Abstract
Rice paddy soil has been shown to have strong denitrifying activity. However, the microbial populations responsible for nitrate respiration and denitrification have not been well characterized. In this study, we performed a clone library analysis of >1,000 clones of the nearly full-length 16S rRNA gene to characterize bacterial community structure in rice paddy soil. We also identified potential key players in nitrate respiration and denitrification by comparing the community structures of soils with strong denitrifying activity to those of soils without denitrifying activity. Clone library analysis showed that bacteria belonging to the phylum Firmicutes, including a unique Symbiobacterium clade, dominated the clones obtained in this study. Using the template match method, several operational taxonomic units (OTUs), most belonging to the orders Burkholderiales and Rhodocyclales, were identified as OTUs that were specifically enriched in the sample with strong denitrifying activity. Almost one-half of these OTUs were classified in the genus Herbaspirillum and appeared >10-fold more frequently in the soils with strong denitrifying activity than in the soils without denitrifying activity. Therefore, OTUs related to Herbaspirillum are potential key players in nitrate respiration and denitrification under the conditions used.
Rice is one of the most important agronomic plants in the world (20). More than 135 million ha are used for rice cultivation worldwide, 88% of which consists of paddy fields (i.e., flooded fields) (16). Since rice paddy soil has limited available oxygen, various anaerobic biochemical processes can occur, including methane production, Mn4+ and Fe3+ reduction, nitrate respiration, and denitrification.
Denitrification is a microbial respiratory process during which soluble nitrogen oxides (NO3− and NO2−) are reduced to gaseous products (NO, N2O, and N2) (14, 43). Reduction of nitrate (NO3−) to nitrite (NO2−) is part of the denitrification process; however, this reaction can also be performed by nondenitrifiers. Reduction of nitrate to nitrite as an end product is called nitrate respiration (43). The emission of N2O from rice paddy soils is less than that from upland crop fields (2), which is probably due to complete nitrate-nitrite reduction to N2, since rice paddy soil is known to have strong denitrifying activity (28). However, the microbes responsible for denitrification in rice paddy soil are not well known.
Denitrifying ability is sporadically distributed among taxonomically diverse groups of bacteria, as well as some archaea and fungi (14, 33, 43). Therefore, it is difficult to identify denitrifying organisms based only on their 16S rRNA gene sequences (33). However, culture-independent 16S rRNA gene analysis can be used to identify microbial populations responsive to denitrification-inducing conditions if they are properly differentiated from background populations. The 16S rRNA gene can provide taxonomic information about organisms which cannot be obtained from analyses targeting nitrite reductase genes (nirS and nirK) alone (34).
One approach to differentiate functionally active populations from background populations is to use stable-isotope probing (SIP) (35). SIP was previously used to identify succinate-assimilating bacterial populations under denitrifying conditions in rice paddy soil, using nitrate and succinate as the electron acceptor and donor, respectively (37). Although SIP analysis can provide solid evidence that links function with taxonomy, it requires assimilation of isotopically labeled substrates. This may limit the application of SIP in studies of dissimilatory processes, such as nitrate respiration and denitrification. For example, previous SIP studies targeted bacteria assimilating 13C-labeled acetate, methanol, or succinate under denitrifying conditions (13, 30, 37).
Another approach is to detect specifically enriched microbial populations under certain conditions by comparative analysis of 16S rRNA gene sequences (9). This approach does not necessarily require addition of isotopically labeled substrates and therefore has the potential to identify microbes performing dissimilatory processes. Furthermore, the community structure of the total population can also be elucidated in this manner (10, 25, 36). However, the usefulness of comparative analysis of 16S rRNA gene sequences has not been thoroughly tested. In addition, this approach has not been used to study nitrate respirators and denitrifiers.
Consequently, the objectives of this study were (i) to characterize the soil bacterial population in rice paddy soil by clone library analysis of >1,000 clones of the nearly full-length 16S rRNA gene and (ii) to identify active bacterial populations under denitrification-inducing conditions by comparing clone libraries.
MATERIALS AND METHODS
Soil microcosms.
Soil samples were obtained from the top 10 cm of a rice paddy field at the Field Production Science Center, Graduate School of Agricultural and Life Sciences, The University of Tokyo (Nishi-Tokyo, Tokyo, Japan) on 8 September 2004. The chemical and physical characteristics of the soil samples have been described elsewhere (37). A previously established reproducible soil microcosm (37) was used in this study. Briefly, 1 g of air-dried soil was placed in a 10-ml glass serum vial (Nichiden-Rika Glass, Kobe, Japan) and submerged in sterile water for 1 week at 30°C (preincubation). After preincubation, 1.8 ml of the upper soil-free water was removed, and 0.1 mg nitrate N and/or 0.5 mg succinate C was added. The vial was tightly sealed using a rubber cap, the air phase was replaced with Ar-C2H2 (90:10) gas, and the vial was incubated at 30°C for 24 h. Five samples were prepared. One sample was incubated with added nitrate and succinate (TSNS sample), one sample was incubated with only added nitrate (TSNI sample), one sample was incubated with only added succinate (TSSU sample), one sample was incubated with no substrates added (TSCO sample), and one sample was not supplemented with any substrate and was not incubated (TSBA sample). Incubation with nitrate and succinate (TSNS sample) was previously shown to greatly enhance denitrification activity (37). Previous studies also showed that succinate could be used by various denitrifiers, whereas there was little utilization of succinate for other functions, such as fermentation and dissimilatory reduction of nitrate to ammonium (DNRA), under the conditions used (37, 38).
Fifteen replicate vials were prepared for each sample. Three vials each were used for (i) DNA extraction, (ii) quantification of CO2, CH4, and N2O gases, as well as anions (NO3−, NO2−, SO42−, and succinate), (iii) quantification of NH4+, (iv) Fe2+ extraction, and (v) Mn2+ extraction (see below).
Soil chemical properties.
After 24 h of incubation, CO2, CH4, and N2O that accumulated in the headspace of the vials were quantified by gas chromatography, as described previously (37). Anions, such as NO3−, NO2−, SO42−, and succinate, were extracted from soil with 5 ml water and quantified by high-performance liquid chromatography (37). An IC-Pak anion column (4.6 by 50 mm; Waters, Milford, MA) was used to separate NO3−, NO2−, and SO42−, and an IC-Pak ion exclusion column (78 by 50 mm; Waters) was used to separate low-molecular-weight carboxylic acids, such as succinate (37). The ammonium ion (NH4+) was extracted from soil by vigorously shaking the soil with 5 ml of a 2 M KCl solution for 30 min, and the NH4+ concentration was measured using a colorimetric method with indophenol blue, as described elsewhere (26). Reduced forms of metals (Fe2+ and Mn2+) were extracted from soil by shaking the soil anaerobically (with Ar headspace gas) with a 1 M ammonium acetate solution (pH 3 for Fe2+ extraction and pH 7 for Mn2+ extraction) for 1 h (7, 42). Atomic absorption was used to quantify manganese (7), while Fe2+ was measured colorimetrically (21).
DNA extraction.
DNA was extracted from the soil using an ISOIL kit for bead beating with 0.8 g of soil as the starting amount. Bead beating was performed using a FastPrep FP120 bead beater (Qbiogene, Carlsbad, CA) at a speed of 6 ms−1 for 90 s. Extracted DNA was purified further using a PowerClean DNA clean-up kit (MoBio Laboratories) to remove PCR inhibitors as much as possible. DNA was extracted from the replicate vials and purified. While denaturing gradient gel electrophoresis (DGGE) and quantitative PCR (qPCR) were performed with replicate DNA samples, clone library analysis was done with pooled DNA samples.
PCR conditions.
For clone library analysis, a nearly full-length bacterial 16S rRNA gene was amplified using modified primers 27F and 1492R (44). For DGGE analysis, a partial 16S rRNA gene (V3 region) was amplified using primers 357F-GC and 520R (27). The reaction mixture (50 μl) contained 1× Ex Taq buffer (Takara Bio, Otsu, Japan), 0.2 μM of each primer, 0.2 mM of each deoxynucleoside triphosphate, 1 U of Ex Taq DNA polymerase (Takara Bio), and 1 μl of DNA template. The PCR was performed using a T1 thermocycler (Biometra, Goettingen, Germany) and the following conditions: initial annealing at 96°C for 5 min, followed by 20 cycles of 96°C for 30 s, 50°C (58°C for DGGE analysis) for 20 s, and 68°C for 90 s (72°C for 30 s for DGGE analysis). After a final extension at 72°C for 10 min, the PCR mixtures were stored at 4°C. The sizes of the final PCR products were confirmed by agarose gel electrophoresis.
DGGE analysis.
DGGE was performed to compare the bacterial population structures for the five samples. Aliquots (500 ng) of the PCR products were loaded onto a 6.5% polyacrylamide gel. After electrophoresis performed as described elsewhere (27), DNA bands unique to the TSNS sample were excised from the gel using a razor blade. DNA was eluted as described previously (37), and 1 μl of supernatant was used as a template for PCR with primers 357F and 520R (27), as described above. Four clones per band were sequenced as described previously (31).
qPCR.
qPCR with primers 357F and 520R was performed to determine total bacterial 16S rRNA gene copy numbers as described previously (12). PCRs were performed using the StepOne real-time PCR system (Applied Biosystems, Foster City, CA). Plasmids containing 16S rRNA genes from Cupriavidus metallidurans JCM 21315T were used to generate standard curves (105 to 1011 copies per reaction mixture). Each reaction mixture (20 μl) contained 1× Power SYBR green PCR master mixture (Applied Biosystems), 0.2 μM of each primer, 0.5 μg μl−1 bovine serum albumin (Wako Pure Chemical Industries, Osaka, Japan), and 2 μl DNA template. The reactions were performed under the following conditions: initial annealing at 95°C for 15 min, followed by 40 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, as described previously (12). Amplification of products that were the correct size was confirmed by dissociation curve analysis and agarose gel electrophoresis.
Cloning and sequencing.
PCR products were ligated with pCR-4-TOPO vectors (Invitrogen, Carlsbad, CA) and transformed into competent Escherichia coli DH12S cells (Invitrogen) using a TOPO-TA cloning kit (Invitrogen) to construct 16S rRNA gene libraries for the five samples. More than 1,000 colonies from each clone library were randomly isolated using the GeneTac G3 picking system (PerkinElmer, Waltham, MA). Colony PCR was performed using primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) to amplify the 16S rRNA insert. PCR products were treated with exonuclease I and shrimp alkaline phosphatase (GE Healthcare, Uppsala, Sweden), and both ends of the DNA were sequenced using a BigDye Terminator v3.1 kit (Applied Biosystems) with primers T7 (5′-TAATACGACTCACTATAGGG-3′) and T3 (5′-AATTAACCCTCACTAAAGGG-3′). The 357F primer was also used for further sequencing of the middle region of the 16S rRNA gene. Sequencing products were cleaned by ethanol precipitation and sequenced with automated ABI 3730xl capillary sequencers (Applied Biosystems).
Phylogenetic and statistical analyses.
Three reads obtained by sequencing with primers T7, T3, and 357F were assembled with the Phred-Phrap program (11) for each 16S rRNA gene to obtain high-quality sequence data. After removal of vector and primer sequences, clone sequences shorter than 1,300 bp were removed from the clone libraries. Chimeric sequences identified using the Mallard program (5) with a 99.9% cutoff were also removed from the libraries. Taxonomic assignment of the 16S rRNA gene sequences was conducted using Naïve Bayesian Classifier, version 2.0 (46). Nucleotide sequences of multiple clones were aligned using ClustalW. Aligned sequences were used to generate neighbor-joining trees. Weighted UniFrac (23, 24) was run using the resulting trees to compare community structures for the five samples.
Operational taxonomic units (OTUs) were calculated using DOTUR (39), and shared OTUs and similarities among the libraries were analyzed using SONS (40). To identify OTUs specifically enriched in some samples, we used the template match method (17, 32) with the genefilter package of R software, version 2.8.0 (http://www.r-project.org/). OTUs with a similarity index of <0.10 with the template pattern were defined as specifically enriched OTUs (17); however, singletons (i.e., OTUs containing only one clone) were removed from the list to minimize false positives, as singletons might have appeared by chance and not due to specific enrichment in response to substrate additions.
All other statistical analyses were performed using R software, version 2.8.0. The digitalized DGGE banding profiles were aligned for samples using a complete linkage clustering algorithm (17) and were used to perform principal component analysis (PCA). The chi-square goodness-of-fit test was used to examine differences in the proportion of clones affiliated with several phyla in the libraries. Analysis of variance and Tukey's honestly significant difference tests were performed to examine soil chemical properties and qPCR results.
Nucleotide sequence accession numbers.
The nonredundant nucleotide sequences of the 16S rRNA genes used in this study (OTUs with 100% similarity) from each library and the sequences of the DGGE bands determined in this study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers AB486034 to AB488390 and AB520689 to AB520704, respectively. The number of clones in each OTU with 100% similarity is shown in Table S1 in the supplemental material.
RESULTS
Soil chemical properties.
The quantities of N2O, CO2, CH4, NO3−, NO2−, SO42−, NH4+, Fe2+, Mn2+, and succinate were measured to establish whether denitrification, DNRA, metal and sulfate reduction, and methanogenesis occurred during the incubation period (Table 1). Denitrification occurred only in the TSNI and TSNS samples, as confirmed by N2O production and decreases in the NO3− concentrations. Initially, approximately 100 mg nitrate N kg−1 soil (0.1 mg nitrate N g−1 soil) was added to the TSNI and TSNS samples. While nearly 20 to 25% of the added nitrate was converted to N2O gas in the TSNI sample, almost all the added nitrate was converted to N2O gas in the TSNS sample. Approximately 70% of the added succinate (500 mg succinate C kg−1 soil) was consumed in the TSNS sample, while ca. 40% was used in the TSSU sample. Other carboxylic acids, such as fumarate, acetate, and propionate, were not detected. The emission of CO2 gas increased as succinate was consumed. The amount of Fe2+ increased by 24 h after inoculation, but addition of substrates did not significantly influence Fe2+ concentrations. Slightly less exchangeable Mn2+ was detected in samples to which nitrate was added (the TSNI and TSNS samples) than in the other samples. Methane gas was not detected in any of the samples. The level of sulfate reduction could not be measured for samples amended with succinate because of a technical issue involving separating peaks derived from sulfate and succinate by a high-performance liquid chromatograph equipped with an IC-Pak anion column (4.6 by 50 mm).
TABLE 1.
Soil chemical properties and numbers of bacterial 16S rRNA gene copies in five samples
| Samplea | Concn ofb: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Succinate (mg C kg−1 soil) | CO2 (mg C kg−1 soil) | NO3− (mg N kg−1 soil) | N2O (mg N kg−1 soil) | NH4+ (mg N kg−1 soil) | Fe2+ (g kg−1 soil) | Mn2+ (mg kg−1 soil) | SO42− (mg kg−1 soil) | Bacterial 16S rRNA genes (1011 copies g−1 soil) | |
| TSBA | <10c | NDd | <5c | ND | 33.1 ± 6.7 | 2.22 ± 0.28 A | 9.51 ± 0.04 A | 87 ± 5 | 6.63 ± 0.31 A |
| TSCO | <10c | 165 ± 21 A | <5c | <0.01c | 27.1 ± 12.4 | 2.95 ± 0.14 B | 9.33 ± 0.31 A | 84 ± 11 | 5.28 ± 0.41 A |
| TSNI | <10c | 151 ± 9 A | 79.1 ± 3.6 | 24.4 ± 1.2 A | 33.4 ± 11.8 | 2.65 ± 0.13 B | 8.77 ± 0.12 B | 99 ± 11 | 7.46 ± 0.58 A |
| TSNS | 150.4 ± 7.0 Ae | 394 ± 40 B | <5c | 115.4 ± 3.9 B | 33.1 ± 5.7 | 2.90 ± 0.35 B | 8.49 ± 0.03 B | ND | 10.9 ± 0.99 B |
| TSSU | 291.3 ± 2.5 B | 253 ± 27 C | <5c | <0.01c | 29.5 ± 1.1 | 3.29 ± 0.22 B | 9.07 ± 0.04 A | ND | 5.33 ± 0.46 A |
TSBA, unincubated control; TSCO, soil sample incubated without substrate; TSNI, soil sample incubated with 100 mg NO3− N kg−1; TSNS, soil sample incubated with 100 mg NO3− N kg−1 and 500 mg succinate C kg−1; TSSU, soil sample incubated with 500 mg succinate C kg−1.
The P values for concentrations of succinate, CO2, N2O, NH4+, Fe2+, Mn2+, SO42−, and bacterial 16S rRNA genes are <0.001, <0.001, <0.001, 0.891, <0.01, <0.001, 0.1867, and <0.05, respectively.
Below the detection limit.
ND, not determined.
Means in the same column followed by the same letter are not significantly different (P > 0.01, n = 3).
Abundance of total bacterial 16S rRNA genes.
qPCR results showed that there were approximately 5.33 × 1011 to 1.1 × 1012 copies of total bacterial 16S rRNA genes per g of soil (Table 1). While the 16S rRNA gene copy numbers were similar for the TSBA, TSCO, TSNI, and TSSU samples, the TSNS sample had higher gene copy numbers than the other samples (P < 0.05).
DGGE analysis.
DGGE banding profiles were similar for three replicates of each sample (Fig. 1A). In addition, DGGE profiles were similar for the TSBA, TSCO, TSNI, and TSSU samples; however, several bands were unique to the TSNS sample. A distinct DGGE profile of the TSNS sample was also observed based on PCA (Fig. 1B). The DGGE profiles of the TSBA, TSCO, TSNI, and TSSU samples were close to each other in the PCA plot, whereas the TSNS sample profile was not close to the other profiles.
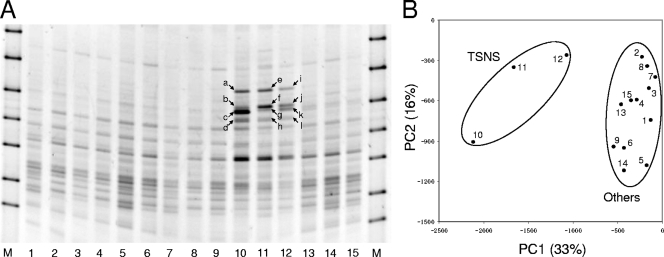
FIG. 1.
Community structures assessed by DGGE analysis. (A) DGGE banding profiles of the five samples (three replicates each). Twelve bands (bands a to l, indicated by arrows) were unique to the TSNS samples and were excised from lanes 10 to 12 and used for sequence analysis. Lanes M, DGGE marker II (Nippon Gene, Tokyo, Japan); lanes 1 to 3, unincubated control (TSBA sample); lanes 4 to 6, soil samples incubated without substrates (TSCO sample); lanes 7 to 9, soil samples incubated with nitrate (TSNI sample); lanes 10 to 12, soil samples incubated with nitrate and succinate (TSNS sample); lanes 13 to 15, soil samples incubated with succinate (TSSU sample). (B) PCA plot based on the DGGE profile. The normalized location and intensity of each DGGE band were used in the PCA. The numbers in the plot correspond to the lanes in panel A. The percentages in parentheses are the percentages of variation explained by the components.
Twelve bands that appeared to be unique in the TSNS sample (Fig. 1A) were excised, and their sequences were determined. Based on phylogenetic analyses, bacteria harboring the sequences for all of the excised bands were considered members of the class Betaproteobacteria.
Clone library analysis.
Since differences in community structure were detected by DGGE analysis, we employed the clone library approach to perform more precise analyses. More than 1,000 sequences of the nearly full-length 16S rRNA gene were obtained for each clone library (Table 2). Using the Mallard program, 8 (0.76%), 15 (1.48%), 34 (3.09%), 18 (1.64%), and 13 (1.22%) sequences were identified as chimeric in the TSBA, TSCO, TSNI, TSNS, and TSSU clone libraries, respectively, and were removed for subsequent analysis. As a result, a total of 5,224 sequences were used in this study. When 3% sequence dissimilarity was used as a cutoff for OTU (OTU0.03), 129 to 195 OTUs were obtained (Table 2).
TABLE 2.
Diversity in each clone library
| Sample | No. of sequenced clones after removal of chimeras | No. of OTU0.03s | Diversity indices for OTU0.03s |
||
|---|---|---|---|---|---|
| Chao1 (LC, HC)a | Simpson (1/D) | Shannon (H′)b | |||
| TSBA | 1,036 | 195 | 441 (343, 604) | 10.28 | 3.42 ± 0.13 |
| TSCO | 995 | 163 | 450 (324, 677) | 8.25 | 3.17 ± 0.12 |
| TSNI | 1,064 | 145 | 308 (308, 438) | 6.76 | 2.92 ± 0.12 |
| TSNS | 1,078 | 129 | 301 (290, 439) | 7.32 | 2.91 ± 0.11 |
| TSSU | 1,051 | 167 | 297 (243, 390) | 8.22 | 3.21 ± 0.12 |
| Total | 5,224 | 799 | |||
LC, lower confidence interval at 95% probability; HC, higher confidence interval at 95% probability.
Sannon index ± confidence interval at 95% probability.
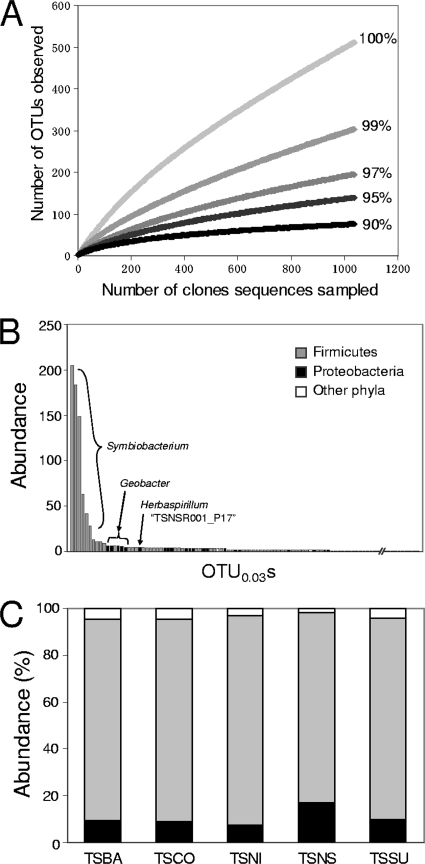
Great diversity was observed in the soil samples. The Chao estimator of species diversity at the 3% sequence dissimilarity level (Table 2), as well as a rarefaction curve analysis (Fig. 2A), suggested that our sampling effort was not sufficient to identify all bacterial species in the soil analyzed (the coverage ranged from 36 to 56%). Rank distribution analysis, as shown in Fig. 2B, suggested that Firmicutes bacteria, especially those belonging to a unique Symbiobacterium clade, were most abundant in the TSBA sample (Fig. 2B). Similar results were obtained for the other samples (data not shown). Abundant Firmicutes bacteria were also observed based on the taxonomic classification of the sequenced clones (Fig. 2C) and the phylogenetic tree (see Fig. S1 in the supplemental material). The proportion of clones belonging to the phylum Proteobacteria was greater in the TSNS library than in the other four libraries (Fig. 2C); however, this tendency was not statistically supported (χ2 goodness-of-fit test).
FIG. 2.
Diversity in the clone libraries. (A) Rarefaction curve analysis performed with sequenced clones obtained from the unincubated control (TSBA sample). Five different sequence dissimilarity values (0, 1, 3, 5, and 10%) were used to define OTUs. Similar results were observed with clones obtained from the other samples (data not shown). (B) Rank distribution analysis performed with OTU0.03s obtained from the TSBA sample. The most abundant OTU0.03s belonged to the Symbiobacterium clade (Firmicutes). (C) Distribution of various phyla in the clone libraries. The other phyla include Acidobacteria, Actinobacteria, Bacteroidetes, the BRC1 clade, Chloroflexi, Cyanobacteria, Gemmatimonadetes, Nitrospira, the OD1 clade, Planctomycetes, Verrucomicrobia, and unclassified bacteria.
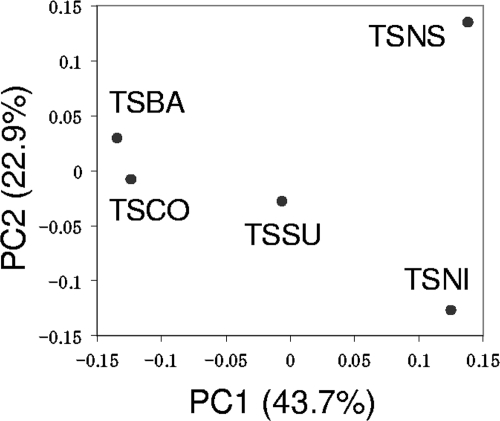
Although OTU0.03s from the five libraries clustered together in the phylogenetic tree (see Fig. S1 in the supplemental material), a distinct population structure of the TSNS clone library became apparent when the number of clone sequences in each OTU0.03 was considered. Figure 3 shows a principal coordinate analysis plot based on weighted UniFrac analysis using the phylogenetic tree of the OTU0.03s. Similar to the PCA plot generated based on DGGE analysis (Fig. 1B), the TSNS sample is plotted distantly from other samples.
FIG. 3.
Principal coordinate analysis plot based on the UniFrac analysis. The first and second principal coordinates (PC1 and PC2) explain 43.7% and 22.9% of the variation, respectively.
Detection of microbial population responsive to denitrifying conditions.
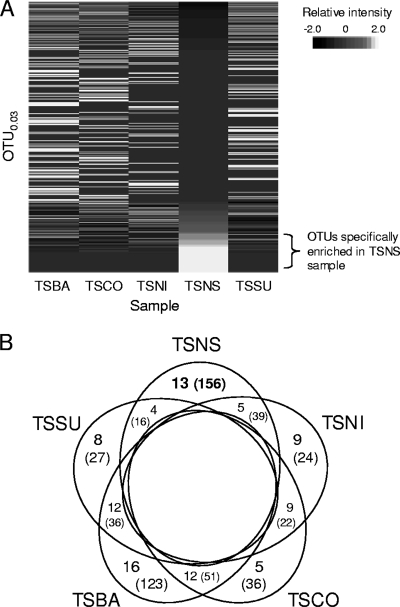
The template match method was used to detect the microbial population responsive to denitrifying conditions. An example of a pseudo-heat-map image generated by the template match method is shown in Fig. 4A. The number of OTU0.03s specifically enriched in each sample is summarized in Fig. 4B. The total numbers of clones belonging to the sample-specific OTU0.03s are also shown. About 6.2% (9/145), 10% (13/129), and 4.8% (8/167) of the total OTU0.03s responded to substrate additions in the TSNI, TSNS, and TSSU samples, respectively. While the proportions of five OTU0.03s specifically increased during incubation (TSCO sample), the proportions of 16 OTU0.03s (identified as TSBA sample-specific OTUs) decreased during incubation. Approximately 80% of the OTUs did not respond to any of the environmental changes. Clone distributions for samples are shown in Fig. S2 in the supplemental material for each sample-specific OTU0.03. Most OTU0.03s identified as specifically enriched populations in the TSNS sample belonged to the phylum Proteobacteria, whereas most of the specific populations in the other samples belonged to the phylum Firmicutes (see Fig. S2 in the supplemental material).
FIG. 4.
Identification of sample-specific OTUs. (A) Example of a pseudo-heat-map image generated based on the relative frequency of clone appearance for samples in each OTU. In this example, OTU0.03s were sorted by similarity to the target TSNS sample-specific pattern [(TSBA, TSCO, TSNI, TSNS, TSSU) = (0, 0, 0, 1, 0)]. Specifically enriched OTU0.03s were identified if they had similarity indices of <0.10 with the target template pattern. (B) Number of OTU0.03s specifically enriched in each sample. The numbers in parentheses are the total numbers of clones belonging to the sample-specific OTU0.03s.
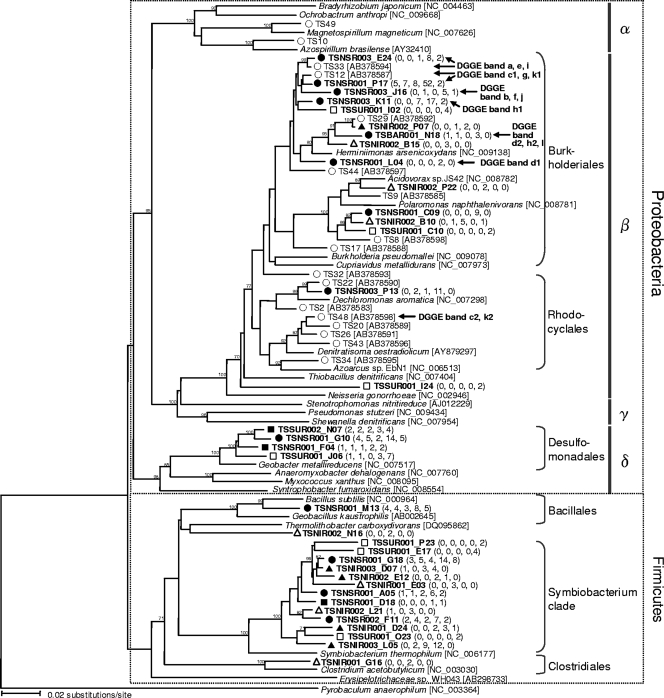
Phylogenetic relationships among the OTU0.03s specifically enriched in the TSNI, TSNS, and TSSU samples and among the commonly enriched OTU0.03s in the TSNI and TSNS samples and the TSNS and TSSU samples are shown in Fig. 5. The 16S rRNA gene sequences of the reference strains and the populations that assimilated [13C]succinate under denitrifying conditions (37) are also included in the tree. Specifically enriched OTU0.03s in the TSNS sample belonging to the phylum Proteobacteria were related to bacteria belonging to the orders Burkholderiales, Rhodocyclales, and Desulfomonadales. Most TSNS-specific OTU0.03s belonging to the orders Burkholderiales and Rhodocyclales appeared with at least 10-fold-greater frequency in the TSNS sample than in the unincubated control (TSBA sample), whereas the TSNS-specific OTU0.03s belonging to the order Desulfomonadales appeared with 3.5-fold-greater frequency in the TSNS sample than in the TSBA sample. The TSNS-specific OTU0.03s belonging to the orders Burkholderiales and Rhodocyclales were also closely related to the clones previously obtained by the SIP approach. Of the TSNS-specific OTU0.03s, those classified in the genus Herbaspirillum (TSNSR001_P17, TSNSR003_E24, and TSNSR003_K11 [see Fig. S2 in the supplemental material]) were most abundant (49% [77/156]).
FIG. 5.
Neighbor-joining tree showing phylogenetic relationships between specifically enriched OTU0.03s (in bold type) and the OTU0.03s previously reported in a SIP study (37). Bootstrap values (expressed as percentages) were generated using 1,000 replicates, and only values greater than 70% are shown. The numbers in parentheses are the numbers of clones in the OTU0.03. The OTU0.03s showing the greatest similarity to the DNA sequences retrieved from DGGE analysis (Fig. 1A) are indicated by arrows. Legend: ▵, specifically enriched OTU0.03s in the TSNI sample; ▴, commonly enriched OTU0.03s in the TSNI and TSNS samples; •, specifically enriched OTU0.03s in the TSNS sample; ▪, commonly enriched OTU0.03s in the TSNS and TSSU samples; □, specifically enriched OTU0.03s in the TSSU sample; ○, clones obtained from the previous SIP study (37). α, Alphaproteobacteria; β, Betaproteobacteria; γ, Gammaproteobacteria; δ, Deltaproteobacteria.
OTU0.03s specific to the other samples were also found in the orders Burkholderiales and Desulfomonadales, but a number of clones in these OTU0.03s were smaller than the clones in the TSNS-specific OTU0.03s. In Firmicutes bacteria, OTU0.03s that were specifically enriched in various samples were clustered together, and most of them were related to the Symbiobacterium clade. TSNS sample-specific OTU0.03s belonging to the phylum Firmicutes appeared in the TSNS sample two- to sixfold more frequently than they appeared in the TSBA sample.
The results obtained by using DGGE were consistent with the results of clone library analyses. Partial 16S rRNA gene sequences obtained from most of the excised DGGE bands (Fig. 2A) were for the most part similar to the sequences of TSNS sample-specific OTU0.03s (Fig. 5).
DISCUSSION
Based on the soil chemical characteristics of the five samples, nitrate respiration and denitrification occurred in samples amended with nitrate (TSNI and TSNS samples). Similar amounts of NH4+ for the five samples suggested that significant DNRA did not occur in the experimental setup used. The presence of NH4+ and nitrate-nitrite may induce anaerobic ammonium oxidation in rice paddy soil. However, this process usually is slow compared with denitrification (3, 18) and therefore may not have occurred at a significant level in this study. Addition of succinate greatly enhanced denitrification activity within 24 h during incubation, supporting previous findings (37, 38); however, our results indicate that significant manganese reduction and iron reduction did not occur during this short incubation period. In addition, evidence for fermentation and evidence for methanogenesis were not obtained in this study. We could not accurately measure SO42− concentrations in the TSNS and TSSU samples. However, since the presence of NO3− or Fe3+ could inhibit SO42− reduction (1), strong sulfate reduction might not have occurred in these samples. Together, nitrate respiration and denitrification were the major respiratory processes in the TSNI and TSNS samples, whereas these processes did not occur in the other samples.
The 16S rRNA gene copy number for total bacteria increased significantly in the TSNS sample, suggesting that the population of some bacteria increased in response to the addition of nitrate and succinate under anoxic conditions. This result, together with strong denitrification activity in the TSNS sample, suggests that some bacteria grew in the TSNS sample, probably by performing denitrification.
Both DGGE and clone library analyses targeting the 16S rRNA gene revealed that the structures of most communities were similar for the samples. Based on clone library analysis, bacteria belonging to the phylum Firmicutes, including a unique Symbiobacterium clade, were the most abundant organisms at the phylum level in the soil samples examined, in contrast to the results of other soil studies (4, 10, 25, 36). This is probably because of the improved DNA extraction protocol, the anoxic nature of rice paddy soils, and/or the use of air-dried soil in this study. Our improved DNA extraction protocol with a strong bead-beating procedure may have been able to destroy cells efficiently, including cells of spore-forming Firmicutes bacteria. Rice paddy field soils in conventional water management systems in Japan face multiple drying-wetting cycles during the growing season (28), potentially selecting the spore-forming Firmicutes bacteria as dominant members of the community in these soils. Use of air-dried soil may also have selected spore-forming bacteria in this study. In addition, differences in rRNA operon copy numbers among bacterial taxa might also have influenced the clone library results, since Firmicutes bacteria generally contain more copies of the rRNA operon than other bacteria (e.g., Alphaproteobacteria) (19). Further research is necessary to investigate whether Firmicutes are dominant in rice paddy soils.
DGGE and comparative clone library analyses showed that several groups of bacteria were specifically enriched under denitrification-inducing conditions in rice paddy soil (TSNS sample). The community structure of the TSNS sample was distinct from that of other samples when diversity was analyzed quantitatively (i.e., when the intensity of each DGGE band and the number of clones in each OTU were considered). Quantitative measures of diversity were previously shown to be suitable when changes in relative taxon abundance were examined (24).
Using the template match method (17, 32), we identified OTU0.03s that were specifically enriched in the TSNS sample as members of the orders Burkholderiales, Rhodocyclales, and Desulfomonadales and the phylum Firmicutes, some of which were previously identified as succinate-assimilating populations under denitrifying conditions (37). Among the TSNS sample-secific OTU0.03s, those classified in the genus Herbaspirillum (Burkholderiales) were probably the key players in nitrate respiration and denitrification under the conditions used, since they appeared >10-fold more frequently in the TSNS sample than in the unincubated control (TSBA sample) and they accounted for almost one-half of the clones belonging to the TSNS sample-specific OTU0.03s. Although 16S rRNA gene analysis alone cannot demonstrate the functional significance of microbes, we believe that the bacteria identified in this study might be involved in nitrate respiration and/or denitrification, since many members of the orders Burkholderiales and Rhodocyclales are known to exhibit denitrifying activity in various environments. Herbaspirillum spp. are also known to perform nitrate respiration (41), and some Herbaspirillum isolates can perform denitrification (N. Ashida, S. Ishii, and K. Senoo, unpublished data).
Several OTU0.03s that were specifically enriched in the TSNI sample or were commonly enriched in the TSNI and TSNS samples were also found to belong to the order Burkholderiales. Since a small amount of denitrifying activity was observed in the TSNI sample, the microbes represented by these OTU0.03s might be able to perform nitrate respiration or denitrification using soil organic materials as electron donors. These bacteria cannot be identified by the SIP approach because this approach requires microbial assimilation of 13C-labeled materials (35, 37). In addition to comparative 16S rRNA gene analysis, other approaches, such as incorporation of bromodeoxyuridine (6) and mRNA-targeted fluorescence in situ hybridization (34), may be used to detect these active populations. Several OTU0.03s specifically enriched in the TSSU sample or commonly enriched in the TSNS and TSSU samples were found to belong to the order Desulfomonadales. Some members of this group were reported to be involved in reduction of nitrate, sulfate, and metals, including Fe3+ and Mn4+ (22), indicating that the OTU0.03s belonging to this order may be enriched by utilizing succinate as an electron donor for nitrate, sulfate, and metal reduction, although the soil chemical properties did not provide solid evidence for sulfate and metal reduction during the 24-h incubation.
In contrast to the specifically enriched OTU0.03s belonging to the phylum Proteobacteria, the specifically enriched OTU0.03s in the phylum Firmicutes were clustered together in different samples, and therefore, it is difficult to speculate on their potential ecological roles. Most OTU0.03s belonging to the phylum Firmicutes were related to the Symbiobacterium clade. Since the genus Symbiobacterium is defined as a symbiont (29), the specific OTU0.03s in the Symbiobacterium clade might have been enriched along with their partners, such as denitrifiers. Genome analysis of Symbiobacterium thermophilum IAM 14863T revealed that this strain harbors the nap and nar nitrate reductase genes (45); therefore, the number of microbes represented by the OTU0.03s in the Symbiobacterium clade may have been increased by nitrate respiration. Furthermore, a recent report suggested that some members of the Firmicutes can generate electricity with acetate as the electron donor (47). Isolation of Firmicutes bacteria represented by the OTUs identified in this study would be necessary to link taxonomic information to the ecological roles of these bacteria.
While it is often claimed that culture-independent analysis based on the 16S rRNA gene is not suitable for studies of denitrifying and nitrate-respiring organisms (33), here we identified potential key players in nitrate respiration and denitrification by comparing community structures of soil with strong denitrifying activity and soil without denitrifying activity. In this study, we demonstrated that the template match method, which has been used frequently to analyze microarray data (32), was useful for identifying specifically enriched populations in clone library data. This approach can also be used to analyze massive sequence reads obtained by pyrosequence technology (8, 9, 15, 36), and this should be tested in the future.
Supplementary Material
Acknowledgments
We thank Masahito Hayatsu for his critical comments on the initial draft of the manuscript. We also thank Erika Iioka, Hiromi Inaba, Keiko Furuya, and Chie Yoshino for technical assistance.
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Biosciences of the Bio-oriented Technology Research Advancement Institution, Tokyo, Japan. Additional financial support was provided by grant-in-aid for scientific research (B) 19380041 from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 18 September 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Achtnich, C., F. Bak, and R. Conrad. 1995. Competition for electron donors among nitrate reducers, ferric iron reducers, sulfate reducers, and methanogens in anoxic paddy soil. Biol. Fertil. Soils 19:65-72. [Google Scholar]
- 2.Akiyama, H., X. Yan, and K. Yagi. 2006. Estimations of emission factors for fertilizer-induced direct N2O emissions from agricultural soils in Japan: summary of available data. Soil Sci. Plant Nutr. 52:774-787. [Google Scholar]
- 3.Amano, T., I. Yoshinaga, K. Okada, T. Yamagishi, S. Ueda, A. Obuchi, Y. Sako, and Y. Suwa. 2008. Detection of anammox activity and diversity of anammox bacteria-related 16S rRNA genes in coastal marine sediment in Japan. Microbes Environ. 22:232-242. [Google Scholar]
- 4.Asakawa, S., and M. Kimura. 2007. Comparison of bacterial community structures at main habitats in paddy field ecosystem based on DGGE analysis. Soil Biol. Biochem. 40:1322-1329. [Google Scholar]
- 5.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman, J. 1999. Culture-independent identification of microorganisms that respond to specified stimuli. Appl. Environ. Microbiol. 65:3398-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cambrell, R. P. 1996. Manganese, p. 665-682. In D. L. Sparks, A. L. Page, P. A. Helmke, R. H. Loeppert, P. N. Soltanpour, M. A. Tabatabai, C. T. Johnson, and M. E. Sumner (ed.), Methods of soil analysis, part 3. Chemical methods. Soil Science Society of America, Madison, WI.
- 8.Christen, R. 2008. Global sequencing: a review of current molecular data and new methods available to assess microbial diversity. Microbes Environ. 23:253-268. [DOI] [PubMed] [Google Scholar]
- 9.Dethlefsen, L., S. Huse, M. L. Sogin, and D. A. Relman. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLOS Biol. 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elshahed, M. S., N. H. Youssef, A. M. Spain, C. Sheik, F. Z. Najar, L. O. Sukharnikov, B. A. Roe, J. P. Davis, P. D. Schloss, V. L. Bailey, and L. R. Krumholtz. 2008. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 74:5422-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 12.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginige, M. P., P. Hugenholtz, H. Daims, M. Wagner, J. Keller, and L. L. Blackall. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70:588-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayatsu, M., K. Tago, and M. Saito. 2008. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 54:33-45. [Google Scholar]
- 15.Huber, J. A., D. B. M. Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 318:97-100. [DOI] [PubMed] [Google Scholar]
- 16.International Rice Research Institute. 2009. Rice production, area, and yield. In IRRI world rice statistics (WRS). Social Sciences Division, International Rice Research Institute, Manila, The Philippines. http://beta.irri.org/solutions/index.php?option=com_content&task=view&id=250.
- 17.Ishii, S., K. Kadota, and K. Senoo. 2009. Application of a clustering-based peak alignment algorithm to analyze various DNA fingerprinting data. J. Microbiol. Methods 78:344-350. [DOI] [PubMed] [Google Scholar]
- 18.Katsuyama, C., N. Kondo, Y. Suwa, T. Yamagishi, M. Itoh, N. Ohte, H. Kimura, K. Nagaosa, and K. Kato. 2008. Denitrification activity and relevant bacteria revealed by nitrite reductase gene fragments in soil of temperate mixed forest. Microbes Environ. 23:337-345. [DOI] [PubMed] [Google Scholar]
- 19.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liesack, W., S. Schnell, and N. P. Revsbech. 2000. Microbiology of flooded rice paddies. FEMS Microbiol. Rev. 24:625-645. [DOI] [PubMed] [Google Scholar]
- 21.Loeppert, R. H., and W. P. Inskeep. 1996. Iron, p. 639-664. In D. L. Sparks, A. L. Page, P. A. Helmke, R. H. Loeppert, P. N. Soltanpour, M. A. Tabatabai, C. T. Johnson, and M. E. Sumner (ed.), Methods of soil analysis, part 3. Chemical methods. Soil Science Society of America, Madison, WI.
- 22.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 23.Lozupone, C., and R. Knight. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228-8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone, C. A., M. Hamady, S. T. Kelley, and R. Knight. 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales, S. E., T. F. Cosart, J. V. Johnson, and W. E. Holben. 2009. Extensive phylogenetic analysis of a soil bacterial community illustrates extreme taxon evenness and the effects of amplicon length, degree of coverage, and DNA fractionation on classification and ecological parameters. Appl. Environ. Microbiol. 75:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulvaney, R. L. 1996. Nitrogen—inorganic forms, p. 1123-1184. In D. L. Sparks, A. L. Page, P. A. Helmke, R. H. Loeppert, P. N. Soltanpour, M. A. Tabatabai, C. T. Johnson, and M. E. Sumner (ed.), Methods of soil analysis, part 3. Chemical methods. Soil Science Society of America, Madison, WI.
- 27.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura, S., T. Sawamoto, H. Akiyama, S. Sudo, and K. Yagi. 2004. Methane and nitrous oxide emissions from a paddy field with Japanese conventional water management and fertilizer application. Glob. Biogeochem. Cycle 18:GB2017. [Google Scholar]
- 29.Ohno, M., H. Shiratori, M. J. Park, Y. Saitoh, Y. Kumon, N. Yamashita, A. Hirata, H. Nishida, K. Ueda, and T. Beppu. 2000. Symbiobacterium thermophilum gen. nov., sp. nov., a symbiotic thermophile that depends on co-culture with a Bacillus strain for growth. Int. J. Syst. Evol. Microbiol. 50:1829-1832. [DOI] [PubMed] [Google Scholar]
- 30.Osaka, T., S. Yoshie, S. Tsuneda, A. Hirata, N. Iwami, and Y. Inamori. 2006. Identification of acetate- or methanol-assimilating bacteria under nitrate-reducing conditions by stable-isotope probing. Microb. Ecol. 52:253-266. [DOI] [PubMed] [Google Scholar]
- 31.Otsuka, S., I. Sudiana, A. Komori, K. Isobe, S. Deguchi, M. Nishiyama, H. Shimizu, and K. Senoo. 2008. Community structure of soil bacteria in a tropical rainforest several years after fire. Microbes Environ. 23:49-56. [DOI] [PubMed] [Google Scholar]
- 32.Pavlidis, P., and W. Noble. 2001. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2:research0042. [DOI] [PMC free article] [PubMed]
- 33.Philippot, L., S. Hallin, and M. Schloter. 2007. Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 96:249-305. [Google Scholar]
- 34.Pratscher, J., C. Stichternoth, K. Fichtl, K.-H. Schleifer, and G. Braker. 2009. Application of recognition of individual genes-fluorescence in situ hybridization (RING-FISH) to detect nitrite reductase genes (nirK) of denitrifiers in pure cultures and environmental samples. Appl. Environ. Microbiol. 75:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 36.Roesch, L. F. W., R. R. Fulthorpe, A. Riva, G. Casella, A. K. M. Hadwin, A. D. Kent, S. H. Daroub, F. A. O. Camargo, W. G. Farmerie, and E. W. Triplett. 2007. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1:283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito, T., S. Ishii, S. Otsuka, M. Nishiyama, and K. Senoo. 2008. Identification of novel betaproteobacteria in a succinate-assimilating population in denitrifying rice paddy soil by using stable isotope probing. Microbes Environ. 23:192-200. [DOI] [PubMed] [Google Scholar]
- 38.Sato, R., Y. Sekine, and H. Wada. 1989. Effect of various organic compounds on nitrate metabolism in waterlogged soils. Jpn. J. Soil Sci. Plant Nutr. 61:134-139. (In Japanese with English abstract.) [Google Scholar]
- 39.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss, P. D., and J. Handelsman. 2006. Introducing SONS, a tool for operational taxonomic unit-based comparisons of microbial community memberships and structures. Appl. Environ. Microbiol. 72:6773-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmid, M., J. I. Baldani, and A. Hartmann. 2006. The genus Herbaspirillum, p. 141-150. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, 3rd ed., vol. 5. Springer, New York, NY. [Google Scholar]
- 42.Shiratori, Y., H. Watanabe, Y. Furukawa, H. Tsuruta, and K. Inubushi. 2008. Effectiveness of a subsurface drainage system in poorly drained paddy fields on reduction of methane emissions. Soil Sci. Plant Nutr. 53:387-400. [Google Scholar]
- 43.Tiedje, J. M. 1994. Denitrifiers, p. 245-267.In R. W. Weaver, J. S. Angle, and P. J. Bottomley (ed.), Methods of soil analysis, part 2. Microbiological and biochemical properties. Soil Science Society of America, Madison, WI.
- 44.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 45.Ueda, K., A. Yamashita, J. Ishikawa, M. Shimada, T. Watsuji, K. Morimura, H. Ikeda, M. Hattori, and T. Beppu. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32:4937-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Q., G. M. Garrity, J. M. Tiedje, and J. R. Cole. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrighton, K. C., P. Agbo, F. Warnecke, K. A. Weber, E. L. Brodie, T. Z. DeSantis, P. Hugenholtz, G. L. Anderson, and J. D. Coates. 2008. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2:1146-1156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.