Abstract
The potential for microbial nitrogen fixation in the anoxic methane seep sediments in a mud volcano, the number 8 Kumano Knoll, was characterized by molecular phylogenetic analyses. A total of 111 of the nifH (a gene coding a nitrogen fixation enzyme, Fe protein) clones were obtained from different depths of the core sediments, and the phylogenetic analysis of the clones indicated the genetic diversity of nifH genes. The predominant group detected (methane seep group 2), representing 74% of clonal abundance, was phylogenetically related to the nifH sequences obtained from the Methanosarcina species but was most closely related to the nifH sequences potentially derived from the anoxic methanotrophic archaea (ANME-2 archaea). The recovery of the nif gene clusters including the nifH sequences of the methane seep group 2 and the subsequent reverse transcription-PCR detection of the nifD and nifH genes strongly suggested that the genetic components of the gene clusters would be operative for the in situ assimilation of molecular nitrogen (N2) by the host microorganisms. DNA-based quantitative PCR of the archaeal 16S rRNA gene, the group-specific mcrA (a gene encoding the methyl-coenzyme M reductase α subunit) gene, and the nifD and nifH genes demonstrated the similar distribution patterns of the archaeal 16S rRNA gene, the mcrA groups c-d and e, and the nifD and nifH genes through the core sediments. These results supported the idea that the anoxic methanotrophic archaea ANME-2c could be the microorganisms hosting the nif gene clusters and could play an important role in not only the in situ carbon (methane) cycle but also the nitrogen cycle in subseafloor sediments.
In the anoxic, methane-abundant marine sediments, there are known to exist microbial ecosystems supported by a high flux of methane derived from the deeper sources of gas and hydrate forms (17, 30). The microbial anoxic oxidation of methane (AOM) in the marine sediments is considered a key process in the global carbon cycle to prevent the emission of methane into the ocean (15, 40). The microorganisms responsible for the AOM in marine sediments have been identified to be anaerobic methanotrophs (ANMEs) (14, 34). To date, marine ANMEs have been classified into three phylogenetic groups (17, 35). ANME-2 and ANME-3 are likely associated with the formation of syntrophic consortia with sulfate-reducing Deltaproteobacteria (2, 17, 19) while it is suggested that ANME-1 operates the AOM singly (35).
Although all the ANME archaea are still resistant to cultivation under laboratory culture conditions, the phylogenetic diversity, biogeographical distribution, and potential methane oxidation mechanisms of the ANMEs have been extensively studied by various culture-independent techniques (11, 12, 18). Nevertheless, little is known about what kinds of substances other than methane and sulfate support the ANMEs' activities and functions in in situ habitats (28). The nitrogen sources for the ANMEs are one of the important questions to be clarified. Recently, fluorescence in situ hybridization-secondary ion mass spectrometer (FISH-SIMS) analysis using 15N-nitrogen compounds has revealed that the ANME-2 archaea have the potential to take up ammonia and amino acids as their nitrogen sources for protein synthesis under microcosm experiment conditions (36). However, the in situ nitrogen sources and assimilation mechanisms of the ANMEs are still unclear. In methane seep sediments, the methane is likely supplied from the potential subseafloor gas and/or hydrate pools that could be accumulated by some of the physical and chemical processes associated with the geological settings, regardless of its biogenic and thermogenic origins. In these processes, it is supposed that molecular nitrogen (N2) represents a transportation behavior that is relatively similar to that of methane in terms of how gaseous components in the subseafloor fluid flow compared to the other dissolved inorganic (ammonium and nitrate) and organic nitrogen compounds. However, as the in situ concentration of the molecular nitrogen in the pore water of subseafloor core sediments is technically difficult to measure due to contamination of air during the sampling process, the biogeochemical relevance of the methane and N2 fluxes in methane-rich marine sediments has been poorly understood. In addition, even with other dissolved nitrogen sources, there have been very few studies on biogeochemical interactions of anoxic methane and nitrogen metabolisms (6, 36, 38).
Nitrogen fixation is the first step of the global nitrogen cycle and converts N2 into ammonia (NH3). But the biological N2-fixing function is limited to several groups of bacteria and methanogenic archaea (39). N2 fixation is catalyzed by a nitrogenase consisting of two components: Fe protein (nitrogenase reductase component) encoded by nifH and MoFe protein (nitrogenase component) encoded by nifDK (42). These nifHDK genes form a cluster in the known nitrogen-fixing prokaryotes (8, 39). Although the Fe protein does not directly react with nitrogen, the amino acid sequences of the Fe proteins are highly conserved among the N2-fixing prokaryotes. In addition, the phylogenetic classification of nifH is highly consistent with the prokaryotic taxonomy by 16S rRNA gene sequences (13, 31, 47). Thus, the nifH gene has been adopted as a useful marker for detection of potential nitrogen-fixing microorganisms in various environments such as oligotrophic oceans (23, 49), marine microbial mats (48), modern marine stromatolites (7), tropical sea grass beds (1), rice roots (5, 20), salt marsh sediments (21, 46), hot springs (10), marine sponges (27), and hydrothermal fields (24, 25).
In the metagenomic analysis of the anoxic methanotrophic microbial communities in the Eel River basin, novel lineages of the nifH gene that was most closely related to the nifH sequences of the Methanosarcina members were identified (37). The cellular level of FISH-SIMS analysis of the microbial communities enriched with 15N2-tracer strongly suggested that some of the ANME-2 members could have both AOM and N2-fixation functions (37). However, the distribution and abundance of the potentially N2-fixing anoxic methanotrophic archaea in the marine methane-rich environments are poorly understood. Therefore, we did not know whether N2 fixation by anoxic methanotrophs affects a global N cycle in the sediments or minor N source uptake in the cell. Knowing the distribution, abundance, and diversity of nif genes can further understanding of the global N cycle in the anoxic methane seep sediments. In this study, we conducted a molecular survey of the nifH genes in the methane seep sediments at a mud volcano, the number 8 Kumano knoll in the Kumano Basin, off shore of Japan, where the active AOM activities were expected by the extensive formation of thioautotrophic endosymbiont-hosting Calyptogena colonies on the seafloor. The phylogenetic characterization of the nifH genes in the 40-cm-deep core sediments revealed the possible existence of the N2-fixing methanotrophic archaea through the depths of the sediments. The predominant nifH phylotype was derived from the intact nifHDK gene cluster, and the genetic components of the gene cluster were expressed at considerable levels in the host microorganisms thriving in the in situ sediments. Based on the distribution and abundance of different molecular markers of the archaeal communities (16S rRNA gene, mcrA, and nifHD) in the sediments, it was determined that the ANME-2 members would be the possible N2-fixing archaeal components.
MATERIALS AND METHODS
Sampling.
A mud volcano, the number 8 Kumano Knoll, is located at 33°37.1′N, 136°33.5′E and at a water depth of 2,050 m in the Nankai Trough. The relative height from the proximal basin is 40 m, and its diameter is about 800 m. The Calyptogena colony was observed with patchy microbial mats at the rim of the mud volcano. Samplings were carried out with the manned submersible Shinkai 6500 (dive 946) during the YK06-03 cruise (May 2006) of R/V Yokosuka (JAMSTEC, Japan). The sediment cores (946C2 and 946C3) from a white microbial mat site and a reference site about 7-m away from the microbial mat, respectively, were sampled by a 50-cm-long push corer. The core sediments were subsampled by sterilized syringes at 5-cm intervals and immediately frozen in liquid nitrogen. After the cruise, samples were stored at −80°C in our laboratory.
Characterization of pore water chemistry.
Pore water was obtained from sediments within 3 h after recovery. The innermost parts of core sediments were taken immediately into 50-ml plastic syringes, and then the pore water was extracted by pressure filtration through a 0.45-μm-pore-size filter by using a stainless steel clamp at 4°C. Some chemical species are very unstable during storage and therefore require shipboard analysis (9). Silica and ammonia concentrations were determined on board by colorimetric techniques (9). Alkalinity was determined by titration with hydrochloric acid using a Gran plot estimation of the endpoint (9). Chloride concentration was determined by the Mohr titration method, with an analytical error of <0.3% (9). For other major dissolved elemental species, analyses were conducted in shore-based laboratories (29). Concentrations of sodium, calcium, and magnesium were determined by inductively coupled plasma-atomic emission spectroscopy (29), and concentrations of sulfate and nitrate were determined by ion chromatography (9). Analytical errors in these instrumental analyses are estimated at ∼5%. Compound-specific concentrations and stable carbon isotopic compositions of total CO2 (ΣCO2), methane, and ethane were measured by an isotope ratio-monitoring gas chromatograph-mass spectrometer (MAT252; Thermo Finnigan, Bremen, Germany) as previously described (45).
DNA extraction and sequencing analysis.
DNA extraction and purification were carried out from 1 g (wet weight) of the core sediments at 5-cm intervals using an UltraClean Soil DNA Isolation kit (MO BIO Laboratories, Solana Beach, CA) and a MagExtractor PCR and Gel Clean-up kit (Toyobo, Osaka, Japan).
The archaeal 16S rRNA gene, mcrA, and nifH-nifD-nifK genes were amplified from the extracted DNA by PCR using LA Taq with GC buffer (Takara Bio, Otsu, Japan). The oligonucleotide primers used in this study are summarized in Table S1 in the supplemental material. All the PCR conditions used in this study were also summarized in Table S2 in the supplemental material.
The amplified DNA fragments were purified by agarose gel electrophoresis using an UltraClean GelSpin DNA extraction kit (MO BIO Laboratories). The purified DNA fragments were ligated into the pCR2.1 vector in a TA cloning kit (Invitrogen, Carlsbad, CA). The clone libraries were constructed by transforming Escherichia coli INVαF′ (Invitrogen) with the ligation mixture, and the cloned fragments were directly sequenced by method of Sanger et al. (41).
Phylogenetic analysis.
The determined sequences were analyzed using the ARB software package (22). The trees in this study were also constructed by the neighbor-joining method using ARB software package.
RNA extraction and RT-PCR analysis.
RNA extraction from the sediments was performed by using an RNA PowerSoil total RNA isolation kit (MO BIO Laboratories). The extracted RNA (500 ng) was treated with 2 μl of 100 U/ml DNase I (Invitrogen) overnight at room temperature. Then, reverse transcription (RT) was carried out by Superscript III (Invitrogen) using random hexamers. The synthesized DNA was subjected to PCR to amplify mcrA, nifH, and methane seep group 2 nifD. The amplification conditions of RT-PCR are summarized in Table S2 in the supplemental material.
qPCR analysis.
All of the quantitative PCR (qPCR) analyses were performed by using a 7500 Real-Time PCR system (PE Applied Biosystems, Foster City, CA) using the DNA samples extracted from the sediments. All of the qPCR programs and primers used are listed in Tables S1 and S2 in the supplemental material. Primers used in these analyses were designed with the ARB software package. Quantification of both the entire prokaryotic 16S rRNA gene population and the archaeal 16S rRNA gene population was performed according to a previously described quantitative fluorescent PCR method (44). Quantification of the mcrA gene population was performed according to a quantitative fluorescent PCR method described previously (33). Other quantifications were performed by SYBR premix EX Taq (Takara bio).
Quantification of the group a-b mcrA genes was conducted by using a dilution series of 1 to 1 × 10−7 ng/ml of an mcrA clone (K8MV-C26mcrA_14) obtained from this study as a standard. Similarly, quantification of the group c-d, group e and group f mcrA genes was tested by using a dilution series of 1 to 1 × 10−7 ng/ml of the mcrA DNA clones (KM-m-1.21, KM-m-1.09, and KM-m-4.04 from Nunoura et al. [32]; KM-m-1.19 and KM-m-4.10 from Nunoura et al., [32]; and K8MV-C22mcrA-19 from this study, respectively) as the standards.
Quantification of the universal nifH gene was done by using 2 to 2 × 10−7 ng/ml of nifH DNA clones obtained from this study (K8MV-C28nifH_06 of methane seep group 1, K8MV-C22nifH_01 of methane seep group 2a, K8MV-C24nifH_04 of methane seep group 2b, K8MV-C28nifH_03 and K8MV-C28nifH_28 of methane seep group 3, and K8MV-C22nifH_05 of methane seep group 4). The DNA mixture of these clones was also utilized as the standard DNA. In addition, quantification of the group 2 of nifD genes was examined by using 1 to 1 × 10−7 ng/ml of cloned nif gene clusters obtained from this study (K8MV-C2nifHK1_08, K8MV-C2nifHK1_40, and K8MV-C2nifHK1_11). The mixture of these clones was also utilized as the standard DNA to obtain standard curves for the quantification of nifD.
The amplification patterns of all qPCRs constructed in this study are shown in Fig. S5 in the supplemental material. The detection limits of these qPCRs were almost 0.1 to 10 μg/ml.
Nucleotide sequence accession numbers.
All of the sequences obtained in this study were deposited into the DDBJ (DNA data bank of Japan) database. The sequences of the nifH-nifI1-nifI2-nifD-nifK clusters were deposited under the accession numbers AB362193 to AB362197. The sequences of the nifH genes, 16S rRNA genes, and mcrA genes obtained in this study were deposited under accession numbers AB362396 to AB362410, AB362542 to AB362552, and AB362198 to AB362208, respectively.
RESULTS
Pore water chemistry.
From the foot of a mud volcano, the number 8 Kumano Knoll, two core samples (946C2 and 946C3) were obtained from a white microbial mat site and a reference site, and the lengths of the cores were 40 and 45 cm, respectively. Immediately after the recovery of the cores, the pore water samples were extracted on board and subjected to the subsequent chemical composition and isotopic analyses (Table 1). Both of the cores consisted of clay- or silt-like sediments, and no apparent stratigraphic layer or hydrate formation was observed.
TABLE 1.
Chemical properties of pore water in the core sediments of 946C2 and 946C3
| Core site and zone depth (cm bsf) | CH4 (μmol/kg) | δ13CCΗ4 (‰ PDBa) | SO42− (mmol/kg) | NH4+ (μmol/kg) | Cl− (mmol/kg) | Ca2+ (mmol/kg) | Mg2+ (mmol/kg) | Na+ (mmol/kg) | ∑CO2 (mmol/kg) | δ13CCΟ2 (‰ PDB) | C2H6 (nM) | δ13CC2Η6 (‰ PDB) | C1/C2b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 946C2c | |||||||||||||
| 0-5 | 11.1 | −26.4 | 27.9 | 5.6 | 535 | 10.2 | 52.2 | 466 | 1.95 | −22.3 | 80.4 | −24.6 | 138 |
| 10-15 | 466.1 | −54.2 | 25.2 | 36.7 | 551 | 2.0 | 52.0 | 474 | 4.40 | −31.0 | 452 | −24.6 | 1,030 |
| 20-25 | 910.0 | −60.3 | 7.9 | 30.1 | 546 | 1.3 | 51.4 | 465 | 4.24 | −27.1 | 490 | −24.4 | 1,860 |
| 30-35 | 26.7 | −57.8 | 2.4 | 63.5 | 548 | 1.2 | 51.7 | 476 | 2.43 | −22.4 | 234 | −24.0 | 115 |
| 946C3c | |||||||||||||
| 0 | 27.8 | 7.8 | 541 | 10.4 | 53.3 | 472 | 1.35 | −2.8 | NDe | ||||
| 5-10 | 0.16 | −48.6 | 27.1 | 28.9 | 532 | 10.5 | 53.1 | 479 | 1.47 | −2.1 | ND | ||
| 15-20 | 0.11 | −43.1 | 27.9 | 36.7 | 533 | 10.3 | 52.3 | 470 | 1.04 | −2.4 | ND | ||
| 25-30 | 0.12 | −38.3 | 27.7 | 36.7 | 537 | 10.6 | 53.3 | 476 | 0.97 | −2.0 | ND | ||
| 35-40 | 0.14 | −35.8 | 26.6 | 37.9 | 537 | 10.4 | 52.9 | 470 |
PDB, PeeDee Belemnite standard.
CH4/C2H6.
Anoxic methane seep sediments. Site is located at 33°36.1′N, 136°33.31′E; depth, 2,059 m.
Reference site; located at 33°36.10′N, 136°33.31′E; depth, 2,058 m.
ND, not detected.
As expected from the existence of the seafloor microbial mat and Calyptogena colony formations, the CH4 concentration in the pore waters was much higher in the microbial mat site than in the reference site (Table 1). However, the vertical profile of CH4 concentration in the microbial mat sediments was very variable. In the shallower zones of the core (0 to 5 cm below the seafloor [bsf]), the pore water CH4 concentration was drastically decreased compared to the concentrations in the deeper zones (Table 1). This was probably due to the diffusion of CH4 from the sediments to the seawater and the activities of the sulfate-reducing anoxic methanotrophic communities in the core sediments down to 25 cm bsf. Through the depths (0 to 25 cm bsf), both the CH4 and sulfate concentrations were inversely changed, and the stable carbon isotope composition of CH4 (δ13CCH4) largely shifted to the heavier values with the decreasing depth (Table 1). On the other hand, the pore water ΣCO2 concentration was more enriched in the microbial mat sediments than in the reference sediments, and the δ13CΣCO2 in the microbial mat sediments was unusually 13C depleted through the depths compared to the δ13CΣCO2 values in the reference sediments (close to the values of the deep-seawater bicarbonate). In addition, the CH4/C2H6 (C1/C2) ratio was changed at the shallower zones of the microbial mat sediments (Table 1). It was also notable that the deepest zone of the core sediments had a much lower concentration of CH4 in the pore water than the overlying sediments (Table 1). The pore water sulfate concentration, the isotopic compositions of CH4 and ΣCO2, and the C1/C2 ratio in the deepest zone suggested the possibility of AOM linked with sulfate reduction (Table 1). Based on all the chemical data described above, the following features in the microbial mat site sediments were delineated: (i) the primary CH4 input occurred at a depth range of 15 to 30 cm bsf, (ii) CH4 was diffused to shallower and deeper zones of sediments, and (iii) the sulfate-reducing anoxic methanotrophic communities could consume the CH4 through the core sediments.
In both of the core sediments, the N2 and nitrate concentrations in the pore waters were not measured. However, the pore water nitrate concentrations in the core sediments (at a depth of approximately 40 cm) collected from a similar microbial mat site in the number 8 Kumano Knoll were below the detection limit (<1 μmol/kg) through the depths (data not shown). Thus, the pore water nitrate concentration may also be quite low in the microbial mat sediments in this study. In contrast, the ammonium was detectable in the core sediments at both the microbial mat and reference sites (Table 1). The vertical profiles of the pore water ammonium concentrations from the microbial mat site and the reference site were generally similar. In the deeper zones of the cores, 30 to 40 μmol/kg ammonium was present while it was depleted in the surface zones (Table 1). However, the ammonium was considerably enriched only in the deepest zone of the microbial mat site (Table 1).
Cloning and diversity of nifH genes.
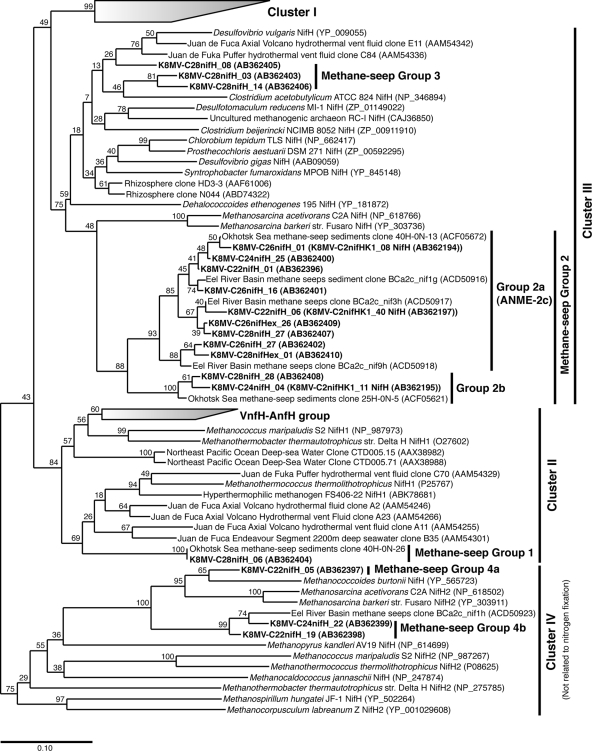
The PCR amplification of nifH genes was conducted with the extracted DNA samples from the sediments at the microbial mat site and reference site. Only from the sediments at the microbial mat site (core 946C2) were the nifH genes successfully amplified. Thus, the subsequent analyses were performed using the samples of the microbial mat site core. In total, 111 of the nifH clones were sequenced and subjected to sequence similarity analysis using the deduced amino acid sequences. We tentatively determined that the nifH clone whose amino acid sequence had <94% similarity to the sequences of all other clones represented an independent phylotype. According to this definition, 16 phylotypes were obtained through the core sediments and were compared with the previously deposited nifH sequences (Table 2). In addition, based on the phylogenetic analysis of the 16 nifH phylotypes, four representative groups of the nifH phylotypes were identified (Fig. 1), as described below.
TABLE 2.
Distribution of representative nifH gene clones obtained from DNA and cDNA in the core sediments of 946C2
| Methane seep group | Phylotype | No. of clones obtained at the indicated depth (cm bsf) from: |
Remarks | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA | cDNA | |||||||||
| 10 | 20 | 30 | 40 | 10 | 20 | 30 | 40 | |||
| 1 | K8MV-C28nifH_06 | 0 | 1 | 0 | 1 | NDa | 0 | 0 | 0 | |
| 2a | K8MV-C22nifH_01 | 12 | 2 | 3 | 4 | ND | 5 | 2 | 4 | |
| K8MV-C22nifH_06 | 2 | 3 | 0 | 1 | ND | 1 | 2 | 4 | Identical with K8MV-C2nifHK1_40 | |
| K8MV-C24nifH_25 | 0 | 2 | 2 | 0 | ND | 0 | 0 | 0 | ||
| K8MV-C26nifH_01 | 5 | 4 | 7 | 1 | ND | 12 | 14 | 9 | Identical with K8MV-C2nifHK1_08 | |
| K8MV-C26nifH_16 | 0 | 0 | 1 | 0 | ND | 0 | 0 | 0 | ||
| K8MV-C26nifH_27 | 0 | 0 | 1 | 0 | ND | 0 | 0 | 0 | ||
| K8MV-C28nifH_27 | 0 | 1 | 1 | 1 | ND | 1 | 0 | 1 | ||
| K8MV-C26nifHex_26 | ND | 0 | 1 | 0 | Detected only by RT-PCR | |||||
| K8MV-C28nifHex_01 | ND | 0 | 0 | 2 | Detected only by RT-PCR | |||||
| 2b | K8MV-C24nifH_04 | 0 | 9 | 8 | 9 | ND | 3 | 7 | 6 | Identical with K8MV-C2nifHK1_11 |
| K8MV-C28nifH_28 | 0 | 2 | 0 | 1 | ND | 0 | 0 | 0 | ||
| 3 | K8MV-C28nifH_03 | 0 | 0 | 1 | 1 | ND | 0 | 0 | 0 | |
| K8MV-C28nifH_08 | 2 | 2 | 2 | 4 | ND | 0 | 2 | 1 | ||
| K8MV-C28nifH_14 | 2 | 0 | 0 | 3 | ND | 0 | 0 | 0 | ||
| 4 | K8MV-C22nifH_05 | 4 | 3 | 0 | 0 | ND | 0 | 0 | 0 | |
| K8MV-C22nifH_19 | 2 | 0 | 0 | 0 | ND | 0 | 0 | 0 | ||
| K8MV-C24nifH_22 | 0 | 1 | 0 | 0 | ND | 0 | 0 | 0 | ||
| Total | 29 | 30 | 26 | 26 | 0 | 22 | 28 | 27 | ||
ND, not detected.
FIG. 1.
Phylogenetic tree of nifH genes including the nifH gene clones obtained from deep-sea methane-rich sediments. The tree was constructed by the neighbor-joining method using the ARB software package (22) based on deduced amino acid sequences (128 amino acid positions). The nifH gene clones obtained in this study are indicated by bold characters. The BchL sequence from Rhodobacter sphaeroides (YP_353362) was used as the outgroup of this tree. Bootstrap analysis was performed with 1,000 resampled data sets.
(i) Methane seep group 1.
Two clones (K8MV-C28nifH_06 is a representative phylotype) of methane seep group 1 were obtained from the sediments at 15 to 20 cm bsf and 35 to 40 cm bsf. These clones had the same amino acid sequences of the amplified fragments and were assigned to cluster II of the nifH gene (39). The phylotype was closely related to the nifH clone sequences (79% similarity of amino acid sequence) obtained from the Juan de Fuca hydrothermal vent fluids (24). This group of nifH sequences was likely hosted by potential methanogens within the order Methanococcales (Fig. 1) (26).
(ii) Methane seep group 2.
Methane seep group 2 is the most abundantly detected group of nifH clones from the microbial mat sediments (core 946C2) (74% of the total clones from all the depths) (Table 2 and Fig. 1). Based on the tree topology, this group could be further classified into methane seep group 2a and 2b (Fig. 1). The nifH sequences most closely related to the group of nifH genes obtained in this study were the sequences from Methanosarcina acetivorans strain C2A and Methanosarcina mazei strain Go1 (75 to 79% similarity). The phylogenetic relationship between the number 8 Kumano Knoll nifH phylotypes of the methane seep group 2 and the Methanosarcina nifH sequences indicated that they were considerably related to but also distinct from each other (Fig. 1). As observed in the methane seep group 2, nifH sequences were likely derived from the host microorganisms potentially classified into the order Methanosarcinales.
(iii) Methane seep group 3.
From the different depths of sediments, we obtained a total of 17 clones (three representative phylotypes) that could be affiliated within cluster III of the nifH gene (39) (Table 2). The amino acid sequences of the phylotypes (K8MV-C28nifH_03 and K8MV-C28nifH_14) were similar to those of the rhizosphere clones of N044 (ABD74322) (21) and HD3-3 (AAF61006) (20). The closest nifH gene of the cultivated microorganism was the one from Clostridium acetobutylicum (NP_346894). The phylotype of K8MV-C28nifH_08 was the most closely related to nifH clone C84 (AAM54336) obtained from the Juan de Fuca deep-sea hydrothermal vent environment (24) (89% identity in amino acid sequence) and the nifH of Desulfovibrio dechloracetivorans as the microorganism of known function (87%). Since all the nifH genes from the cultured microorganisms related to the methane seep group 3 were derived from anaerobic bacteria such as the Clostridia and sulfate-reducing Deltaproteobacteria, the methane seep group 3 nifH may be derived from the bacterial components of the communities in the methane seep site sediments.
(iv) Methane seep group 4.
A total of 10 clones (three representative phylotypes) that were phylogenetically affiliated with cluster IV nifH were obtained from the relatively shallower zones (0 to 20 cm bsf) of the microbial mat sediments (Table 2 and Fig. 1). These phylotypes were related to nifH sequences from methanogenic archaea such as Methanococcoides and Methanosarcina members, and the amino acid identity to the several nifH genes from the methanogens was more than 70%.
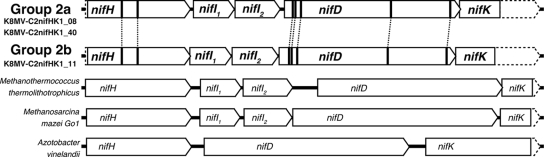
Cloning, sequencing and phylogenetic analysis of nif gene cluster (nifH-nifI1-nifI2-nifD-nifK cluster).
Since the nifH was one of the key gene components in microbial nitrogen fixation, we sought to obtain the other genetic components directly from the DNA samples of the sediments to estimate the N2-fixing potential of the in situ microbial communities. Several degenerate primers targeting the nif gene cluster including the nifD and nifK genes were constructed based on the multiply aligned sequences of various nifH, nifD, and nifK genes. Finally, a primer set of nifHfw and nifK1rv successfully produced the amplified fragments of the nif gene clusters from the sediments (see Table S2 in the supplemental material).
The nifH sequences included in the gene clusters (32 clones of the gene cluster from sediments at various depths) were sequenced. The 32 clones contained two predominant and two less abundant nifH phylotypes, all of which were 100% identical to the nifH clones classified as either methane seep group 2a or 2b obtained from the nifH clone analysis (the phylotype of K8MV-C2nifHK1_08 and K8MV-C2nifHK1_40 was group 2a and the phylotype of K8MV-C2nifHK1_11 was group 2b) (Table 2). Thus, we determined the whole sequences of these three predominant types of nif gene clusters. These clones consisted of potential nifH, nifI1, nifI2, nifD, and nifK genes. These gene structures were found in the typical N2-fixing prokaryotes having cluster II or III nifH genes (Fig. 2). The conserved amino acid residues for the [4Fe-4S] cluster of the nifH coding Fe protein and for the P and MoFe clusters of the nifD coding the MoFe protein were identified in both of the clusters, except for the residue arginine 97 of the Fe protein from K8MV-C2nifHK1_40, whose original residue was cysteine (Fig. 2). In addition, the phylogenetic analyses of the genetic components (nifI1, nifI2, and nifD) were conducted by using each of the deduced amino acid sequences (see Fig. S1 and S2 in the supplemental material). These phylogenetic analyses including the nifH tree revealed that some of the gene components of both types were related to the entities of Methanosarcina spp. (nifH and nifI2) (Fig. 1; see also Fig. S1 in the supplemental material) while some were related to the genes of the Methanococcales members (nifI1 and nifD) (see Fig. S1 and S2 in the supplemental material). These results suggested that both of the nif gene clusters were derived from the previously unidentified N2-fixing archaea phylogenetically associated with the known methanogens.
FIG. 2.
The nif gene cluster structures including clones from deep-sea methane-rich sediments. The sequences of nif genes from Methanothermococcus thermolithotrophicus (X13830), Methanosarcina mazei Gö1 (NC_003901), and Azotobacter vinelandii (M20568) were obtained from the public database. The bold lines in the genes indicate the corresponding conserved active and metal-binding sites of the enzymes.
RT-PCR analyses of nifH, nifD, and mcrA in the methane seep sediments.
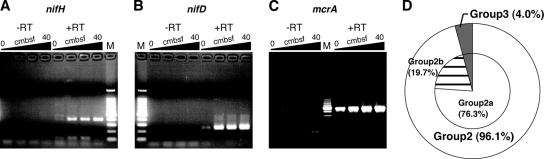
The successful amplification of various types of nifH genes and at least two types of nif gene clusters pointed to the possible expression and operation of these genes and their products in the in situ microbial communities occurring in methane-rich sediments. Thus, the expression of the nifH and nifD genes was examined by RT-PCR using the RNA samples directly extracted from the core sediments. As a control, the expression of the archaeal methanogenic and methantrophic mcrA gene (a gene encoding methyl-coenzyme M reductase α subunit) was checked using the same samples as used for the expression of nif genes. In an examination of the samples without the RT process, no positive fragment was detected in any instance of the amplification of the nifH, nifD, and mcrA genes, indicating that there was negligible DNA contamination in the samples (Fig. 3). From the cDNA samples at all depths of the core, the expression of mcrA was identified (Fig. 3C). As described below, as all the mcrA genes obtained from the methane seep sediments were phylogenetically identified as the ones hosted by the ANME, it was demonstrated that the key gene component of AOM was consistently expressed through the depths of the core sediments. In contrast, RT-PCR of the nifH and nifD genes provided the expressed products from the sediments below 10 cm bsf. Although it was not exactly quantified, the fluorescence signatures of the expressed mcrA and the nif genes in the agarose gel electrophoresis were increased with increases in the depth of the sediments.
FIG. 3.
Transcription patterns of nifH (A), group 2-specific nifD (B), and mcrA (C) at different depths of the sediment core (946C2) and the clonal phylotype composition of the expressed nifH genes at all the depths of the sediments (D). (A, B, and C) Ten microliters of PCR solution was loaded on agarose gel. M, 100-bp ladder marker. PCRs were conducted with (+) and without (−) RT. (D) In total, 96.1% of the expressed nifH genes were assigned to methane seep group 2.
The vertical profile in phylotype composition of the expressed nifH genes is shown in Table 2. The predominantly expressed nifH gene phylotypes were highly consistent with the predominant phylotypes identified in the nifH clone analysis using the extracted DNA samples (Table 2). Two new phylotypes of the nifH genes were found in the RT-PCR clone analysis (K8MV-C26nifHex26 and K8MV-C28nifHex01) (Table 2). The expressed nifH genes were dominated by methane seep group 2, particularly by group 2a, in the sediments (Fig. 3D). The expressed nifD products were also cloned and sequenced. All the expressed nifD sequences were closely related to the nifD sequences included in the nif gene cluster clones of K8MV-C2nifHK1_08 and K8MV-C2nifHK1_11. However, this was because the RT-PCR primers for the nifD gene were very specific to the sequences of nifD in the clones K8MV-C2nifHK1_08, K8MV-C2nifHK1_40, and K8MV-C2nifHK1_11, which were the only nifD sequences other than those of the known methanogens. In addition, the expression of the methane seep group 3 nifH phylotype was also detected in the deeper zones of the sediments (Table 2). Concomitant expression of the ANME mcrA and the methane seep group 2 nifH and nifD genes in the methane seep sediments also suggested the possible N2-fixing function of the ANME archaea in the deeper zones of the core sediments.
Diversity of archaeal 16S rRNA genes and mcrA genes at the Kumano number 8 mud volcano.
In order to know what kinds of archaea potentially hosted the operative nif gene components in the anoxic methane seep sediments, archaeal 16S rRNA and mcrA gene clone analyses were conducted. A total of 150 clones of the 16S rRNA genes were sequenced (approximately 940 bp) (see Table S3 and Fig. S3 in the supplemental material). At all the depths of the core sediments, the ANME (ANME-1, -2a, -2c, and -3) phylotypes dominated the archaeal 16S RNA gene communities (representing 93% clonal frequency of 150 clones) (see Table S3 and Fig. S3 in the supplemental material). Other than the ANME phylotypes, the rRNA gene sequences of the deep-sea archaeal group (marine benthic group B) (43), miscellaneous crenarchaeotic group (16), and marine benthic group D were detected as minor components in the deeper zones of the sediments, and marine group I phylotypes were found in the surface of the sediments (see Table S3 and Fig. S3 in the supplemental material). None of the rRNA gene sequences related to the known N2-fixing methanogens was observed at any depths of the sediments (see Table S3 in the supplemental material).
Predominance of anoxic methanotrophic archaeal phylotypes was also demonstrated by the mcrA clone analysis (see Table S4 and Fig. S4 in the supplemental material). A total of 67 clones were sequenced, and all the mcrA sequences were associated with potential ANME mcrA genes (see Fig. S4 in the supplemental material). The most abundantly obtained mcrA phylotypes were classified into group c-d (84% clonal frequency of the total) (11). Other phylogenetic groups of mcrA such as the group a-b (11), group e (11), and group f (19) represent 3%, 9%, and 4% clonal abundance of 67 clones, respectively (see Table S4 in the supplemental material).
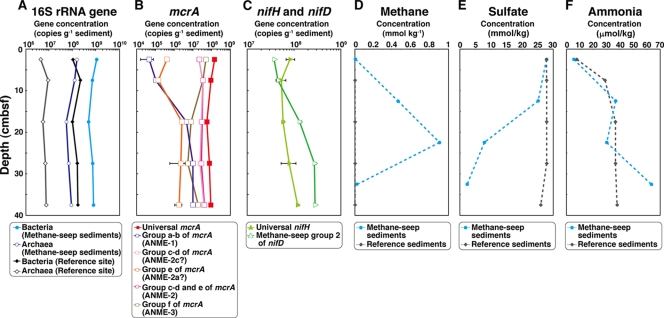
qPCR analyses.
To estimate the depth-dependent distribution and abundance of prokaryotic and archaeal 16S rRNA genes, archaeal nifH and nifD genes, and group-specific mcrA phylotypes, qPCR analyses were conducted with the DNA samples extracted from the core sediments (Fig. 4). The qPCR methods for the nifH and nifD genes were newly established in this study, and the qPCR techniques for the group-specific mcrA phylotypes were modified from a previously reported method (31) to detect the various mcrA phylotypes obtained in this study. The newly developed techniques provided sensitive and confident quantification statistics when the representative mcrA, nifH, and nifD clones found in this study were used as standards (data not shown). For the qPCR for prokaryotic and archaeal 16S rRNA genes, not only the methane seep sediments but also the reference core sediments were examined (Fig. 4).
FIG. 4.
Depth profiles of numbers of the entire microbial and archaeal 16S rRNA gene population, mcrA genes, and nifH and group 2-specific nifD genes in the DNA assemblages and the methane, sulfate, and ammonia concentrations in the sediment core(s). (A) The numbers of the entire microbial 16S rRNA gene population and archaeal 16S rRNA genes in the methane-rich sediment core and reference sediment core are shown, as indicated. Open symbols indicate the proportion of archaeal 16S rRNA genes in the microbial 16S rRNA gene population in the both cores. (B) Red filled squares indicate the number of all the groups (universal) of mcrA genes in the methane-rich sediment core. Numbers of mcrA genes by group are as identified on the figure. (C) The numbers of universal nifH genes and group 2-specific nifD genes in the methane seep sediment core are indicated as shown. (D, E, and F) Data from the methane-rich sediment core and the reference core are shown, as indicated on the figure.
The number of the prokaryotic 16S rRNA genes detected was about 1 order of magnitude higher in the methane seep sediments and than in the reference sediments while the difference in the abundance of archaeal 16S rRNA genes was greater (2 orders of magnitude) than that in the abundance of universal 16S rRNA genes (Fig. 4A). However, the numbers significantly changed in either case through the depths (Fig. 4A). These results indicated that the archaeal rRNA gene populations (potentially the archaeal cellular populations) were enriched in the methane-rich sediments.
In addition, the mcrA genes of the entire microbial population were also consistently distributed at all the depths of the methane seep sediments (Fig. 4B). In the shallower zones, a little increase in the mcrA archaeal genes was observed (Fig. 4B). Among the depth profile patterns of various group-specific mcrA genes, the profile of the group f mcrA gene was very similar to the profile of the mcrA genes of the entire microbial population in the near-surface zones, and the group f mcrA was the most abundant mcrA component in the shallower zones (down to ∼10 cm bsf) (Fig. 4B). However, below about 10 cm bsf, the most abundant mcrA population became group c-d mcrA (Fig. 4B). The number of group c-d mcrA genes was very consistent through the depths (Fig. 4B). In contrast, the numbers of the group a-b and group e mcrA genes were highly depleted in the surface zones (Fig. 4B).
The depth profiles of the various groups of mcrA genes provided an important insight to elucidating the relevance between the ANME 16S rRNA gene and mcrA phylotypes. It is not yet clear which of the ANME groups (ANME-2a, -2b, -2c, or -3, based on the 16S rRNA gene phylotypes) hosts a specific mcrA phylotype (mcrA group a-b, c-d, e, or f). In the clone analysis and the qPCR experiment of the archaeal 16S rRNA genes, the ANME-3 members were always enriched in the methane seep sediments near the surface while the ANME-2c members showed relatively consistent abundance through the depths (Table 2 and Fig. 4B). The similarity between the depth-dependent abundance profiles of ANME-3 and mcrA group f phylotypes or of ANME-2c and mcrA group c-d phylotypes may specify the previously unidentified link between the host methanotrophic archaea and their mcrA phylotypes.
On the other hand, the distribution and abundance of the nifH and nifD genes were relatively constant through the depths (Fig. 4C) although it was shown by the RT-PCR experiment that the expression levels of both genes were increased with increasing depths of the core (Fig. 3). The numbers of the nifH and nifD genes were also relatively comparable for the archaeal 16S rRNA genes and the group c-d mcrA.
DISCUSSION
In this study, we obtained multiple lines of evidences that the anoxic methanotrophic microbial communities coupled with sulfate reduction were functionally active and that the potential N2 fixation could be operated by some of the ANME components in the methane seep sediments of the number 8 Kumano Knoll, the Nankai Trough. Based on the quantitative estimation of archaeal rRNA genes, group-specific mcrA genes, and the methane seep group 2 of nifH-nifD genes in the DNA assemblages obtained from the methane seep sediments, it was suggested that the methane seep group 2 of nifH-nifD components could be hosted by the ANME members that had been classified into the ANME-2c as the 16S rRNA gene phylogroup and were probably linked with the of the mcrA group c-d phylogroup.
By the metagenomic analysis and the FISH-SIMS analysis using a 15N2 tracer of the anoxic methane-oxidizing microbial communities in the Eel River basin, Pernthaler et al. (37) have suggested that some of the ANME-2c members were capable of operating both AOM and N2 fixation in the in situ sediments. The anoxic methane-oxidizing consortia magnetically separated by using ANME-2c-specific oligonucleotide probes and enriched with 15N in the cellular nitrogen possessed the archaeal nifH sequences named BCa2c-nif1g, BCa2c-nif3h, and BCa2c-nif9h (37). These sequences were closely related to the nifH clones of methane seep group 2a (80 to 90% identity) obtained from the methane seep sediments in the number 8 Kumano Knoll (Fig. 1). As Pernthaler et al. show that ANME-2 takes up 15N2 and that nifH genes were cloned from ANME-2 consortia, the methane seep group 2 of nifH genes (or nifHIDK clusters) are hosted by some members of the ANME-2c. In addition, recently several environmental nifH gene clones that are closely related to our methane seep group 2 nifH gene sequences have been obtained from the deep-sea methane seep sediments in the Sea of Okhotsk (4). Since it has been known that the ANME-2c members are one of the predominant ANME groups distributed in global marine sedimentary environments (17), the N2-fixing potentials of the ANME-2c members would provide a great insight into understanding the biogeochemical nitrogen cycle in the global marine sedimentary environment. N2 fixation by ANME-2 may also support the syntrophic relationship between ANME-2 and sulfate-reducing Deltaproteobacteria.
Based on our results found in the methane seep core sediments, it is also suggested that not all the ANME-2c members would adopt the N2 fixation as their primary nitrogen assimilation mechanism (Fig. 3A and B and Fig. 4). In fact, Orphan et al. (36) have recently demonstrated by using FISH-SIMS analysis that the ANME-2 archaea are able to assimilate ammonia and amino acids as the nitrogen sources for protein synthesis. It remains very uncertain whether the N2 fixation capability of ANME-2c is constrained by the genetic level (e.g., the incidence of the genetic components such as the nifHIDK gene cluster in the genomes), by the expression level (e.g., regulation mechanisms of transcription and expression of the genetic components), or by both with respect to their alternative nitrogen anabolisms. In this study, the qPCR estimation of the nifH-nifD genes demonstrated that the abundance of methane seep group 2 nifH-nifD genes was relatively constant through the depth of the core (Fig. 4C). The depth-dependent distribution pattern and copy number were comparable to those of the archaeal 16S rRNA genes and the group c-d of mcrA (Fig. 4B). Nevertheless, semiquantitative analysis of nifH-nifD expression by RT-PCR indicated that the methane seep group 2 nifH-nifD genes were more abundantly expressed in the deeper zones of the core sediments than near the seafloor surface although the archaeal methanotrophic mcrA genes were relatively constantly expressed at all depths (Fig. 3). These results suggest that most of the ANME-2c members may have the potential genetic components of N2 fixation but could regulate the expression and operation of the N2-fixing capability. In the pore water chemistry analysis, the deepest zone of the sediments represented a considerable enrichment of ammonium (Table 1 and Fig. 4F). The qPCR and the RT-PCR estimations also pointed to the highest abundant occurrence and expression of the methane seep group 2 nifH-nifD genes in the deepest core sediments (Fig. 3 and 4C). The enriched amount of ammonium in the pore water of the deepest core sediments may be accumulated by functions of the actively N2-fixing ANME-2c populations.
Other than the methane seep group 2, we also obtained several different phylogenetic groups of nifH genes from the methane seep sediments (Table 2 and Fig. 1). The methane seep group 1 nifH genes were related to cluster II of the nifH genes (39). The phylogenetic analysis suggests that this group of nifH sequences could be hosted by potential methanogens within the Methanococcales (Fig. 1) (24, 26). However, the downstream region of this group of nifH genes was not identified from any of the DNA samples in the core sediments, and further clarification was unsuccessful. We also obtained nifH sequences phylogenetically affiliated with cluster III of the nifH genes (39) consisting of the nifH genes from the Firmicutes, Clorobi, and some of the Deltaproteobacteria. This group of nifH sequences was retrieved from the DNA and even cDNA samples taken from the deeper zones of the methane seep sediments. Thus, the host bacteria may be involved in N2 fixation together with the N2-fixing ANME-2c members. Since this group of nifH gene sequences has also been detected from the anoxic methane-oxidizing consortia magnetically separated by using ANME-2c-specific oligonucleotide probes (37), both the microbial populations in the anoxic methane-oxidizing interdomains (archaeal-bacterial) consortia may be capable of assimilating N2 as their own nitrogen sources. The cluster IV nifH genes are widely found in many methanogens and even in the methanogens that cannot fix N2. Recent investigations have suggested that cluster IV of nifH is not associated with the N2-fixing function in the methanogens and may be involved in the biosynthesis of factor 430 (3, 26). In the hyperthermophilic N2-fixing Methanocaldococcus sp. strain FS406-22, only cluster II of nifH was expressed in the cells grown under the N2-fixing growth conditions (26). Thus, cluster IV of nifH obtained from the methane-rich sediments may be independent of the N2-fixing potential of the in situ microbial communities.
In this study, the potential for microbial nitrogen fixation in the anoxic methane seep sediments in the number 8 Kumano Knoll was explored, and the significance of the ANME-2c archaea in in situ N2 fixation was clarified. The emerging patterns of different functional genes indicated that the abundance of different groups of ANME archaea is variable at different depths of the sediments, which might be linked with the emerging patterns of different ecophysiological functions including carbon and nitrogen metabolisms in the methane seep sediments. However, it is still completely unknown how much the N2 fixation of ANME communities supports the biogeochemical nitrogen requirement of the community activities in the in situ environments. A detailed geochemical characterization will be required for not only carbon compounds but also nitrogen compounds such as N2, nitrate, nitrous oxide, ammonium, and organic nitrogens in the pore water of the deep-sea sedimentary environments. In addition, compound-specific stable isotope characterization and tracer experiments using 15N-labeled substrates will be conducted with a combination of various molecular ecological techniques.
Supplementary Material
Acknowledgments
We are grateful to the crews of R/V Yokosuka and the operation team of DSV Shinkai 6500 for helping us to collect samples.
Part of this study was supported by a Grant-in-Aid for Scientific Research (no. 19880041) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 25 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bagwell, C. E., J. R. La Rocque, G. W. Smith, S. W. Polson, M. J. Friez, J. W. Longshore, and C. R. Lovell. 2002. Molecular diversity of diazotrophs in oligotrophic tropical seagrass bed communities. FEMS Microbiol. Ecol. 39:113-119. [DOI] [PubMed] [Google Scholar]
- 2.Boetius, A., K. Ravenschlag, C. J. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 3.Christopher, R. S., S. Lahiri, J. Raymond, L. V. Herbulis, B. Mukhophadhyay, and R. E. Blankenship. 2007. Expression and association of group IV nitrogenase NifD and NifH homologs in the non-nitrogen-fixing archaeon Methanocaldococcus jannaschii. J. Bacteriol. 189:7392-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang, H., X. Luan, J. Zhao, and J. Li. 2009. Diverse and novel nifH and nifH-like gene sequences in the deep-sea methane-seep sediments of the Okhotsk Sea. Appl. Environ. Microbiol. 75:2238-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demba Diallo, M., B. Reinhold-Hurek, and T. Hurek. 2008. Evaluation of PCR primers for universal nifH gene targeting and for assessment of transcribed nifH pools in roots of Oryza longistaminata with and without low nitrogen input. FEMS Microbiol. Ecol. 65:220-228. [DOI] [PubMed] [Google Scholar]
- 6.Ettwig, K. F., S. Shima, K. T. Van De Pas-Schoonen, J. Kahnt, M. H. Medema, H. J. Op den Camp, M. S. Jetten, and M. Strous. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10:3164-3173. [DOI] [PubMed] [Google Scholar]
- 7.Falcón, L. I., R. Cerritos, L. E. Eguiarte, and V. Souza. 2007. Nitrogen fixation in microbial mat and stromatolite communities from Cuatro Cienegas, Mexico. Microb. Ecol. 54:363-373. [DOI] [PubMed] [Google Scholar]
- 8.Fani, R., R. Gallo, and P. Liò. 2000. Molecular evolution of nitrogen fixation: the evolutionary history of the nifD, nifK, nifE, and nifN genes. J. Mol. Evol. 51:1-11. [DOI] [PubMed] [Google Scholar]
- 9.Gieskes, J. M., T. Shaw, T. Brown, A. Sturz, and A. Campbell. 1991. Chemical methods for interstitial water analysis aboard Joides Resolution. Ocean Drilling Program technical note 15. Texas A&M University, College Station, TX. http://www-odp.tamu.edu/publications/tnotes/tn15/f_chem1.htm.
- 10.Hall, J. R., K. R. Mitchell, O. Jackson-Weaver, A. S. Kooser, B. R. Cron, L. J. Crossey, and C. D. Takacs-Vesbach. 2008. Molecular characterization of the diversity and distribution of a thermal spring microbial community by using rRNA and metabolic genes. Appl. Environ. Microbiol. 74:4910-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 311:1457-1462. [DOI] [PubMed] [Google Scholar]
- 13.Hennecke, H., K. Kaluza, B. Thony, M. Fuhrmann, W. Ludwig, and E. Stackebrandt. 1985. Concurrent evolution of nitrogenase genes and 16S rRNA in Rhizobium species and other nitrogen fixing bacteria. Arch. Microbiol. 142:342-348. [Google Scholar]
- 14.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 15.Hinrichs, K. U., R. E. Summons, V. Orphan, S. P. Sylva, and J. M. Hayes. 2000. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 31:1685-1701. [Google Scholar]
- 16.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knittel, K., T. Lösekann, A. Boetius, R. Kort, and R. Amann. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krüger, M., A. Meyerdierks, F. O. Glöckner, R. Amann, F. Widdel, M. Kube, R. Reinhardt, J. Kahnt, R. Böcher, R. K. Thauer, and S. Shima. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878-888. [DOI] [PubMed] [Google Scholar]
- 19.Lösekann, T., K. Knittel, T. Nadalig, B. Fuchs, H. Niemann, A. Boetius, and R. Amann. 2007. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73:3348-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lovell, C. R., Y. M. Piceno, J. M. Quattro, and C. E. Bagwell. 2000. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66:3814-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovell, C. R., P. V. Decker, C. E. Bagwell, S. Thompson, and G. Y. Matsui. 2008. Analysis of a diverse assemblage of diazotrophic bacteria from Spartina alterniflora using DGGE and clone library screening. J. Microbiol. Methods 73:160-171. [DOI] [PubMed] [Google Scholar]
- 22.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Man-Aharonovich, D., N. Kress, E. B. Zeev, I. Berman-Frank, and O. Béjà. 2007. Molecular ecology of nifH genes and transcripts in the eastern Mediterranean Sea. Environ. Microbiol. 9:2354-2363. [DOI] [PubMed] [Google Scholar]
- 24.Mehta, M. P., D. A. Butterfield, and J. A. Baross. 2003. Phylogenetic diversity of nitrogenase (nifH) genes in deep-sea and hydrothermal vent environments of the Juan de Fuca ridge. Appl. Environ. Microbiol. 69:960-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta, M. P., J. A. Huber, and J. A. Baross. 2005. Incidence of novel and potentially archaeal nitrogenase genes in the deep Northeast Pacific Ocean. Environ. Microbiol. 7:1525-1534. [DOI] [PubMed] [Google Scholar]
- 26.Mehta, M. P., and J. A. Baross. 2006. Nitrogen fixation at 92°C by a hydrothermal vent archaeon. Science 314:1783-1786. [DOI] [PubMed] [Google Scholar]
- 27.Mohamed, N. M., A. S. Colman, Y. Tal, and R. T. Hill. 2008. Diversity and expression of nitrogen fixation genes in bacterial symbionts of marine sponges. Environ. Microbiol. 10:2910-2921. [DOI] [PubMed] [Google Scholar]
- 28.Moran, J. J., E. J. Beal, J. M. Vrentas, V. J. Orphan, K. H. Freeman, and C. H. House. 2008. Methyl sulfides as intermediates in the anaerobic oxidation of methane. Environ. Microbiol. 10:162-173. [DOI] [PubMed] [Google Scholar]
- 29.Murray, R. W., D. J. Miller, and K. A. Kryc. 2000. Analysis of major and trace elements in rocks, sediments, and interstitial waters by inductively coupled plasma-atomic emission spectrometry. Ocean Drilling Program technical note 29. Texas A&M University, College Station, TX. http://www-odp.tamu.edu/publications/tnotes/tn29/TNOTE_29.PDF.
- 30.Nauhaus, K., A. Boetius, M. Krüger, and F. Widdel. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 31.Normand, P., and J. Bousquet. 1989. Phylogeny of nitrogenase sequences in Frankia and other nitrogen-fixing microorganisms. J. Mol. Evol. 29:436-447. [DOI] [PubMed] [Google Scholar]
- 32.Nunoura, T., H. Oida, T. Toki, J. Ashi, K. Takai, and K. Horikoshi. 2006. Quantification of mcrA by quantitative fluorescent PCR in sediments from methane seep of the Nankai Trough. FEMS Microbiol. Ecol. 57:149-157. [DOI] [PubMed] [Google Scholar]
- 33.Nunoura, T., H. Oida, J. Miyazaki, A. Miyashita, H. Imachi, and K. Takai. 2008. Quantification of mcrA by fluorescent PCR in methanogenic and anaerobic methanotrophic microbial communities. FEMS Microbiol. Ecol. 64:240-247. [DOI] [PubMed] [Google Scholar]
- 34.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 35.Orphan, V. J., C. H. House, K. U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orphan, V. J., K. A. Turk, A. M. Green, and C. H. House. 2009. Patterns of 15N assimilation and growth of methanotrophic ANME-2 archaea and sulfate-reducing bacteria within structured syntrophic consortia revealed by FISH-SIMS. Environ. Microbiol. 11:1777-1791. [DOI] [PubMed] [Google Scholar]
- 37.Pernthaler, A., A. E. Dekas, C. T. Brown, S. K. Goffredi, T. Embaye, and V. J. Orphan. 2008. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc. Natl. Acad. Sci. USA 105:7052-7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raghoebarsing, A. A., P. A. Pol, K. T. Van De Pas-Schoonen, A. J. Smolders, K. F. Ettwig, W. I. Rijpstra, S. Schouten, J. S. Damsté, H. J. Op den Camp, M. S. Jetten, and M. Strous. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918-921. [DOI] [PubMed] [Google Scholar]
- 39.Raymond, J., J. L. Siefert, C. R. Staples, and R. E. Blankenship. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 21:541-554. [DOI] [PubMed] [Google Scholar]
- 40.Reeburgh, W. S. 1996. ‘Soft spots’ in the global methane budget, p. 334-342. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 41.Sanger, F., E. F. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin, H., C. Kisker, J. L. Schlessman, J. B. Howard, and D. C. Rees. 1997. Structure of ADP·AIF4−-stabilized nitrogenase complex and its implications for signal transduction. Nature 387:370-376. [DOI] [PubMed] [Google Scholar]
- 43.Takai, K., and K. Horikoshi. 1999. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152:1285-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takai, K., and K. Horikoshi. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66:5066-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsunogai, U., N. Yoshida, J. Ishibashi, and T. Gamo. 2000. Carbon isotopic distribution of methane in deep-sea hydrothermal plume, Myojin Knoll Caldera, Izu-Bonin arc: implications for microbial methane oxidation in the oceans and applications to heat flux estimation. Geochim. Cosmochim. Acta 64:2439-2452. [Google Scholar]
- 46.Welsh, A., D. J. Burke, and D. Hahn. 2007. Analysis of nitrogen-fixing members of the ɛ subclass of Proteobacteria in salt marsh sediments. Appl. Environ. Microbiol. 73:7747-7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young, J. P. W. 1992. Phylogenetic classification of nitrogen-fixing organisms, p. 43-86. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, NY.
- 48.Zehr, J. P., M. T. Mellon, S. Braun, W. Litaker, T. Steppe, and H. W. Paerl. 1995. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl. Environ. Microbiol. 61:2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.