Abstract
In the commercial food industry, demonstration of microbiological safety and thermal process equivalence often involves a mathematical framework that assumes log-linear inactivation kinetics and invokes concepts of decimal reduction time (DT), z values, and accumulated lethality. However, many microbes, particularly spores, exhibit inactivation kinetics that are not log linear. This has led to alternative modeling approaches, such as the biphasic and Weibull models, that relax strong log-linear assumptions. Using a statistical framework, we developed a novel log-quadratic model, which approximates the biphasic and Weibull models and provides additional physiological interpretability. As a statistical linear model, the log-quadratic model is relatively simple to fit and straightforwardly provides confidence intervals for its fitted values. It allows a DT-like value to be derived, even from data that exhibit obvious “tailing.” We also showed how existing models of non-log-linear microbial inactivation, such as the Weibull model, can fit into a statistical linear model framework that dramatically simplifies their solution. We applied the log-quadratic model to thermal inactivation data for the spore-forming bacterium Clostridium botulinum and evaluated its merits compared with those of popular previously described approaches. The log-quadratic model was used as the basis of a secondary model that can capture the dependence of microbial inactivation kinetics on temperature. This model, in turn, was linked to models of spore inactivation of Sapru et al. and Rodriguez et al. that posit different physiological states for spores within a population. We believe that the log-quadratic model provides a useful framework in which to test vitalistic and mechanistic hypotheses of inactivation by thermal and other processes.
Thermal processes for the food industry are typically developed and validated through laboratory-based experimentation. A common experimental format involves isothermal inactivation to determine the decimal reduction time (DT), which is the time required for a 10-fold (1-log10 or decimal) reduction in the number of microorganisms at a given temperature (T), according to the primary model:
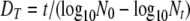 |
(1) |
where Nt is the number of microorganisms (usually per unit of volume or weight) at time t and N0 is the number of microorganisms at time zero (5). When DT values are determined at different temperatures, the relative lethality of these temperatures can be determined if we assume the secondary model:
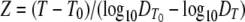 |
(2) |
where T0 is a reference temperature and z is the temperature difference required for a 10-fold change in the DT of the target microorganism compared with the DT at T0 (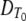 )(5, 24).
)(5, 24).
The z value concept has been popular in commercial food processing settings (for example, when cans of food are retorted) as it allows dynamic description of inactivation during a nonisothermal process (i.e., when the temperature varies with the processing time) and across space, as heat penetrates the food (10). The thermal equivalence of nonisothermal processes is often calculated by integration (9) using
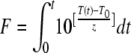 |
(3) |
where F is the accumulated lethality expressed as the equivalent time at a specific reference temperature (T0) for a specific z value (4). F0 is accepted shorthand for the case where T0 is 121.1°C and z is 10°C. For low-acid foods (pH >4.6), the minimum microbiologically safe thermal process is generally regarded as an F0 value of 2.8 min at the slowest heating point of the food (9). This minimum F0 value targets the most heat-resistant relevant pathogen for low-acid foods (proteolytic Clostridium botulinum) and is calculated on the basis of a D value at 121.1°C of 0.23 min and a targeted reduction of 12D (19).
Implicit in the concept of log-linear inactivation kinetics are two assumptions: (i) that the length of time that a microorganism is exposed to a treatment has no effect on the probability that it will be inactivated in the next unit of time (15) and (ii) that all of the cells or spores in a population have the same resistance (3). Despite the ubiquity of log-linear kinetics in the thermal processing literature, frequent observations of non-log-linear inactivation of microorganisms are well documented, indicating that inactivation is exposure time dependent.
Geeraerd et al. (11) described nine different commonly observed shapes for inactivation curves when the log10 number of microorganisms is plotted against time: linear, linear with a preceding shoulder, linear with tailing, sigmoid-like, concave, convex, convex-concave with tailing, biphasic, and biphasic with a shoulder. The most common explanation for deviations from log-linear inactivation kinetics invokes the vitalistic theory, which holds that the resistance of individual cells in a population is not the same but follows a distribution (3). Several models have been used to describe inactivation of bacterial populations where death is obviously non-log linear, including the log-normal (26), log-logistic (3, 7), biphasic (6), and Weibull (1, 12, 13, 16, 18) models.
To illustrate models commonly used to describe curvature in microbial survival, we considered the biphasic and Weibull models in further detail. The biphasic model can describe upward concavity (tailing), embodying the assumption that a given microbial population is not homogeneous but is instead composed of two subpopulations that decay independently according to log-linear kinetics (27). It has the form
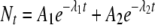 |
(4) |
where A1 and A2 are the initial numbers (or concentrations) of spores from the two populations and λ1 and λ2 are the rate parameters (equivalent to D values) for the populations.
While the biphasic model usefully preserves the physical meaning of DT in its parameterization, fitting the model directly is usually a difficult nonlinear optimization problem. (Numerical difficulties arise in fitting sums of exponentials like those in equation 4 because of indeterminacies in the parameters; similar Nt curves can be obtained with a relatively broad range of A and λ values, making it difficult to obtain precise parameter estimates.)
The Weibull model is able to describe concave, convex, and linear inactivation curves and has the form (in our notation):
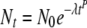 |
(5) |
where the rate parameter λ is no longer equivalent to a D value (except when p = 1) because it is multiplied by time (t) raised to the power p. Upward concavity (tailing) in this model embodies the idea that sensitive members of the population perish rapidly, leaving behind progressively sturdier survivors that, as the process continues, take longer and longer to decimate (15). Survival curves are often concave upward when microorganisms are exposed to a relatively low-intensity treatment (14). In contrast, downward concavity (shoulders) indicates that continued exposure results in accumulated damage that lowers the heat resistance of the surviving cells or spores, and thus progressively shorter times are necessary to decimate survivors as the process continues (15). Survival curves with downward concavity often indicate harsher treatment conditions (14). Unfortunately, despite these links to physiological effects, the major drawback to the Weibull model remains the difficulty of interpreting the biological significance of its parameters (8); despite the fact that a rate-like (DT-like) parameter appears in equation 5, the meaning of this parameter is altered depending on the value of a second descriptive parameter, p, which describes the extent of curvature in the inactivation kinetics.
Introduction of the DT, z, and F0 value concepts occurred at a time when computers were not routinely used, and there was a strong need for linear methods to simplify prediction of inactivation kinetics and therefore process safety (15). Current computing technology enables us to handle nonlinear semilogarithmic mathematical models and their underlying distributions to describe microbial inactivation (15). However, despite proposals to replace the traditional F0 concept with alternate methodology to validate process safety (14, 17), the clear interpretability of DT, z, and F0 values remains attractive; provision of these values to describe microbial inactivation by thermal processing is expected by food safety regulators, including the United States Food and Drug Administration (25). We note that some authors have argued that a first-order model may be the best generally applicable model to describe microbial inactivation even for emerging and innovative technologies, such as high-pressure processing (8).
The frequent observation of non-log-linear microbial inactivation kinetics and the current state of end user preferences and regulatory expectations motivated our development of a relatively simple modeling routine to extract DT-like values from thermal inactivation data that show obvious curvature. Here we also demonstrate how non-log-linear models of microbial inactivation, such as the Weibull model, can fit into a statistical linear model framework that simplifies their solution and provides additional useful information, such as confidence intervals about fitted values. It is not our intent to propose that the quadratic model is better in all circumstances. However, we believe that the selection procedure that we have used (analysis of variance) should give rise to the best model among the models that are available. The quadratic model is an additional model.
MATERIALS AND METHODS
Development of a statistical linear model framework.
Laboratory-based thermal inactivation experiments typically consist of preheating (or heating rapidly to a fixed temperature) a quantity of medium inoculated with viable microorganisms, the approximate concentration of which (N0) is known, and, at a number of sampling times, removing a quantity of medium (either a small subsample or a separate heated volume, sampled destructively) from which the concentration of viable microorganisms remaining in the heated sample is estimated. It is generally assumed that the medium removal has a negligible effect on the remainder of the samples withdrawn in the run and that estimated counts are therefore independent once an overall run effect is accounted for.
To establish a statistical linear model framework, we rearranged equation 1 as follows:
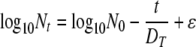 |
(6) |
adding the “noise” term ɛ to represent random variation in the estimate of the log10 count of surviving spores. The use of an additive, normally distributed noise term enables the standard “least-squares” approach to fit equation 6.
In statistical terms, and switching to natural logarithms, equation 6 can be rewritten as
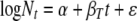 |
(7) |
If the initial concentrations (N0) were known, then α would be known. However, while estimates of initial spore concentrations are generally known for inactivation studies, the exact viable count is generally unknown. In this approach, α is treated as an unknown and can differ in each replicate run. In equation 7, βT would be expected to be negative, indicating that the numbers of survivors decrease with increasing thermal process exposure time.
Equation 7 is regarded as a statistical linear model because it is linear in parameters α and βT (equation 7 is also linear in time). Estimation in this statistical setting is particularly straightforward. A separate intercept parameter α is used for each replicate; a separate β parameter is used for each temperature; and the random variation ɛ is assumed to be normally distributed with constant variance across all replicates and temperatures. Introducing some further notation, equation 7 can be rewritten
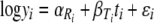 |
(8) |
where yi is the i-th measurement of spore concentration, Ri is the replicate number of measurement i, Ti is the temperature of measurement i, and ti is the time of measurement i. A great advantage of this statistical linear model approach is that, for a typical series of isothermal inactivation experiments, equation 8 can be fitted to data from all replicates and temperatures at once, alleviating the need (for the moment) for any secondary model of the dependence of βT (equivalent to DT) on temperature. This approach allows all model parameters to be jointly optimized; fitting a model in two or more steps means that secondary model parameter values are conditional on primary model parameters (but not vice versa). One-step fitting eliminates this asymmetric treatment of parameters and allows us to state that the fitted parameters are jointly optimal. Furthermore, confidence intervals about fitted values are readily obtained under the assumed normal distribution of random variation.
Extension of the statistical linear model framework to a model describing curvature, the Weibull model.
Using the notation of equation 8, the Weibull model can be written
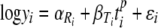 |
(9) |
or
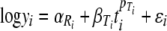 |
(10) |
where 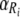 and
and 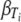 are as described above and p (or
are as described above and p (or 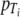 ) is a curvature parameter (15). Unfortunately, p and
) is a curvature parameter (15). Unfortunately, p and 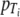 are nonlinear parameters in this statistical model and require more complex model-fitting methods. However, in the statistical linear model framework it is possible to fit the same p for all temperatures and to allow only β (equivalent to the D value) to vary by temperature (equation 9). The second form allows a different Weibull exponent for each temperature (equation 10).
are nonlinear parameters in this statistical model and require more complex model-fitting methods. However, in the statistical linear model framework it is possible to fit the same p for all temperatures and to allow only β (equivalent to the D value) to vary by temperature (equation 9). The second form allows a different Weibull exponent for each temperature (equation 10).
Development of the log-quadratic model to describe curvature in a statistical linear model framework.
Equations 9 and 10 allow some types of curvature in isothermal inactivation, but there are other, simpler models that also do this. The simplest extension of equation 8 to allow curvature is
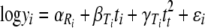 |
(11) |
which, while quadratic in time, is linear in all its parameters, permitting standard linear modeling approaches.
Superficially, equation 11 may appear to be purely empirical, but there are motivating links to the mechanistically based biphasic model. On the count scale, the log-linear model corresponds to
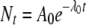 |
(12) |
where −λ0 replaces β (equivalent to the D value) in previous equations. If the microbial population actually consists of two subpopulations (i.e., is biphasic) with corresponding λ parameters, λ1 and λ2, and if the inoculum contains A1 and A2 of each subpopulation, respectively, the model corresponds to equation 4.
As mentioned above, fitting the biphasic model (equation 4) directly is usually a difficult nonlinear optimization problem. We propose an alternate, simpler approximation.
Provided that λ1 and λ2 are sufficiently close to each other, it can be shown that, to first order, equation 4 corresponds to
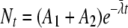 |
(13) |
where λ̄ is a weighted average of λ1 and λ2 (equivalent to the average D value):
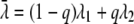 |
(14) |
where
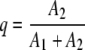 |
(15) |
This is exactly the form of the log-linear model and so does not capture curvature. However, a second-order approximation of equation 4 is
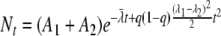 |
(16) |
where counts are log quadratic in time, as in equation 11, and thus capture certain types of curvature. Specifically, since the coefficients β and γ in equation 11, have the forms
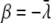 |
(17) |
and
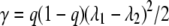 |
(18) |
the parameter γ is positive in equation 11 and so can represent only concave upward curvature. In comparison, the Weibull model (equation 5) can represent concave (p > 1) and convex (p < 1) curvature. Of course, when a model that has the form of equation 11 is fitted, the γ parameter(s) does not need to be positive and convex curvature can be represented, but mixed population interpretability (approximating distribution of D values) is lost.
A similar approximation of the Weibull model can be made using (the remainder form of) Taylor's theorem
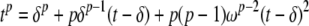 |
(19) |
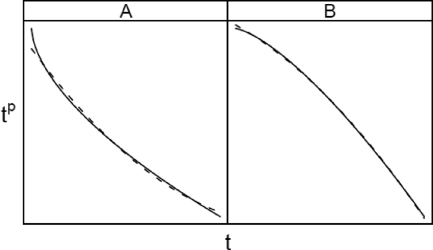
where δ is a small positive number less than t and ω is between δ and t. Some rearrangement, neglecting ω's dependence on t, gives us a quadratic approximation which should work well for p ≤ 2. (Readers unfamiliar with Taylor series expansions need only understand that it provides a way to express tp in terms of t, t2, t3, etc. for small deviations of δ around t. Figure 1 shows the quadratic approximation of two idealized Weibull curves as examples.)
FIG. 1.
Plots of −tp for p value of 0.6 (A) and 1.4 (B) (solid lines) and a least-squares quadratic approximation (dashed lines).
In summary, the log-quadratic model (equations 11 and 16) can be considered an approximation of both the biphasic (equation 4) and Weibull (equation 5) models. Both the biphasic and Weibull models impose constraints on the parameters of the log-quadratic model, but we ignore these constraints in favor of ensuring the best possible fit to the data. Naturally, the log-quadratic model admits the possibility of a fit that shows increasing microbial counts over time for t greater than some large threshold (i.e., when γ is positive). This situation is unlikely to arise when data that show decreasing microbial counts over time are interpolated, but it does mean that the log-quadratic model is unsuitable for extrapolation (i.e., for determining the thermal processing time required to achieve a specific log reduction outside the experimentally observed range of processing times). It is conceivable that the minimum value of a fitted log-quadratic model could occur within the range of the data, and for this reason (as with all models) we advise that the adequacy of the fitted model be reviewed. For extrapolation, it may be necessary to convert to the equivalent biphasic or Weibull form of the model, and more research is required in this area. (We mention this because specifications of thermal processing times to achieve 12D reductions are generally based on extrapolations that assume log-linear inactivation kinetics.)
Illustrative isothermal inactivation data.
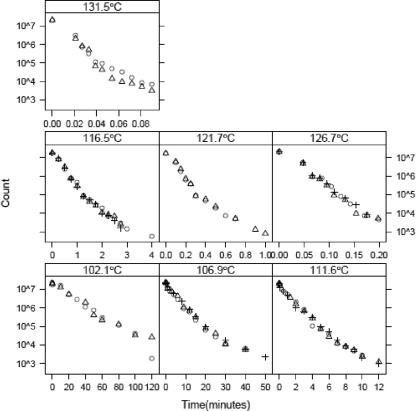
To illustrate the value of the proposed statistical linear model framework, various models were applied to kinetic thermal inactivation data for C. botulinum 213B (2). Complete details concerning the methodology used have been described by Anderson (2). Briefly, inactivation experiments were performed using a thermoresistometer with either manual or automated sampling (2). Following heating, spores of C. botulinum 213B were recovered initially in MPA3679 broth supplemented with sodium thioglycolate, sodium bicarbonate, and lysozyme and then pour plated on MPA3679 agar and incubated at 30°C for 48 h prior to enumeration (2). The data set consists of isothermal kinetic inactivation data for seven temperatures, 102.1, 106.9, 111.6, 116.5, 121.7, 126.7, and 131.5°C, and kinetic samples were withdrawn at 10 or more time points for each temperature. Generally, there were two or three replicates for each temperature, although replicates were not always sampled at the same time intervals (Fig. 2).
FIG. 2.
Data of Anderson (2) for thermal inactivation of C. botulinum 213B. The panels show data for different temperatures. Replicates are indicated by different symbols.
Model fitting.
Log-linear (equation 8), Weibull (equation 10), and log-quadratic (equation 11) models were fitted to the data using the lm (linear model) or nls (nonlinear least squares) fitting functions in R, as appropriate. (R is a popular statistical analysis language that is freely available [20].) Each model was fitted to data for all replicates at all temperatures simultaneously. Each model allowed a different intercept for each replicate and (where relevant) a different slope (rate parameter, equivalent to D value) for each temperature. Analysis of variance of the models was carried out using the anova function in R.
RESULTS
Comparison of model fits to describe the thermal inactivation of C. botulinum 213B.
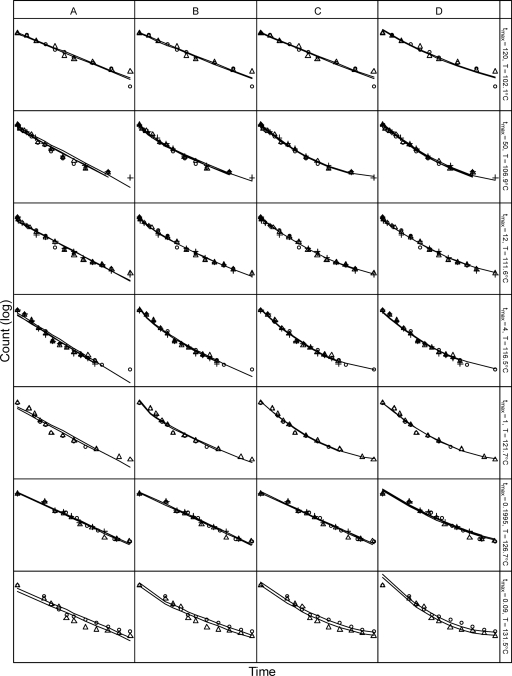
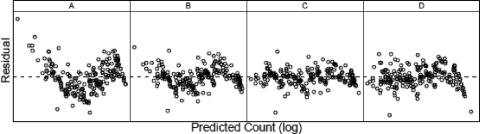
Figure 3A shows the fit of the log-linear model to the thermal inactivation of C. botulinum 213B data of Anderson (2), and Fig. 4A shows a plot of the residuals (on a log scale) against the predicted counts. There did not appear to be a strong relationship between the size of the residuals and the predicted means; this supports the model's assumption of constant noise variance (on the log scale) for all replicates and temperatures. However, the roughly “V-shaped” distribution of the residuals (Fig. 4A) indicates structure in the data that was not captured by the log-linear model, suggesting that the log-linear model was not an ideal fit to the data. Similarly, there was evidence of residual structure in the fit of the Weibull model to the data (Fig. 4B). In comparison, the quadratic model fit to the data (Fig. 3C) seemed better than the fit of both the log-linear and Weibull models, and its residuals showed much less structure (Fig. 4C).
FIG. 3.
Fits of the log-normal log-linear model (equation 8) (A), Weibull model with p varying by temperature (equation 10) (B), log-quadratic model (equation 11) (C), and quadratic model including a log-linear model for temperature dependence of the rates (equation 26) (D) to the data of Anderson (2) for thermal inactivation of C. botulinum 213B. At each temperature, time and count axes are rescaled so the data points span the plot area.
FIG. 4.
Residuals of the model fits for the log-normal log-linear model (equation 8) (A), Weibull model with p varying by temperature (equation 10) (B), log-quadratic model (equation 11) (C), and quadratic model including a log-linear model for temperature dependence of the rates (equation 26) (D) to the data of Anderson (2) for thermal inactivation of C. botulinum 213B.
Analysis of variance among the models (Table 1) showed increasing improvement in fit from the log-linear model to the Weibull model with fixed p to the Weibull model with p varying by temperature. The log-quadratic model gave a much improved fit compared to the log-linear model. Formal comparison of the Weibull and log-quadratic models was not possible, as these models were not nested, but the residual sum of squares for the log-quadratic model was much lower than that for the Weibull model with the same degrees of freedom, indicating a better fit.
TABLE 1.
Analysis of variance for the log-linear model, the Weibull model with fixed p, the Weibull model with p varying by temperature, and the log-quadratic modela
| Model | Residual df | Residual sum of squares | df | Sum of squares | F statistic | P value |
|---|---|---|---|---|---|---|
| Log-linear | 181 | 67.335 | ||||
| Weibull (fixed p) | 180 | 38.229 | 1 | 29.106 | 137.0448 | <0.001 |
| Weibull (varying p) | 174 | 30.879 | 6 | 7.350 | 6.9025 | <0.001 |
| Log quadratic | 174 | 19.396 | 7 | 47.939 | 61.437 | <0.001 |
The Weibull model with fixed p was compared with the log-linear model; the Weibull model with varying p was compared with the Weibull model with fixed p; and the log-quadratic model was compared with the log-linear model.
Determination of rate parameters.
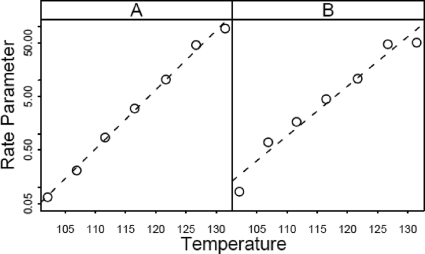
The rate parameter (equivalent to D value) estimates for the log-linear model, Weibull model with varying p, and log-quadratic model are shown in Fig. 5A, 5B, and 6A, respectively. Recall that the dependence of D on temperature is commonly modeled as a log-linear relationship, which gives rise to the z value. These plots could be used to verify this type of dependence.
FIG. 5.
Rate parameters (equivalent to D values) for the data of Anderson (2) for thermal inactivation of C. botulinum 213B estimated using the log-linear model (equation 8) (A) and using the Weibull model (equation 10) (B).
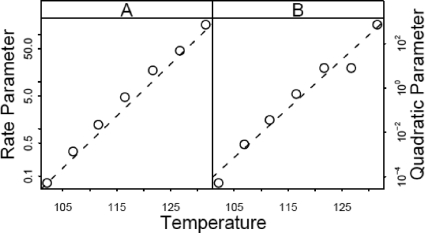
FIG. 6.
Rate parameters (equivalent to D values) (A) and quadratic term parameters (B) for the data of Anderson (2) for thermal inactivation of C. botulinum 213B estimated using the log-quadratic model (equation 11).
Figure 5A shows a plot of the equivalent of D versus temperature, with D on a log scale. There may be some slight curvature on the upper right. Figure 5B shows the same plot for the Weibull model, with p also allowed to vary with temperature; there appears to be more variability in the estimated D values than there is with the log-linear model or the coefficient of the linear term in the log-quadratic model (Fig. 6A). The addition of the quadratic term seems to have less impact on estimation of the D value equivalent than it does in the Weibull model.
Figure 6B shows a plot of the coefficients of the quadratic terms on a log scale against temperature. Interestingly, these coefficients also seem to have a linear relationship to temperature, and we discuss a possible secondary model to capture this relationship below.
DISCUSSION
Having reviewed the conventional “D-z-F” approach for modeling microbial inactivation, we showed how this approach can be cast as a statistical linear model that can be fitted to data from all replicates and temperatures at once. We also showed how the quadratic extension of this model can be related to the Weibull and biphasic models of inactivation. Fitting these models to C. botulinum inactivation data showed that the log-quadratic model best explained the observed microbial counts.
Thus far, we have not attempted to relate isothermal inactivation curves obtained at different temperatures. The log-linear model underpinning the “D-z-F” approach relates DT at different temperatures by the secondary model of equation 2, which in turn allows derivation and comparison of the accumulated lethality of the nonisothermal processes typically employed by the food industry via equation 3. In effect, this approach characterizes the thermal inactivation of a microbe at different temperatures with just two parameters, 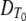 and z, but in doing so it makes strong assumptions about inactivation over time that are at odds with experimental evidence in many cases. Below, we discuss ways to relax these assumptions using the log-quadratic model.
and z, but in doing so it makes strong assumptions about inactivation over time that are at odds with experimental evidence in many cases. Below, we discuss ways to relax these assumptions using the log-quadratic model.
Equation 2 implies that there is a log-linear relationship between DT and T. In terms of equation 8 this is equivalent to
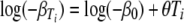 |
(20) |
and rewriting equation 8 gives
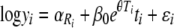 |
(21) |
which is now nonlinear in one of its parameters (θ) and must be fitted using nonlinear least-squares methods.
In Fig. 5A, the almost straight-line relationship between log-scale DT estimates at different temperatures suggests that the log-linear relationship between DT and temperature holds well in Anderson's data (2), although the higher temperatures may show some departure. Fitting equation 21 directly involves nonlinear procedures for which the straight-line fit in Fig. 5A provides useful starting values.
Incorporating this kind of temperature dependence into the better-fitting quadratic model is a little more complex because of the quadratic term. Figure 6 shows the negative of the plot of the linear and quadratic term parameters (β and γ) against temperature. On a log scale, both parameters show a reasonably linear relationship with temperature. In terms of the biphasic model that motivated this quadratic form, each of the temperature-dependent rate parameters λ1T ≡ λ1 and λ2T ≡ λ could be thought of as satisfying a log-linear dependence on temperature:
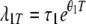 |
(22) |
and
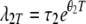 |
(23) |
This could further be simplified by setting θ1 equal to θ2 (equivalent to z), which corresponds to the two subpopulations having the same z value but different D values at the reference temperature. Now the approximate log-quadratic relationship between counts and time (equation 16) can be written as
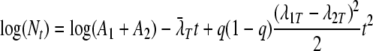 |
(24) |
(without the i subscripts for clarity). Substituting λ1T and λ2T gives
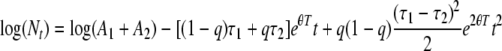 |
(25) |
which is equivalent to
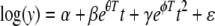 |
(26) |
which is linear in all the parameters except θ and φ. In addition, the linear and quadratic terms in this model display a log-linear relationship with temperature consistent with Fig. 6. Recall that α can vary by replicate but β and γ do not vary by replicate, implying that the same proportions of each subpopulation are in the inoculum for each replicate but not necessarily the same total spore count.
Figures 3D and 4D show fitted curves and residuals, respectively, from equation 26. Analysis of variance indicates that this four-parameter (β, γ, θ, φ) model is a better fit than the conventional two-parameter (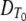 , z) log-linear model with secondary model structure but is a worse fit than the 14-parameter (βT, γT) log-quadratic model with no secondary model structure. This is to be expected given that we are imposing fewer assumptions than the conventional (DT0, z) log-linear model but more assumptions than the log-quadratic model with no secondary model structure.
, z) log-linear model with secondary model structure but is a worse fit than the 14-parameter (βT, γT) log-quadratic model with no secondary model structure. This is to be expected given that we are imposing fewer assumptions than the conventional (DT0, z) log-linear model but more assumptions than the log-quadratic model with no secondary model structure.
However, the merit of secondary structure is that it may give us clues about possible mechanisms of microbial inactivation at different temperatures. If two subpopulations have the same z value but different D values at the reference temperature, then φ ≈ 2θ. We found this to be the case for Anderson's data, where φ was 0.5273 and θ was 0.2634, giving a φ value of 2.002θ. This is consistent with a mixed-population (e.g., biphasic) model where each microbe has the same z value but a different D value at the reference temperature.
In terms of biological mechanisms, this is consistent with Anderson's C. botulinum data that behaved according to the model of spore inactivation of Rodriguez et al. (21, 22). This is actually a special case of the model of Sapru et al. (23) which posits that spores may assume three different physiological states, dormancy, inactivation, and activation, and that inactivation may occur from either an activated state or a dormant state (effectively, two subpopulations) according to two independent rate constants, Kd1 and Kd2. The model of Rodriguez et al. hypothesizes that the rate constants of activated and dormant spores are close enough to be considered equal (Kd1 = Kd2 = Kd). Determining all three rate constants for the full model of Sapru et al. appears to be challenging using traditional viability estimation methodology. In fact, all of the models of Sapru et al. and Rodriguez et al. can be expressed as sums of exponential terms in an isothermal setting and can be approximated by a quadratic model on a log scale.
Thus, as stated by other workers, deciding whether a population is really a mixture cannot be done on the basis of the survival curve's shape alone (17). Still, the log-quadratic model that we have proposed provides an opportunity to explore mechanistic and vitalistic hypotheses because (unlike the log-linear model) it can capture certain kinds of curvature in microbial inactivation kinetics and (unlike the Weibull model) its parameterization allows a biologically interpretable secondary model to capture the dependence of these kinetics on temperature.
Acknowledgments
We thank W. Anderson for access to C. botulinum inactivation data and A. Trajstman, T. Ross, and particularly M. Bull for stimulating discussions.
Footnotes
Published ahead of print on 18 September 2009.
REFERENCES
- 1.Albert, I., and P. Mafart. 2005. A modified Weibull model for bacterial inactivation. Int. J. Food Microbiol. 100:197-211. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, W. 1997. Non-log-linear thermal inactivation of Clostridium botulinum spores. Ph.D. thesis. University of Reading, Reading, United Kingdom.
- 3.Anderson, W., P. J. McClure, A. C. Baird-Parker, and M. B. Cole. 1996. The application of a log-logistic model to describe the thermal inactivation of Clostridium botulinum 213B at temperatures below 121.1°C. J. Appl. Bacteriol. 80:283-290. [DOI] [PubMed] [Google Scholar]
- 4.Bakalis, S., P. W. Cox, and P. J. Fryer. 2001. Modelling particular thermal technologies, p. 113-137. In P. Richardson (ed.), Thermal technologies in food processing. Woodhead Publishing, Cambridge, England.
- 5.Ball, C. O. 1943. Short-time pasteurization of milk. Ind. Eng. Chem. 35:71-84. [Google Scholar]
- 6.Cerf, O. 1977. Tailing of survival curves of bacterial spores: a review. J. Appl. Bacteriol. 42:1-19. [DOI] [PubMed] [Google Scholar]
- 7.Cole, M. B., K. W. Davies, G. Munro, C. D. Holyoak, and D. C. Kilsby. 1993. A vitalistic model to describe the thermal inactivation of Listeria monocytogenes. J. Ind. Microbiol. 12:232-239. [Google Scholar]
- 8.Diels, A., I. Van Opstal, B. Masschalck, and C. W. Michiels. 2007. Modelling of high-pressure inactivation of microorganisms in foods, p. 161-197. In S. Brul, S. van Gerwen, and M. Zwietering (ed.), Modelling microorganisms in foods. CRC Press LLC, Boca Raton, FL, and Woodhead Publishing Limited, Cambridge, England.
- 9.Food Science Australia and D. Warne. 2005. Australian Quarantine Inspection Service approved persons course for thermal processing of low-acid foods. Food Science Australia, Werribee, Australia.
- 10.Geeraerd, A. H., C. H. Herremans, and J. F. Van Impe. 2000. Structural model requirements to describe microbial inactivation during a mild heat treatment. Int. J. Food Microbiol. 59:185-209. [DOI] [PubMed] [Google Scholar]
- 11.Geeraerd, A. H., V. P. Valdramidis, and J. F. Van Impe. 2005. GInaFit, a freeware tool to assess non-log-linear microbial survivor curves. Int. J. Food Microbiol. 102:95-105. (Erratum, Int. J. Food Microbiol. 110:297, 2006.) [DOI] [PubMed] [Google Scholar]
- 12.Mafart, P., O. Couvert, S. Gaillard, and I. Leguerinel. 2002. On calculating sterility in thermal preservation methods: application of the Weibull frequency distribution model. Int. J. Food Microbiol. 72:107-113. [DOI] [PubMed] [Google Scholar]
- 13.Peleg, M. 1996. Evaluation of the Fermi equation as a model of dose response curves. Appl. Microbiol. Biotechnol. 46:303-306. [Google Scholar]
- 14.Peleg, M. 1999. On calculating sterility in thermal and non-thermal preservation methods. Food Res. Int. 32:271-278. [Google Scholar]
- 15.Peleg, M. 2006. Advanced quantitative microbiology for foods and biosystems: models for predicting growth and inactivation. CRC Series in Contemporary Food Science. CRC Press, Boca Raton, FL.
- 16.Peleg, M., and M. B. Cole. 1998. Reinterpretation of microbial survival curves. Crit. Rev. Food Sci. 38:353-380. [DOI] [PubMed] [Google Scholar]
- 17.Peleg, M., M. D. Normand, and M. G. Corradini. 2005. Generating microbial survival curves during thermal processing in real time. J. Appl. Microbiol. 98:406-417. [DOI] [PubMed] [Google Scholar]
- 18.Peleg, M., M. D. Normand, and E. Dramrau. 1997. Mathematical interpretation of dose-response curves. Bull. Math. Biol. 59:747-761. [Google Scholar]
- 19.Pflug, I. J. 1987. Factors important in determining the heat process value, FT, for low-acid canned food. J. Food Prot. 50:528-533. [DOI] [PubMed] [Google Scholar]
- 20.R Development Core Team. 22 December 2008, accession date. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/.
- 21.Rodriguez, A. C., G. H. Smerage, A. A. Teixeira, and F. F. Busta. 1988. Kinetic effects of lethal temperatures on population dynamics of bacterial spores. Trans. ASAE (Am. Soc. Agric. Eng.) 31:1594-1606. [Google Scholar]
- 22.Rodriguez, A. C., G. H. Smerage, A. A. Teixeira, J. A. Lindsay, and F. F. Busta. 1992. Population model of bacterial spores for validation of dynamic thermal processes. J. Food Sci. 57:1248-1257. [Google Scholar]
- 23.Sapru, V., G. H. Smerage, A. A. Teixeira, and J. A. Lindsay. 1993. Comparison of predictive models for bacteria spore population response to sterilisation temperatures. J. Food Sci. 58:223-228. [Google Scholar]
- 24.Stumbo, C. R., J. R. Murphy, and J. Cochran. 1950. Nature of thermal death time curves for P.A. 3679 and Clostridium botulinum. Food Technol. 4:321-326. [Google Scholar]
- 25.United States Food and Drug Administration. 22 December 2008, accession date. Code of Federal Regulations 21CFR108.35, Title 21, Food and Drugs, vol. 2, chapter 1, Food and Drug Administration Department of Health and Human Services, subchapter B, Food for human consumption. http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=108.35.
- 26.Withell, E. R. 1942. The significance of the variation on shape of time-survivor curves. J. Hyg. 42:124-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong, R., G. Xie, A. E. Edmondson, and M. A. Sheard. 1999. A mathematical model for bacterial inactivation. Int. J. Food Microbiol. 46:45-55. [DOI] [PubMed] [Google Scholar]