Abstract
The worldwide decline in honeybee colonies during the past 50 years has often been linked to the spread of the parasitic mite Varroa destructor and its interaction with certain honeybee viruses. Recently in the United States, dramatic honeybee losses (colony collapse disorder) have been reported; however, there remains no clear explanation for these colony losses, with parasitic mites, viruses, bacteria, and fungal diseases all being proposed as possible candidates. Common characteristics that most failing colonies share is a lack of overt disease symptoms and the disappearance of workers from what appears to be normally functioning colonies. In this study, we used quantitative PCR to monitor the presence of three honeybee viruses, deformed wing virus (DWV), acute bee paralysis virus (ABPV), and black queen cell virus (BQCV), during a 1-year period in 15 asymptomatic, varroa mite-positive honeybee colonies in Southern England, and 3 asymptomatic colonies confirmed to be varroa mite free. All colonies with varroa mites underwent control treatments to ensure that mite populations remained low throughout the study. Despite this, multiple virus infections were detected, yet a significant correlation was observed only between DWV viral load and overwintering colony losses. The long-held view has been that DWV is relatively harmless to the overall health status of honeybee colonies unless it is in association with severe varroa mite infestations. Our findings suggest that DWV can potentially act independently of varroa mites to bring about colony losses. Therefore, DWV may be a major factor in overwintering colony losses.
Deformed wing virus (DWV), acute bee paralysis virus (ABPV), and black queen cell virus (BQCV) are single-stranded positive-sense RNA viruses of the order Picornavirales and are regularly detected in honeybee populations in the United Kingdom (1). ABPV has been assigned to the family Dicistroviridae and is known to follow a classic acute-type infection strategy since relatively low loads (103 to 106 viruses per honeybee) can rapidly translate into overt symptoms of paralysis and ultimately death for the honeybee, depending on the mode of transmission (6, 33). ABPV shares >92% sequence homology with other members of the family Dicistroviridae, Kashmir bee virus and Israeli acute paralysis virus, across the eight conserved domains of the RNA-dependent RNA polymerase gene, and it has been proposed that these viruses have recently diverged and are variants of each other (7). Advances in the study of this proposed ABPV complex is revealing the significant impact these viruses may have on honeybee colonies on a global scale. For example, a recent study in the United States has observed a correlation between Israeli acute paralysis virus and colony collapse disorder (17). That said, other agents, including bacteria and microsporidia, have also been proposed as important factors in the onset of colony loss (25, 27).
BQCV is similar to ABPV in that it, too, follows a typical acute infection strategy. This virus is known to infect honeybee queen cell larvae, causing the larvae to discolor and die (5). It has been shown to be associated with the microsporidian Nosema apis (4) although whether N. apis has a direct role in the transmission of this virus still needs to be determined. Both ABPV and BQCV have been detected in worker honeybees and pupae (38), and the viruses are transmitted orally, via food and feeding activities (14). BQCV has also been detected in queen honeybees (13), suggesting that vertical transmission is also important for this virus. Both BQCV (12) and ABPV (38) have been detected within the varroa mite; however, only ABPV (9) has been shown to be vectored by varroa mites and has been found associated with dead colonies infested with varroa mites in Germany, Russia, and the United States (1). Later modeling work (33) indicated that very large (10,000+) mite populations are required to kill a colony since it is difficult for ABPV to become established among the bee population due to its high virulence.
DWV is currently designated as a member of the unassigned genus Iflavirus within the order Picornavirales. It is generally considered as less virulent than ABPV or Kashmir bee virus, but it is known to cause overt symptoms of wing deformities in developing honeybees, resulting in emerging honeybees that are unable to fly and die shortly (5). It is also speculated that a cloud of DWV sequence variants exists that have evolved from a common ancestor. This is due to the high sequence similarities DWV isolates share with Kakugo virus and Varroa destructor virus within the RNA-dependent RNA polymerase gene, yet differences in virus epidemiology and pathological effects distinguish them from each other (29). DWV has been detected in worker honeybees, pupae, larvae, drones, and queens (15, 18, 20) as well as within the varroa mite (38, 43) and more recently the mite Tropilaelaps mercedesae (21), implying a range of horizontal and vertical transmission routes. Despite their global occurrence, it is generally accepted that DWVs play a secondary role in the causes of honeybee disease compared to their parasitic and bacterial counterparts as the viruses routinely reside at low levels in colonies, with symptomatic infections being rare (5). Moreover, multiple variants with differing infection strategies can account for a lack of discernible symptoms.
Whether these viruses follow a persistent, latent, inapparent, or progressive infection strategy still remains unclear. Persistent (often called chronic) infections imply that the rate of infection within a host is in balance with the reproduction rate of the infected cell type or host itself. This is achieved through a combination of changing virus replication and host immune responses. Latent infections occur when the virus lies dormant within the host (replication inactive) until activation by defined stimuli. Progressive infections are caused by viruses that enter the host cell and replicate undetected for many cellular generations over many years before manifesting overt or acute symptoms. These three infection strategies all evade the host immune system, which results in the inability of the host to fully expel the virus, and this inability is often lethal. Inapparent (often referred to as covert) infections are indicative of a highly evolved relationship between the virus and natural host. Moreover, these infections are distinct in that the natural host can eventually clear itself from this short-term infection (19). Infections of DWV are often described as inapparent (15); however, Yue et al. (44) have suggested that a distinction should be made between “true inapparent” and their newly defined “covert infection” based on the long-term nature of DWV infection in honeybee colonies and on the nature of its transmission. This conclusion is congruent with current knowledge that traditional serological screening methods for DWV have limitations in their sensitivity (20). Therefore, the presence and duration of DWV within colonies have often been underestimated using serological assays as the overt symptoms of the deformed wing phenotype (>1011 virions per honeybee) are short-lived. Advances in virus detection methodologies have enabled the development of more sensitive techniques, such as PCR, and this has demonstrated that DWV persists for longer periods within colonies (38). However, based on the current research evidence, a case could be made that DWV actually follows the classic persistent infection strategy.
DWV is thought to have an intricate relationship with varroa mites such that immunosuppression of the honeybee pupae by the mites results in increased DWV amplification when the honeybees are exposed to other pathogens (42). It has additionally been shown that the number of mites parasitizing honeybee pupae is positively correlated with the probability of their developing malformed wings (10). Other findings indicate that DWV replication within the mite and subsequent transmission to developing honeybees lead to the increased likelihood of the bees' emerging with wing deformities (24, 43). Taken together, the expectation is that DWV-associated colony collapse would typically occur in the presence of a large (>2,000) varroa mite infestation carrying high levels of DWV and with a high proportion of deformed honeybees. While the effect of varroa mite-induced DWV disease is well recognized, i.e., wing deformities coupled with downregulation of immunity-related genes and antimicrobial peptides (36, 42) and impaired learning behavior (28), the impact of non-varroa mite-vectored DWV within asymptomatic honeybees still needs to be realized. Moreover, it was recently reported that varroa mite-free bumblebees that tested positive for DWV actually showed symptoms of DWV infection (23). Even though these bumblebees were in close proximity to DWV-infected and varroa mite-infested honeybee colonies, it is evidence that the dependency of DWV on varroa mite vectoring for a symptomatic infection (manifested as classic wing deformities or other symptoms) may not be as critical as previously thought.
The purpose of this study was to investigate asymptomatic viral dynamics within husbanded honeybee colonies over an annual cycle. We set out to observe the relationship, if any, between virus infections, varroa mite parasitism and vectoring, honeybee colony health, and colony longevity. For the first time, a quantitative analysis of three picorna-like honeybee viruses over the course of a year was undertaken for DWV, ABPV, and BQCV.
MATERIALS AND METHODS
Sample collection.
Three asymptomatic colonies from each of five apiaries (n = 15 colonies), all known to have a history of varroa mite infestation, from Devon in the southwest of England (Shute, Honiton, Plymouth, Ashburton, and Newton Abbot) were sampled over a year (bimonthly between May and October 2006, monthly between November 2006 and March 2007, and bimonthly in April 2007). Three colonies from the same apiary located on the Scilly Island (St. Agnes) off the British Isles was similarly sampled (bimonthly between June and October 2007 and monthly between November 2007 and May 2008). This apiary is confirmed to have always been varroa mite free (United Kingdom National Bee Inspectorate). These colonies served as an important control group to those from Devon (varroa mites present since the early 1990s). Twenty asymptomatic worker honeybees were collected from each of the 18 colonies and stored at −20°C before shipment to the laboratory, where the 20 worker honeybees were pooled for analysis (hereafter referred to as pooled worker honeybees).
An extra sample of 30 worker honeybees was collected from one of the three colonies of the apiary of Shute in May 2007, and a sample of four worker honeybees was collected from the apiary of St. Agnes in February 2008 in order to analyze each individual separately (hereafter referred to as individual worker honeybees). An additional sample of four worker honeybees with deformed wings (hereafter referred to as symptomatic honeybees) was collected from an additional colony from an apiary in Postbridge, Devon, in September 2005. A record was kept of the sampling date, any swarming events, queen supercedure, information on varroa mite drop, other pathogens detected, and any chemical treatments that were undertaken. All study colonies were maintained using standard beekeeping practices.
Sample preparation.
The pooled honeybees were ground into a fine homogenous powder in liquid nitrogen and then stored at −80°C. Each individual honeybee from the extra samples (38 worker honeybees in total) was ground up separately in liquid nitrogen and also stored at −80°C. Total RNA was extracted from 30 mg of ground honeybee material using an RNeasy minikit (Qiagen) according to the manufacturer's instructions, with the exception that the elution volume was 30 μl. One microgram of RNA extracted was treated with DNase I (Promega) according to the manufacturer's instructions and was quantified using an Agilent Bioanalyser Nano Assay Series II (Agilent Technologies).
In vitro transcription and quantification of cRNA standards.
Quantitative PCR (QPCR) standards were prepared for each virus and the housekeeping gene actin by amplifying PCR products from extracted total RNA using the primers detailed in Table 1. A total of 100 ng of the gel-purified PCR product was ligated into the pCR2.1 cloning vector containing the T7 promoter sequence (Invitrogen) and was transformed according to the manufacturer's instructions. Positive clones were picked and confirmed for the presence of inserts by M13 PCR. Orientations of the inserts were determined by restriction endonuclease digestion according to the manufacturer's instructions. One microgram of the correctly orientated M13 PCR product was used for in vitro transcription using an mMessage mMachine T7 kit (Ambion) according to the manufacturer's instructions. DNA was removed from the reaction mixture by incubation with Turbo DNase (Ambion), and cRNA was recovered using an Ambion MEGA Clear Kit according to the manufacturer's instructions. cRNA was quantified using the Agilent 6000 Nano Assay Series II (Agilent Technologies) and was verified by sequence analysis. Virus and actin copy numbers were determined using the following equation: N = C/K × 182.5 × 1013 (where N is molecules per μl, C is the concentration of cRNA [μg/μl], and K is fragment size [bp]) (22). Independent dilution series of the cRNA standards were prepared for ABPV, DWV, BQCV, and actin. These were analyzed in triplicate as per the samples for every corresponding QPCR plate screened.
TABLE 1.
Primers used in this study
| Target | Primer name | Sequence (5′-3′) | Size of product (bp) |
|---|---|---|---|
| DWV | DWVQ_F1 | TAG TGC TGG TTT TCC TTT GTC | 145 |
| DWVQ_R1 | CTG TGT CGT TGA TAA TTG AAT CTC | ||
| ABPV | ABPVQ_F2 | GGA TGA GAG AAG ACC AAT TG | 169 |
| ABPVQ_R2 | CCA TAG GAA CTA ATG TTT ATT CC | ||
| BQCV | BQCVQ_F1 | CCA ATA GTA GCG GTG TTA TCT GAG | 177 |
| BQCVQ_R1 | AGC GTA TAA TAT GTC GGA CTG TTC | ||
| Actin | Actin_F1 | CCT GGA ATC GCA GAT AGA ATG C | 120 |
| Actin_R1 | AAG AAT TGA CCC ACC AAT CCA TAC |
SYBR green QPCR.
Total RNA was analyzed in triplicate for each annual cycle sample and individual honeybee sample using a One-Step SensiMix with SYBR green kit (Quantace) and the primers detailed in Table 1. Preliminary experiments were undertaken using 5, 12.5, 25, 50, 100, and 200 ng of RNA as the starting template. Optimized QPCR reaction mixtures contained 50 ng of RNA template, 1× Quantace One Step enzyme mix, 2.5 mM MgCl2, 1× SYBR green, 5 units of RNase inhibitor, and 10 pmol of each primer for BQCV or 7.5 pmol of each primer for actin, ABPV, and DWV. Reactions were run on a Quantica QPCR cycler (Techne) and proceeded with an initial reverse transcription stage at 49°C for 30 min and a denaturation step at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 54°C for 20 s for ABPV and DWV or at 58°C for BQCV and actin, and extension at 72°C for 20 s. The SYBR green signal was measured after each extension step (150 ms; 10% integration). A final dissociation curve was performed between 72°C and 95°C with 0.5°C increments, each with a 10-s hold, to ensure that a single product had been amplified and that no contamination was present in the reverse transcription negative controls or in the no-template controls. Sample copy numbers were determined using Quantica analysis software; the threshold cycle (CT) number was determined for each sample run in triplicate, and the average was taken. If the replicates were greater than 1 CT of the mean, the anomaly was removed, and the average was taken from duplicates. A total of 95.4% of samples were analyzed in triplicate, and 4.6% were analyzed in duplicate. The appropriate cRNA standards were run on each plate analyzed with QPCR, and standard curves were generated by the Quantica software by plotting the CT values against the logarithm of the calculated initial copy numbers. The sample copy numbers were generated by using the CT values and comparing them to the cRNA standard curve and then by normalizing values to the housekeeping gene actin. Viral loads per honeybee were calculated by averaging the amount of RNA extracted from 30 mg of a single honeybee; by taking into account the elution volume and the average weight of a honeybee, we determined the average amount of RNA in a single honeybee and consequently the viral load.
Statistical analysis.
Viral load values obtained from the pooled honeybees collected at each sampling date were tested to identify whether the values were representative of individual honeybees. DWV loads were compared between the pooled samples of 20 honeybees sampled in May 2006 (n = 12) and 12 individual honeybees randomly selected from the 30 individual samples collected on the same day in May but the following year, 2007. One-way comparisons were conducted using a permutational analysis of variance (PERMANOVA). This analysis presents the advantage of being distribution free but requires data independence (2, 34). The latter assumption was met for all the tests conducted in this study. Data were fourth-root transformed to down-weight the effect of larger or smaller fluctuations in the virus loads (16), and a Bray Curtis dissimilarity distance measure (11) was used for the analysis. The alpha level was determined as 5% for all statistical tests. For each of the defined seasonal periods, overwintering, spring, and summer, DWV load values obtained from pooled honeybees were compared between colonies recorded by the beekeepers as collapsed or surviving using a one-way PERMANOVA. When the number of colonies that had survived was higher than the number that collapsed, virus load samples were randomly selected from the surviving colonies to allow a balanced amount of data for virus level in both collapsed and surviving colonies (winter, n = 24; spring, n = 9; summer, n = 36). Similar analyses were conducted for ABPV and BQCV loads for the summer only (n = 36) as generally no virus loads were detected in the overwintering period. Comparison between asymptomatic (data randomly selected from the pooled bee samples of September 2006, where the virus load did not differ between apiaries; PERMANOVA, F4,10 = 1.60 and P = 0.21) and symptomatic worker honeybees infected with DWV collected on the same date the previous year, September 2005, was done using one-way PERMANOVA (n = 4).
Varroa mite analysis.
An integrated pest management plan (IPM) for the varroa mite was administered to all Devon colonies. As the St. Agnes colonies were confirmed to be varroa mite free, no IPM was required. The IPM involved at least two different mite control methods to be used each year. These included the use of Apistan (active ingredient, tau-fluvalinate), Apivar (active ingredient, amitraz), Apiguard (active ingredient, thymol), oxalic acid in sugar syrup, or drone uncapping. Where applicable, drone removal was carried out during May and June; July and August saw the pretreatment of the colonies, the autumn treatment was in August and September, and the winter treatment was in December and January (Table 2). The pretreatment estimates were calculated by multiplying the daily mite drop by 30 in July or 100 in August (32). The autumn and winter figures are the 6- and 1-week cumulative posttreatment mite drop counts, respectively.
TABLE 2.
Estimated varroa mite populations during the summer and the numbers of mites knocked down during the autumn and winter mite treatments
| Colonya | Drone removal (May-June) | Mite population (no.)b |
||
|---|---|---|---|---|
| Pretreatment estimatec | Drop at 6 wks post-autumn treatment | Drop at 1 wk post-winter treatment | ||
| JH1* | No | No data | Colony dead | Colony dead |
| JH2 | Yes | 300-1,000 | 1,600 | ≥725 |
| JH3 | Yes | 900-3000 | 3,638 | ≥269 |
| CT1 | Yes | 30 | 222 | 64 |
| CT2 | Yes | 4-14 | 166 | 87 |
| CT3 | Yes | 3-10 | 95 | 71 |
| GD1* | No | >1 | No data | No data |
| GD2 | No | No data | No data | No data |
| GD3* | No | 390 | 330 | No data |
| PW1 | No | >1 | 46 | 934 |
| PW2* | No | >1 | 35 | 198 |
| PW3* | No | >1 | 7 | 65 |
| DM1* | Yes | 120 | ∼500 | No treatment |
| DM2 | Yes | 120 | ∼500 | No treatment |
| DM3 | Yes | 120 | ∼500 | No treatment |
*, colony collapsed during the study.
Data are from the following periods: pretreatment, July and August; autumn treatment, August and September (with Apiguard, Apivar, or Apistan); winter treatment, December and January (with oxalic acid). No data, no mite data available although the colonies were treated.
The pretreatment estimates were calculated by multiplying the daily mite drop by 30 in July or by 100 in August (33).
RESULTS
QPCR assay.
cRNA standards (dilution series of cloned PCR amplicons for all three viruses plus the housekeeping gene actin) were assessed using a Quantica Techne QPCR thermocycler (Techne) using a SYBR green assay. The cRNA standard curves were generated using Quantica software by plotting the CT values against the logarithm of the calculated initial copy numbers (Fig. 1). The QPCR assays revealed copy number sensitivities in the range of 104 to 107, which were subsequently normalized to actin, giving detection limits of approximately 102 for DWV and ABPV and 104 for BQCV.
FIG. 1.
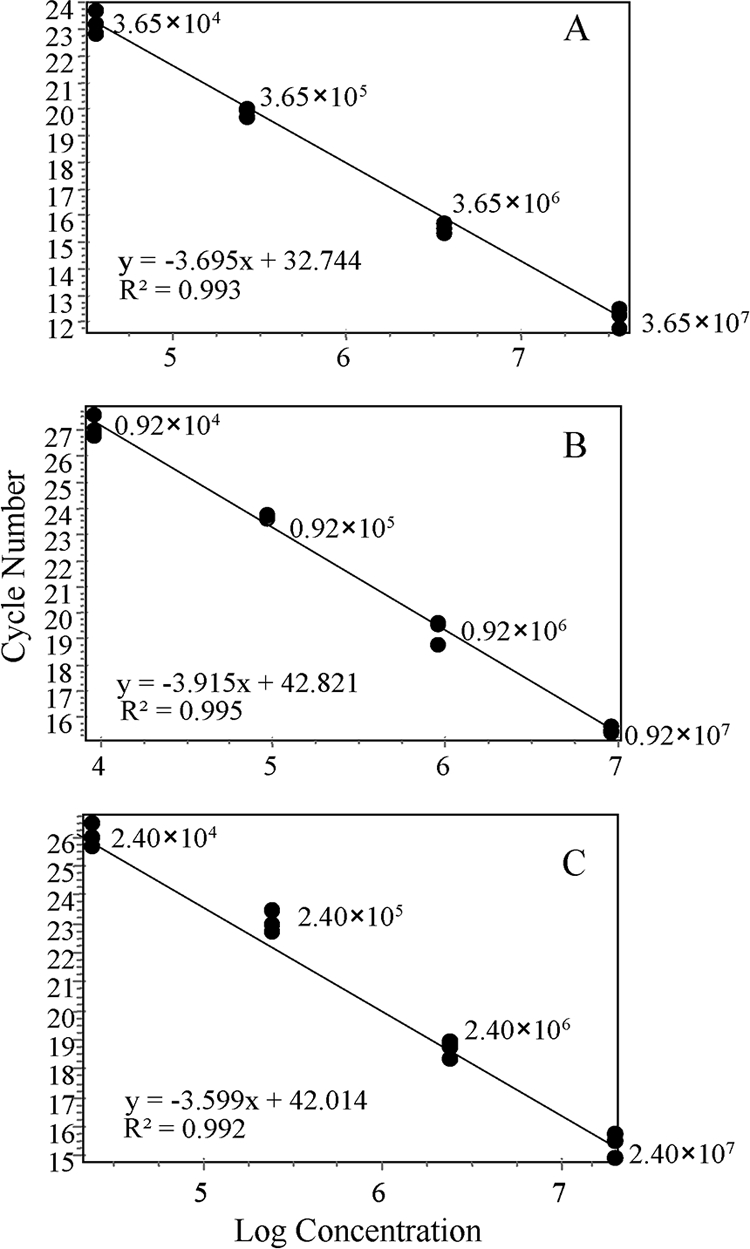
Examples of standard curves generated using cRNA standards of DWV (A), ABPV (B), and BQCV (C) where the values indicate nonnormalized virus copy numbers, R2 is the correlation coefficient, and the equation corresponds to the slope (m) and the intercept (c) according to the equation y = mx + c.
Optimization of QPCR assay based on DWV detection.
Initially, four individual honeybees were analyzed in triplicate for DWV using different initial quantities (5 ng, 50 ng, 100 ng, and 200 ng of RNA) of total RNA as a template. In this initial screen 33% of the reactions using 5 ng of starting material (5-ng reactions) failed, while all 50-ng reactions were successful; however, the 100-ng and 200-ng reactions were variable, with replicates often being inconsistent with each other (greater than 1 CT apart). A further test using 12.5 ng, 25 ng, and 50 ng of total RNA was performed, revealing a high success rate for all three of the concentrations: 95% for 12.5 ng, 98% for 25 ng, and 97% for 50 ng. As before, all of the concentrations tested were performed in triplicate, and the reliability test of >1 CT between triplicates was also assessed. Reliabilities of 73% for the 12.5-ng, 56% for the 25-ng, and 66% for the 50-ng triplicate reactions were obtained. Finally, dissociation curves of PCR products were done to ensure that only single peaks were being generated, corresponding to a single product, and that there were no nonspecific amplification products. The majority of the samples analyzed gave a single definitive dissociation peak; however, PCR products from 12.5 ng of RNA often generated broader peaks, suggesting some nonspecific products (data not shown). Taken together, all future QPCRs used 50 ng of total RNA starting material based on a 97% success rate for detecting DWV, yielding specific amplicons with 66% reliability.
DWV infection in individual asymptomatic and symptomatic honeybees.
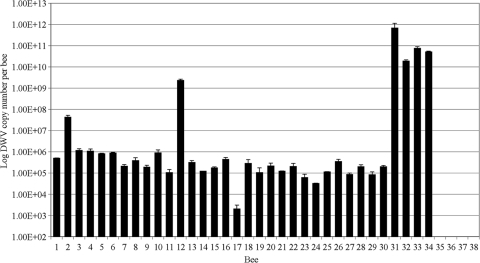
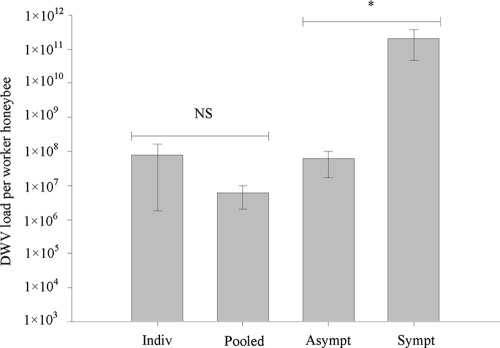
The level of DWV in individual bees was quantified and normalized to actin, with the DWV load ranging from 1.4 × 103 to 2.4 × 109 genome equivalents per asymptomatic honeybee (Fig. 2). Quantification of DWV load in individual asymptomatic honeybees collected from apiaries with a known history of varroa mite infestation confirmed that there was no significant difference in the load detected in 12 randomly selected individuals from 30 assayed asymptomatic worker honeybees versus 12 independent sets of 20 pooled asymptomatic worker honeybees (Fig. 3) (PERMANOVA, F1,22 = 1 and P = 0.39). Significant differences in DWV load were seen between randomly selected asymptomatic pooled bee samples collected in September 2006 and individually screened symptomatic worker honeybees collected in September 2005 (PERMANOVA, F1,6 = 8.91 and P = 0.04). The DWV load in symptomatic honeybees was 3 orders of magnitude higher in symptomatic honeybees: 1.8 × 1010 to 6.9 × 1011 DWV per worker honeybee (Fig. 2). The asymptomatic individual honeybees collected from colonies verified as always being varroa mite free were confirmed to be DWV free or below the limits of detection (Fig. 2).
FIG. 2.
DWV loads measured in 30 individual asymptomatic worker honeybees from a varroa mite-infested colony (bees 1 to 30), four individual symptomatic worker honeybees from a varroa mite-infested colony (bees 31 to 34), and four individual asymptomatic worker honeybees from a varroa mite-free colony (bees 35 to 38) where no DWV was detected.
FIG. 3.
DWV loads were measured in individual asymptomatic worker honeybees (Indiv; n = 30) and in sets of 20 pooled asymptomatic worker honeybees (Pooled; n = 12); samples from were from early May in both cases. DWV loads were also measured in asymptomatic pooled worker honeybees (Asympt) and symptomatic individual worker honeybees (Sympt); samples were from September 2 in both cases (n = 4). Data are mean DWV loads (± standard error). Differences in DWV loads between the individual bees and the 20 pooled bees were not significant (NS). Differences between asymptomatic and symptomatic bees were significant at a P value of < 0.05 (*).
Quantitative determination of DWV, ABPV, and BQCV infection in honeybee colonies during 1 year.
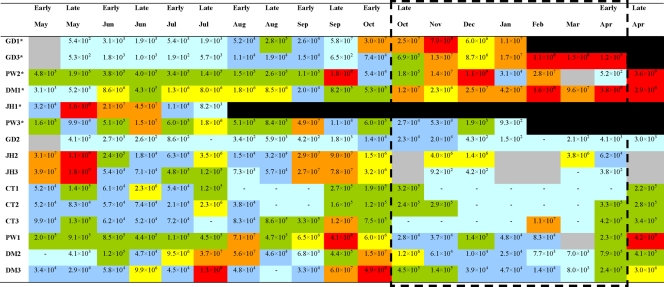
All of the viruses screened for were below the limits of detection in the three colonies on the varroa mite-free island of St. Agnes throughout the study. DWV infection within the five apiaries in Devon was seen to occur throughout the year, with numbers fluctuating between undetectable levels (<102) and 4.2 × 109 copies per asymptomatic worker honeybee (Fig. 4). Six of the 15 colonies sampled in Devon were lost during the study (GD1 collapsed in February 2007, GD3 collapsed in April 2007, PW2 collapsed in July 2007, PW3 collapsed in February/March 2007, DM1 collapsed in May 2007, and JH1 collapsed in August 2006) (Fig. 4). Of these collapsed colonies 83% were lost during the overwintering period (late October-early April) or shortly thereafter, and these colonies (with the exception of PW3) showed DWV loads exceeding 1 × 108 copies per asymptomatic worker honeybee at some stage during the overwintering period. The queen of colony PW3 ran out of sperm and so became a drone layer, with the result that the colony collapsed as no new queens or workers were produced; this colony was thus excluded from the study. The difference in DWV load during the overwintering period between collapsed (GD1, GD3, PW2, and DM1) and surviving colonies was statistically significant (PERMANOVA, F1, 46 = 15.62 and P = 0.001). No differences were observed during the spring and summer periods (spring, F1,16 = 1.08 and P = 0.37; summer, F1,70 = 1.51 and P = 0.20, both by PERMANOVA).
FIG. 4.
DWV load per asymptomatic worker honeybee during the sample period of May 2006 to April 2007. Asterisks, colonies that collapsed during the experiment; black boxes, no sample collected due to colony loss; gray boxes, no sample collected; red boxes, >1 × 108 copies of DWV per honeybee; orange boxes, 1 × 107 to 1 × 108 copies of DWV per honeybee; yellow boxes, 1 × 106 −1 × 107 copies of DWV per honeybee; green boxes, 1 × 105 to 1 × 106 copies of DWV per honeybee; blue boxes, 1 × 104 to 1 × 105 copies of DWV per honeybee; light-blue boxes, below the limits of detection, <102 (-), to 1 × 104 copies of DWV per honeybee. The dashed box indicates the overwintering period.
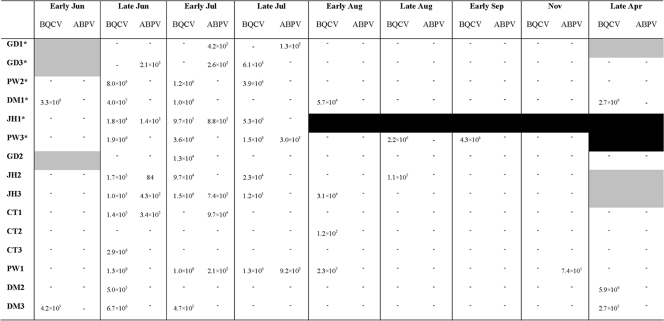
BQCV and ABPV were detected in Devon colonies mainly between June and September (Fig. 5) with the exception of one occurrence of ABPV in November in colony PW1 and three occurrences of BQCV in April 2007 in colonies DM1, DM2, and DM3. BQCV loads fluctuated both in colonies that survived and those that collapsed, and BQCV was detected at its highest level of 3.3 × 108 copies per asymptomatic worker honeybee in June. There was no significant difference observed between BQCV load in surviving and collapsed colonies (PERMANOVA, F1,70 = 0.79 and P = 0.35) during the summer period. ABPV was either absent from colonies or present at low levels, with the exception of one colony, CT1, where it was detected at its highest load of 3.4 × 105 copies per asymptomatic worker honeybee in June (Fig. 5). There was no significant difference between ABPV loads in surviving and collapsed colonies in the summer period (PERMANOVA, F1,70 = 0.92 and P = 0.34).
FIG. 5.
BQCV and ABPV load per asymptomatic worker honeybee for samples collected from May 2006 to April 2007. Asterisks, colonies that collapsed during the experiment; black boxes, no sample collected due to colony loss; gray boxes, no sample collected; −, below the limits of detection. If neither virus was detected, the sample date has been omitted from the figure.
Colony monitoring.
Although varroa mites were present in all the Devon colonies throughout the study, the mite drop analysis indicated that, with the exception of one colony (JH3), all of the colonies had mite populations in autumn well below the economic threshold of 2,000 to 3,600 mites (32) (Table 2).
DISCUSSION
Currently, there is no standardized methodology for the sampling of honeybees for viral screening (which may vary according to the purpose of the study), with some studies screening individual bees (15, 18) and others pooling sets of bees before screening/quantification, with pools of 10 (17), 12 (42), and 100 (38) bees being reported. In our study, no significant difference in DWV loads was detected between the randomly selected individuals from 30 assayed asymptomatic worker honeybees versus 12 independent sets of 20 pooled asymptomatic worker honeybees. This result validated our method of quantifying the virus load from 20 pooled bees to determine the level of viral infection in a colony rather than screening sets of individual bees. Moreover, DWV loads in individual asymptomatic worker honeybees collected from a varroa mite-infested colony were in the range of 103 to 109 copies per worker honeybee, values comparable to those reported in previous studies employing a similar methodology (20). Compared to asymptomatic worker honeybees, DWV loads in symptomatic bees were significantly higher, confirming previous findings of higher viral loads in symptomatic bees (20). Data from the annual cycle of DWV load in pooled asymptomatic worker honeybees also support this, where none of the honeybees showed any symptoms and, accordingly, had lower levels of DWV than loads reported in symptomatic honeybees. While this proposed QPCR assay methodology is highly sensitive and representative, a word of caution is nonetheless warranted here. Practical limitations of screening a limited subset of a dynamic population constrain the degree to which a comparative analysis can be made between sampling points. That said, broad trends can nonetheless be gleaned by comparing the levels of virus present over months and seasons.
All of the colonies analyzed on St. Agnes survived throughout the sample period (2007 to 2008), and neither viruses nor varroa mites were detected within these colonies. Six of the colonies monitored in Devon (2006 to 2007) did not survive into the following summer. Many factors are considered imperative for the continued persistence of honeybee colonies, particularly over the winter period. One of the colonies (PW3) that collapsed is thought to have done so due to queen failures, a factor often reported to be a cause of colony losses (39). Therefore, this colony was excluded from the statistical analysis. Colony JH1 collapsed during July 2006 and was also excluded from the analysis since only six data points were collected. It is thought that the remaining four colonies that collapsed were “true” overwintering colony losses; i.e., the colonies collapsed during the winter or shortly after with no obvious trigger for their decline. These four colonies were used for virus load comparisons with those that survived over the winter.
BQCV and ABPV were typically detected in Devon colonies during the period of June to October. This seasonal occurrence has been previously reported; however, the exact explanation for this has still to be determined (3, 40). It has been proposed that the short-term occurrence of ABPV in a colony can be attributed to its virulent nature, with infected pupae quickly dying and infected adult honeybees suffering paralysis and death (33). It has additionally been reported that worker honeybees can be infected with up to 106 ABPV particles without showing any symptoms of disease (6). The highest load detected in any of the colonies studied was lower than this value, which may be why no symptoms associated with this disease were reported in the colonies studied. There was no link between ABPV occurrence and load in surviving and collapsed colonies. ABPV seasonality occurs when there is a very rapid turnover of worker honeybees (37). Therefore, ABPV is not directly involved in overwintering colony loss, and for the most part, the colony potentially has a mechanism to cope with its presence.
BQCV has been shown to be associated with the microsporidian N. apis (4). N. apis is known to cause dysentery in honeybees; however, as is often the case with viral infections, honeybees can be infected with high levels of N. apis spores, yet no symptoms are observed (26). Although the presence of this parasite was not monitored throughout this study, it is known that N. apis has a regular annual cycle, with population peaks in spring/summer (40). There is a clear summer incidence for BQCV which could reflect a peak in the N. apis population during these times and could play a possible role of virus vector between honeybees although this cannot be verified. High levels of BQCV are experienced in the summer period, but viral loads do not correspond with colony death; i.e., no difference was observed in virus load between collapsed and surviving colonies. Again, it is thought that the rapid turnover of worker honeybees, the continuous egg-laying by the queen, and the short life span of workers during the productive months indicate that the virus is quickly purged from the colony (37).
As with the two aforementioned viruses, colonies are able to endure levels of DWV of up to 1.8 × 109 copies per asymptomatic worker honeybee through the spring/summer months. During this period, there is an increasing level of worker turnover, with workers having a maximum age of approximately 38 days, and all the overwintering worker honeybees have died. Both colonies that survived and those that collapsed in Devon experienced similarly high levels of DWV in the summer months, suggesting that a high level of DWV infection during these months does not dictate whether a colony will survive through the following winter. Elevated DWV loads during the overwintering period, however, are strongly associated with colony loss, with the four colonies which showed typical overwinter colony loss traits having significantly higher DWV load during this period than surviving colonies. In winter, honeybee populations have been shown to decrease significantly to less than 104 workers, the queen has ceased egg production, and the worker honeybees can live up to 200 days (37). Due to the aged worker honeybees and static population structure, the colony is more susceptible to any pathogenic agent.
The varroa mite-DWV association in our study does not appear to match the classical relationship previously reported in the literature (24, 33, 44). First, all colonies studied were positive for the presence of varroa mites at various levels, yet the colonies that collapsed were surprisingly not necessarily the colonies with the highest estimated varroa mite populations. What is striking with this data set are the low levels of DWV detected in some colonies over winter even though they have previously experienced significant varroa mite populations. For example, colony JH3, which had a posttreatment autumn varroa mite drop of 3,638 and a winter varroa mite drop in excess of 269, experienced DWV levels from undetectable to 102 per worker honeybees over winter; in contrast, colony PW2 experienced a much lower estimated varroa mite population and lower varroa mite drops after treatment yet saw DWV levels in excess of 109 per worker honeybees over winter. This is also seen within apiaries, with both colonies PW1 and PW2 estimated to have relatively low varroa mite populations; following winter treatment PW1 had a much higher varroa mite drop than PW2, yet PW2 is the colony that experienced high levels of DWV during the overwintering months and eventually collapsed. These observations could arguably be attributed to the varroa mite treatments being ineffective in killing the varroa mites in certain colonies. However, within-apiary comparisons, where the colonies have been treated similarly, negate this factor. The classic varroa mite-DWV relationship would predict that a colony with high levels of varroa mites would have high levels of DWV and that a colony with low levels of varroa mites would have low levels of DWV. Second, the IPM for treating the varroa mites involved at least two different mite control methods that were used during the study period. All of these control methods have a proven effectiveness of removing around 90% of the varroa mite population when administered correctly (30, 31, 35, 41). Consequently, throughout the study beekeepers maintained the varroa mite populations well below the economic threshold of 2,000 to 3,600 mites in autumn (33), with the possible exception of JH3. Third, no deformed winged honeybees were analyzed throughout the study period, and this is consistent with finding that the levels of DWV did not exceed 109 genome equivalents per honeybee. As such, a definitive link between varroa mite infestation (DWV associated or not) and colony collapse cannot be identified.
Although we cannot be certain of the exact level of infestation, the approximate intensity of the scale of the varroa mite infestation can be surmised. It has been suggested that if mite populations are kept low over winter and the honeybee population is large enough, colonies are highly likely to persist into the following spring (33). Our data implicate an alternate non-varroa mite-vectored DWV effect in asymptomatic worker honeybees in the form of overwintering colony loss. It is evident from the QPCR data that DWV can persist independently of varroa mite infestations and that DWV-associated colony loss is not necessarily always dependent on varroa mite interaction with the virus and vectoring. Moreover, the data point to an additional factor that may be critical for the manifestation of asymptomatic DWV-associated overwintering colony losses. It is possible that another pathogen is acting synergistically with DWV within the worker honeybees, triggering rapid proliferation of the virus in overwintering honeybees. Certainly, Yang and Cox-Foster (42) reported that in varroa mite-infested asymptomatic honeybees, DWV replication is increased upon injection of Escherichia coli. It was concluded that at least two agents, in this case varroa mites and E. coli, were required for enhanced replication of DWV in adult honeybees. Therefore, varroa mite infestation acting alone in a colony may not be as imperative to DWV-associated colony collapse as once thought. Our results certainly place DWV high on the list of associated causative agents. Whether the increase in DWV is responsible for eventual overwintering colony losses or is actually a product of other pathogenic interactions still needs to be established; however, DWV is clearly associated with 67% of the overwintering losses seen here. Moreover, all three viruses were below detection limits in colonies sampled in the varroa mite-free apiary across the annual cycle, with no virus-like symptoms reported or any colony losses in the apiary. This further supports the supposition that DWV is an integral component of overwintering colony losses; however, as varroa mites are also absent from these colonies, a reliable conclusion behind the survival success in these colonies cannot be ascertained.
The infection strategy of DWV over the annual cycle in asymptomatic colonies is more consistent with a persistent rather than an inapparent or covert infection strategy. First, DWV appears to be prevalent in honeybee colonies, especially those exposed to varroa mites, and persists for long periods undetected. Second, DWV in itself does not induce cell death/lysis. Third, DWV acts solely or synergistically with an agent that has not yet been identified to induce death of the colony. This does not occur via the previously described routes of deformed wing abnormalities and overpowering varroa mite infestations but, rather, more likely by a subtle and persistent alteration in the behavior of key members within the colony. Furthermore, since DWVs have an underlying breadth of genetic diversity (8), it is likely that certain genotypes or variants could be responsible for these overwintering colony losses. We therefore propose that analysis of DWV load in overwintering asymptomatic honeybees is an important diagnostic parameter for assessing whether a colony will persist into the following year. Clearly, there are still complex associations of different pathogens within the honeybee that require resolving; however, DWV monitoring will be a key factor for apiary management, highlighting colonies that will require observation in order for scientists and beekeepers alike to further resolve the causes of overwintering colony losses.
Acknowledgments
We thank the C. B. Dennis Beekeepers Research Trust for funding this research. D.C.S. is a Marine Biological Association of the United Kingdom Research Fellow funded by a grant-in-aid from the Natural Environmental Research Council of the United Kingdom.
We thank the beekeepers G. Davis, P. West, J. Hewson, C. Turner, D. Milford, D. Pratley, R. Aitken, and M. Hicks for their invaluable assistance in collecting the bees. We also thank Donald Smith for critically reviewing the manuscript.
Footnotes
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Allen, M. F., and B. V. Ball. 1996. The incidence and world distribution of the honey bee viruses. Bee World 77:141-162. [Google Scholar]
- 2.Anderson, M. J. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26:32-46. [Google Scholar]
- 3.Bailey, L., B. V. Ball, and J. N. Perry. 1981. The prevalence of viruses of honey bees in Britain. Ann. Appl. Biol. 97:109-118. [Google Scholar]
- 4.Bailey, L., B. V. Ball, and J. N. Perry. 1983. Association of viruses with two protozoal pathogens of the honey bee. Ann. Appl. Biol. 103:13-20. [Google Scholar]
- 5.Bailey, L., and B. V. Ball. 1991. Honeybee pathology, 2nd ed. Academic Press, London, United Kingdom.
- 6.Bailey, L., and A. J. Gibbs. 1964. Acute infection of bees with paralysis virus. J. Insect Pathol. 6:395-407. [Google Scholar]
- 7.Baker, A. C., and D. C. Schroeder. 2008. The use of RNA-dependent RNA polymerase for the taxonomic assignment of picorna-like viruses (order Picornavirales) infecting Apis mellifera L. populations. Virol. J. 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baker, A. C., and D. C. Schroeder. 2008. Occurrence and genetic analysis of Picorna-like viruses infecting worker bees of Apis mellifera L. populations in Devon, south west England. J. Invertebr. Pathol. 98:239-242. [DOI] [PubMed] [Google Scholar]
- 9.Batuev, Y. M. 1979. New information about virus paralysis. Pchelovodstvo 7:10-11. [Google Scholar]
- 10.Bowen-Walker, P. L., S. J. Martin, and A. Gunn. 1999. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasite mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 73:101-106. [DOI] [PubMed] [Google Scholar]
- 11.Bray, J. R., and J. T. Curtis. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325-349. [Google Scholar]
- 12.Chantawannakul, P., L. Ward, N. Boonham, and M. Brown. 2006. A scientific note on the detection of honeybee viruses using real-time PCR (TaqMan) in Varroa mites collected from a Thai honeybee (Apis mellifera) apiary. J. Invertebr. Pathol. 91:69-71. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Y., J. S. Pettis, and M. F. Feldlaufer. 2005. Detection of multiple viruses in queens of the honey bee Apis mellifera L. J. Invertebr. Pathol. 90:118-121. [DOI] [PubMed] [Google Scholar]
- 14.Chen, Y., J. D. Evans, and M. Feldlaufer. 2006. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebr. Pathol. 92:152-159. [DOI] [PubMed] [Google Scholar]
- 15.Chen, Y. P., J. A. Higgins, and M. F. Feldlaufer. 2005. Quantitative real time reverse transcription-PCR analysis of deformed wing virus infection in honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 71:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke, K. R., and R. M. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation. Primer-E Ltd., Plymouth, United Kingdom.
- 17.Cox-Foster, D. L., S. Conlan, E. C. Holmes, G. Palacios, J. D. Evans, N. A. Moran, P-L. Quan, T. Briese, M. Hornig, D. M. Geiser, V. Martinson, D. van Englesdorp, A. L. Kalkstein, A. Drysdale, J. Hui, J. Zhai, L. Cui, S. K. Hutchison, J. F. Simons, M. Egholm, J. S. Pettis, and W. I. Lipkin. 2007. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318:283-287. [DOI] [PubMed] [Google Scholar]
- 18.de Miranda, J. R., and I. Fries. 2008. Venereal and vertical transmission of deformed wing virus in honeybees (Apis mellifera L.). J. Invertebr. Pathol. 98:184-189. [DOI] [PubMed] [Google Scholar]
- 19.Dimmock, N. J., and S. B. Primrose. 1987. Introduction to modern virology. Blackwell Science, Cambridge, MA.
- 20.Fievet, J., D. Tentcheva, L. Gauthier, J. de Miranda, F. Cousserans, M. E. Colin, and M. Bergoin. 2006. Localisation of deformed wing virus infection in queen and drone Apis mellifera L. Virol. J. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forsgren, E., J. R. de Miranda, and M. Isaksson. 2009. Deformed wing virus associated with Tropilaelaps mercedesae infesting European honey bees (Apis mellifera). Exp. Appl. Acarol. 47:87-97. [DOI] [PubMed] [Google Scholar]
- 22.Fronhoffs, S., G. Totzke, S. Stier, N. Wernert, M. Rothe, T. Bruning, B. Koch, A. Sachinidis, H. Vetter, and Y. Ko. 2002. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol. Cell. Probes 16:99-110. [DOI] [PubMed] [Google Scholar]
- 23.Genersch, E., C. Yue, I. Fries, and J. R. de Miranda. 2006. Detection of deformed wing virus, a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J. Invertebr. Pathol. 91:61-63. [DOI] [PubMed] [Google Scholar]
- 24.Gisder, S., P. Aumeier, and E. Genersch. 2009. Deformed wing virus: replication and viral load in mites (Varroa destructor). J. Gen. Virol. 90:463-467. [DOI] [PubMed] [Google Scholar]
- 25.Hansen, H., and C. J. Brødsgaard. 1999. American foulbrood: a review of its biology, diagnosis and control. Bee World 80:5-23. [Google Scholar]
- 26.Higes, M., F. Esperon, and J. M. Sanchez-Vizcaino. 2007. First report of black queen cell virus detection in honeybee (Apis mellifera) in Spain. Span. J. Agric. Res. 3:322-325. [Google Scholar]
- 27.Higes, M., R. Martin-Hernandez, C. Botias, E. Garrido Ballon, A. V. Gonzalez-Porto, L. Barrios, M. Jesus del Nozal, J. L. Bernal, J. J. Jimenez, P. Garcia Palencia, and A. Meana. 2008. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10:2659-2669. [DOI] [PubMed] [Google Scholar]
- 28.Iqbal, J., and U. Mueller. 2007. Virus infection causes specific learning deficits in honeybee foragers. Proc. Biol. Sci. 274:1517-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanzi, G., J. R. de Miranda, M. B. Boniotti, C. E. Cameron, A. Lavazza, L. Capucci, S. M. Camazine, and C. Rossi. 2006. Molecular and biological characterisation of deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 80:4998-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodesani, M., S. Bergomi, A. Pellacani, E. Carpana, and T. Rabitti. 1990. A comparative study on the efficacy of some products for controlling Varroa, and determinations of their residues. Apicoltura 6:105-130. [Google Scholar]
- 31.Marchetti, S., and R. Barbattini. 1984. Comparative effectiveness of treatments used to control Varroa jacobsoni Oud. Apidologie 15:363-377. [Google Scholar]
- 32.Martin, S. J. 1999. Population modelling and the production of a monitoring tool for Varroa jacobsoni an ectoparasitic mite of honey bees. Asp. Appl. Biol. 53:105-112. [Google Scholar]
- 33.Martin, S. J. 2001. The role of Varroa and viral pathogens in the collapse of honey bee colonies: a modelling approach. J. Appl. Ecol. 38:1082-1093. [Google Scholar]
- 34.McArdle, B. H., and M. J. Anderson. 2001. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290-297. [Google Scholar]
- 35.Nanetti, A. 1999. Oxalic acid for mite control—results and review, p. 6-11. In Coordination in Europe of research on integrated control of Varroa mites in honey bee colonies. Commission of the European Communities, Merelbeke, Belgium.
- 36.Navajas, M., A. Migeon, C. Alaux, M. L. Martin-Magniette, G. E. Robinson, J. D. Evans, S. Cros-Arteil, D. Crauser, and Y. Le Conte. 2008. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genomics 9:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Page, R. E., and C. Y.-S. Peng. 2001. Aging and development in social insects with emphasis on the honeybee, Apis mellifera L. Exp. Gerontol. 36:695-711. [DOI] [PubMed] [Google Scholar]
- 38.Tentcheva, D., L. Gauthier, N. Zappulla, B. Dainat, F. Cousserans, M. Colin, and M. Bergoin. 2004. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 70:7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Englesdorp, D., J. Hayes, Jr., R. M. Underwood, and J. Pettis. 2008. A survey of honey bee colony losses in the U.S., fall 2007 to spring 2008. PLoS ONE 3:e4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varis, A. L., B. V. Ball, and M. Allen. 1992. The incidence of pathogens in honey bee (Apis mellifera L) colonies in Finland and Great Britain. Apidologie 23:133-137. [Google Scholar]
- 41.Wilkinson, D., and G. C. Smith. 2002. Modelling the efficiency of sampling and trapping Varroa destructor in the drone brood of honey bees (Apis mellifera). Am. Bee J. 142:209-212. [Google Scholar]
- 42.Yang, X., and D. L. Cox-Foster. 2005. Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA 102:7470-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yue, C., and E. Genersch. 2005. RT-PCR analysis of deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 86:3419-3424. [DOI] [PubMed] [Google Scholar]
- 44.Yue, C., M. Schroder, S. Gisder, and E. Genersch. 2007. Vertical-transmission routes for deformed wing virus of honeybees (Apis mellifera). J. Gen. Virol. 88:2329-2336. [DOI] [PubMed] [Google Scholar]