,671-bp open reading frame that encodes a protein of 556 amino acids with a deduced molecular mass of 58,296 Da and a calculated pI of 4.59. It was predicted that the signal sequence cleavage site of premature XylK1 was between Ala33 and Ala34, which may generate a mature XylK1 of 523 amino acids with a deduced molecular mass of 54,843 Da and a calculated pI of 4.49 (Fig. 1). The results of a protein BLAST survey revealed that XylK1 was a unique modular xylanase composed of an N-terminal catalytic GH10 (Leu38-to-Asp330) domain, an Fn3 (Pro359-to-Gly430) domain, and a C-terminal CBM 2 (Cys454 to Cys553), which was very comparable to the domain architectures of other related GH10 enzymes (Fig. 2). To the best of our knowledge, no xylanase with domain architecture identical to that of XylK1 with an Fn3 domain has been reported to date, although an uncharacterized modular xylanase (GenBank accession no. ABQ06877) consisting of an N-terminal Fn3 domain and a C-terminal GH10 domain from Flavobacterium johnsoniae UW101 was previously identified through a genome survey. As shown in Fig. 1, the catalytic GH10 domain of XylK1 showed the highest sequence identity (67%) with that of the Cellulomonas fimi xylanase (AAA56792) among other GH10 enzymes available in the NCBI database. However, its CBM 2 was 64% identical to that of Cellulomonas fimi GH6 cellulase (AAC36898). The highest sequence identity (70%) of the Fn3 domain in XylK1 was obtained when it was compared to that of the Acidothermus cellulolyticus 11B GH48 enzyme (ABK52390), which degrades cellulose. The two conserved residues of Glu161 (acid/base catalyst) and Glu266 (catalytic nucleophile) were predicted in the active site of premature XylK1.
FIG. 1.
Alignment of the deduced amino acid sequence of GH10 xylanase from Cellulosimicrobium sp. strain HY-13 with those of other GH10 xylanases. Shown are sequences (GenBank accession numbers) of Cellulosimicrobium sp. strain HY-13 (Csp) xylanase (FJ859907), Cellulomonas fimi (Cfi) xylanase (AAA56792), Streptomyces coelicolor (Sco) A3 xylanase (CAB61191), Streptomyces ambofaciens (Sam) xylanase (CAJ88420), Acidothermus cellulolyticus 11B (Ace) xylanase (ABK51955), and Thermobifida alba (Tal) xylanase (CAB02654). The identical and similar amino acids are shown by black and gray boxes, respectively. The predicted signal peptide is indicated by a black bar. The internal peptide sequences used in the design of degenerate oligonucleotides for PCR are marked by arrows. Highly conserved amino acid residues that play an essential role in the catalytic reaction are indicated by asterisks. GH10, Fn3, and CBM 2 domains are outlined by solid, dashed, and dotted lines, respectively.
FIG. 2.
Domain architectures of Cellulosimicrobium sp. strain HY-13 xylanase and the following related bacterial GH10 xylanases (GenBank accession numbers): Cellulosimicrobium sp. strain HY-13 xylanase (FJ859907) (A), Streptomyces thermocarboxydus HY-15 xylanase (EU880430) (B), Cellulomonas fimi xylanase (AAZ76373) (C), Streptomyces thermoviolaceus xylanase (BAD02382) (D), Pseudomonas fluorescens xylanase (P23030) (E), and Cellvibrio japonicus xylanase (YP 001982932) (F).
The molecular mass of the purified enzyme was estimated to be approximately 42.0 kDa, which was smaller than the deduced molecular mass (57,138 Da) of intact His-tagged rXylK1, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). In addition, MALDI-TOF MS analysis revealed that the purified His-tagged rXylK1 with a calculated molecular mass of 45,169 Da was a smaller protein than the intact rXylK1. These results indicate that rXylK1 was formed by proteolytic cleavage at the C terminus region because the enzyme was able to tightly bind to a His tag column. Based on the calculated molecular mass (45,169 Da) of the truncated rXylK1, it is assumed that the intact rXylK1 was processed at the Val439-Thr440 site in a hinge region between the Fn3 domain and the C-terminal CBM 2 of the premature XylK1. The deduced molecular mass (45,179 Da) of rXylK1 with the Val439 residue at the C-terminal extremity was very close to the molecular mass (45,169 Da) of the enzyme calculated by MALDI-TOF MS analysis. A similar C-terminal processing of some modular xylanases with a cellulose-binding domain by proteases has also been observed when they are expressed in E. coli (8, 14). It is likely that the C-terminal truncation does not induce a significant alteration of the binding affinity of rXylK1 with an Fn3 domain to carbohydrate polymers since the truncated enzyme could still bind to both Avicel and insoluble oat spelt xylan (Table 1). In this case, only weak catalytic activity of the rXylK1 (<3% of its original activity [0.5 IU]) was recovered from an enzyme solution after binding. It was of great interest that no rXylK1ΔFn3 was bound to Avicel, although the enzyme could still not only bind to insoluble oat spelt xylan but also catalyze the hydrolysis of xylosic polymers. The specific activity (27 IU/mg) of rXylK1ΔFn3 for birch wood xylan was evaluated to be approximately 19% of that (143 IU/mg) of rXylK1 with the Fn3 domain for the same substrate. Taken together, the binding ability of the C-terminal CBM 2-lacking rXylK1 to Avicel and insoluble oat spelt xylan clearly suggests that the Fn3 domain plays an important role in enzyme-substrate binding because rXylK1ΔFn3 was not bound to Avicel. A significant decrease in the catalytic activity of rXylK1ΔFn3 induced by deletion of the Fn3 domain also suggests that the Fn3 domain may take part in the promotion of the catalytic hydrolysis of xylosic substrates by modifying their surfaces, as shown in other GHs (10, 17). The maximum catalytic activity of rXylK1 toward birch wood xylan was observed at pH 6.0 and 55°C, and it maintained over 80% of its highest activity at a relatively broad pH range of 5.0 to 9.0 during the reaction period of 15 min. These high activities of rXylK1 in alkaline pH ranges suggest that it is a peculiar enzyme that can be distinguished from xylanases of other invertebrate-symbiotic microorganisms, which showed weak hydrolytic activities toward xylan at the same alkaline pHs (4, 11, 15). At 55°C, the half-life of rXylK1 was approximately 10 min, which indicates that it is a typical mesophilic enzyme. Compared to many other xylanases that were completely inhibited by Hg2+ (11, 13), rXylK1 was partially inhibited (by 40% relative to its original activity) by 1 mM Hg2+. In addition, the enzyme was relatively suppressed by <25%, relative to its original activity, in the presence of some divalent cations at a concentration of 1 mM, in the order of Ca2+ > Cu2+ > Ba2+. No significant alterations of rXylK1 activity by Mn2+ and Co2+ were interesting to note because the xylanases from Streptomyces sp. strain S9 [12] and Aeromonas caviae ME-1 [14] have been negatively affected by the compound. In this study, the catalytic activity of rXylK1 increased by approximately 1.4-fold when the reaction was conducted in the presence of 1 mM Fe2+. The promotion of rXylK1 activity by Fe2+ was comparable to previous observations of xylanase inhibition by the metal ion (6, 15). The rXylK1 was relatively unaffected by sulfhydryl reagents (5 mM) such as sodium azide, iodoacetamide, and N-ethylmaleimide, while the enzyme lost 68% of its original activity when preincubated with 5 mM EDTA for 10 min. The complete inhibition of rXylK1 by 5 mM N-bromosuccinimide was in good agreement with the fact that three Trp residues in the highly conserved region of the GH10 enzymes are critically involved in enzyme-substrate interaction, as shown for Streptomyces lividans (16) and Geobacillus stearothermophilus T-6 (18) GH10 xylanases. It was predicted that the three residues Trp118, Trp306, and Trp314 in premature XylK1 might be responsible for catalysis and substrate binding of the enzyme. It is also noteworthy that the catalytic activity of His-tagged rXylK1 increased significantly, by approximately 1.8-fold, when the reaction was conducted in the presence of Tween 80 or Triton X-100 at a concentration of 0.5%. It is assumed that the nonionic-detergent-induced activation of His-tagged rXylK1 might be due to the direct interaction of the recombinant enzyme with the Tween 80 or Triton X-100 molecule, which may lead to an alteration of the enzyme-substrate interaction. Indeed, the stimulation of His-tagged rXylK1 activity was insignificant when the enzyme reaction was conducted in the presence of the detergents without preincubation with the same compounds for 10 min. As with rXylK1, it has been reported that the catalytic activity of a His-tagged esterase from Bacillus megaterium 20-1 expressed in E. coli is greatly stimulated by various nonionic detergents (9).
TABLE 1.
Binding of rXylK1 and rXylK1ΔFn3 to hydrophobic polysaccharides
| Substrate | Residual xylanase activity after binding (total IU)a |
|
|---|---|---|
| rXylK1 | rXylK1ΔFn3 | |
| Control | 0.50 | 0.50 |
| Avicel | ≤0.01 | 0.49 ± 0.02 |
| Insoluble oat spelt xylan | 0.05 ± 0.01 | ≤0.01 |
Residual xylanase activity was assayed using birch wood xylan.
Of the evaluated xylosic materials, oat spelt xylan was most efficiently hydrolyzed by rXylK1; however, the enzyme was not capable of degrading glucose-based polysaccharides, which is indicative of a lack of other GH activities (Table 2). It should also be noted that the catalytic activity of rXylK1 toward PNP-cellobioside was approximately 1.7-fold higher than that (193 IU/mg) of the enzyme toward oat spelt xylan. In this study, the cleavage activity of rXylK1 for PNP-cellobioside was approximately 48 IU/mg, which is much higher than the activity (<10 IU/mg) of other known xylanases for the same substrate (6, 11). However, LC-MS analysis revealed that cellooligosaccharides of cellobiose to cellotetraose were not susceptible to rXylK1 (data not shown). Taken together, these results indicate that rXylK1 is a true endo-β-1,4-xylanase that lacks cellulase activity. Interestingly, rXylK1 was found to have transxylosylation activity (approximately 7.5% of its maximum hydrolytic activity for oat spelt xylan) that enabled the cleavage of PNP-xylopyranoside (Table 2). A similar transxylosylation reaction by rXylK1 was also observed when xylotriose (X3) and xylotetraose (X4) were subjected to hydrolytic reaction by the enzyme (Table 3). Specifically, a series of xylooligosaccharides (X4 to X7) were produced after the enzymatic hydrolysis of X3 for 3 h at 37°C, although X2 and X3 were identified as the major products. Similarly, the hydrolysis of X4 by rXylK1 resulted in the production of a mixture that contained longer xylooligosaccharides (42.3%) of X5 to X8, which suggests that these xylooligomers were produced by an rXylK1-catalyzed transxylosylation reaction. However, no X1 was detected as the hydrolysis product of X2, X3, or X4. The ability of rXylK1 to catalyze the synthesis of longer xylooligosaccharides from X3 or X4 was of interest because microbial xylanases generally produced shorter xylooligosaccharides, such as X2 and/or X3, from the same substrates (2, 15). Additionally, rXylK1 primarily degraded birch wood xylan to X2 (65.1%) and X3 (29.5%) together with small amounts of X4 (5.4%) when the enzyme reaction was conducted for 6 h at 37°C.
TABLE 2.
Hydrolysis activity of rXylK1 for different substrates
| Substrate | Relative activity (%)a |
|---|---|
| Birch wood xylan | 74.1 ± 2.8 |
| Beech wood xylan | 85.8 ± 3.5 |
| Oat spelt xylan | 100.0 |
| Soluble starch | ND |
| Avicel | ND |
| Carboxy methylcellulose | ND |
| PNP-cellobioside | 171.7 ± 4.9 |
| PNP-glucopyranoside | <0.5 |
| PNP-xylopyranoside | 7.5 ± 0.6 |
Relative activity was obtained from three repeated experiments. ND, not detected.
TABLE 3.
LC analysis of the hydrolysis products of xylosic materials by rXylK1
| Substrate | Composition (%)a of products formed by hydrolysis reaction |
||||||
|---|---|---|---|---|---|---|---|
| X2 | X3 | X4 | X5 | X6 | X7 | X8 | |
| X2 | 100.0 | ||||||
| X3 | 27.8 | 45.4 | 16.8 | 5.6 | 3.9 | 0.5 | |
| X4 | 12.8 | 26.3 | 18.6 | 18.1 | 14.5 | 8.4 | 1.3 |
| Birch wood xylan | 65.1 | 29.5 | 5.4 | ||||
LC area percent.
In conclusion, the novel gene (xylK1) encoding a modular GH10 xylanase that consists of three putative functional domains (an N-terminal GH10 domain, an Fn3 domain, and C-terminal CBM 2) was identified from an earthworm-symbiotic bacterium, Cellulosimicrobium sp. strain HY-13. The molecular architecture of XylK1 indicates that it is a unique GH10 enzyme with an Fn3 domain that has not previously been reported. In addition, the relatively high cleavage activity of rXylK1 toward PNP-cellobioside and its transxylosylation activity that enables it to produce longer xylooligosaccharides from X3 or X4 differentiate it from other known GH10 xylanases.
Acknowledgments
This work was supported by a grant from the KRIBB Research Initiative Program (KGS2330911) and the 21C Frontier Microbial Genomics and Applications Center Program (MGM0900837) of the Korean Ministry of Education, Science, and Technology.
Footnotes
Published ahead of print on 18 September 2009.
REFERENCES
- 1.Black, G. W., G. P. Hazlewood, S. J. Millward-Sadler, J. I. Laurie, and H. J. Gilbert. 1995. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem. J. 307:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan, Y., W. N. Callen, L. Christoffersen, P. Dupree, F. Goubet, S. Healey, M. Hernández, M. Keller, K. Li, N. Palackal, A. Sittenfeld, G. Tamayo, S. Wells, G. P. Hazlewood, E. J. Mathur, J. M. Short, D. E. Robertson, and B. A. Steer. 2004. Unusual microbial xylanases from insect guts. Appl. Environ. Microbiol. 70:3609-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brune, A. 1998. Termite guts: the world's smallest bioreactors. Trends Biotechnol. 16:16-21. [Google Scholar]
- 4.Cazemier, A. E., J. C. Verdoes, A. J. J. van Ooyen, and H. J. M. Op den Camp. 1999. Molecular and biochemical characterization of two xylanase-encoding genes from Cellulomonas pachnodae. Appl. Environ. Microbiol. 65:4099-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins, T., C. Gerday, and G. Feller. 2005. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 29:3-23. [DOI] [PubMed] [Google Scholar]
- 6.Haga, K., M. Kitaoka, Y. Kashiwagi, T. Sasaki, and H. Taniguchi. 1991. Purification and properties of a xylanase from Cellvibrio gilvus that hydrolyzes p-nitrophenyl cellooligosaccharides. Agric. Biol. Chem. 55:1959-1967. [PubMed] [Google Scholar]
- 7.Heo, S. Y., J. Kwak, H. W. Oh, D. S. Park, K. S. Bae, D. H. Shin, and H. Y. Park. 2006. Characterization of an extracellular xylanase in Paenibacillus sp. HY-8 isolated from an herbivorous longicorn beetle. J. Microbiol. Biotechnol. 16:1753-1759. [Google Scholar]
- 8.Ito, Y., T. Tomita, N. Roy, A. Nakano, N. Sugawara-Tomita, S. Watanabe, N. Okai, N. Abe, and Y. Kamio. 2003. Cloning, expression, and cell surface localization of Paenibacillus sp. strain W-61 xylanase 5, a multidomain xylanase. Appl. Environ. Microbiol. 69:6969-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung, Y. J., J. K. Lee, C. G. Sung, T. K. Oh, and H. K. Kim. 2003. Nonionic detergent-induced activation of an esterase from Bacillus megaterium 20-1. J. Mol. Catal. B 26:223-229. [Google Scholar]
- 10.Kataeva, I. A., R. D. Seidel III, A. Shah, L. T. West, X.-L. Li, and L. G. Ljungdahl. 2002. The fibronectin type 3-like repeat from the Clostridium thermocellum cellobiohydrolase CbhA promotes hydrolysis of cellulose by modifying its surface. Appl. Environ. Microbiol. 68:4292-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, D. Y., M. K. Han, J. S. Lee, H. W. Oh, D. S. Park, D. H. Shin, K. H. Son, K. S. Bae, and H. Y. Park. 2009. Isolation and characterization of a cellulase-free endo-β-1,4-xylanase produced by an invertebrate-symbiotic bacterium, Cellulosimicrobium sp. HY-13. Proc. Biochem. 44:1055-1059. [Google Scholar]
- 12.Kulkarni, N., A. Shendye, and M. Rao. 1999. Molecular and biotechnological aspects of xylanases. FEMS Microbiol. Rev. 23:411-456. [DOI] [PubMed] [Google Scholar]
- 13.Li, N., K. Meng, Y. Wang, P. Shi, H. Luo, Y. Bai, P. Yang, and B. Yao. 2008. Cloning, expression, and characterization of a new xylanase with broad temperature adaptability from Streptomyces sp. S9. Appl. Microbiol. Biotechnol. 80:231-240. [DOI] [PubMed] [Google Scholar]
- 14.Liu, C. J., T. Suzuki, S. Hirata, and K. Kawai. 2003. The processing of high-molecular-weight xylanase (XynE, 110 kDa) from Aeromonas caviae ME-1 to 60-kDa xylanase (XynE60) in Escherichia coli and purification and characterization of XynE60. J. Biosci. Bioeng. 95:95-101. [DOI] [PubMed] [Google Scholar]
- 15.Oh, H. W., S. Y. Heo, D. Y. Kim, D. S. Park, K. S. Bae, and H. Y. Park. 2008. Biochemical characterization and sequence analysis of a xylanase produced by an exo-symbiotic bacterium of Gryllotalpa orientalis, Cellulosimicrobium sp. HY-12. Antonie van Leeuwenhoek 93:437-442. [DOI] [PubMed] [Google Scholar]
- 16.Roberge, M., F. Shareck, R. Morosoli, D. Kluepfel, and C. Dupont. 1999. Characterization of active-site aromatic residues in xylanase A from Streptomyces lividans. Protein Eng. 12:251-257. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe, T., Y. Ito, T. Yamada, M. Hashimoto, S. Sekine, and H. Tanaka. 1994. The roles of the C-terminal domain and type III domains of chitinase A1 from Bacillus circulans WL-12 in chitin degradation. J. Bacteriol. 176:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zolotnitsky, G., U. Cogan, N. Adir, V. Solomon, G. Shoham, and Y. Shoham. 2004. Mapping glycoside hydrolase substrate subsites by isothermal titration calorimetry. Proc. Natl. Acad. Sci. USA 101:11275-11280. [DOI] [PMC free article] [PubMed] [Google Scholar]