Abstract
Chlamydia trachomatis is the trachoma agent and causes most bacterial sexually transmitted infections worldwide. Its major outer membrane protein (MOMP) is a well-known porin and adhesin and is the dominant antigen. So far, investigation of MOMP variability has been focused mainly on molecular epidemiological surveys. In contrast, we aimed to evaluate the impact of the host pressure on this key antigen by analyzing its evolutionary dynamics in 795 isolates from urogenital infections, taking into account the MOMP secondary structure and the sizes/positions of antigenic regions. One-third of the specimens showed a mutational drift from the corresponding genotype, where ∼42% of the mutations had never been described. Amino acid alterations were sixfold more frequent within B-cell epitopes than in the remaining protein (P = 0.027), and some mutations were also found within or close to T-cell antigenic clusters. Interestingly, the two most ecologically successful genotypes, E and F, showed a mutation rate 60.3-fold lower than that of the other genotypes (P < 10−8), suggesting that their efficacy may be the result of a better fitness in dealing with the host immune system rather than of specific virulence factors. Furthermore, the variability exhibited by some genetic variants involved residues that are known to play a critical role during the membrane mechanical movements, contributing to a more stable and flexible porin conformation, which suggests some plasticity to deal with environmental pressure. Globally, these MOMP mutational trends yielded no mosaic structures or important phylogenetic changes, but instead yielded point mutations on specific protein domains, which may enhance pathogen's infectivity, persistence, and transmission.
Chlamydia trachomatis is an obligate intracellular human pathogen that can be classified into 18 serovariants based on the immunoreactivity of its major outer membrane protein (MOMP) or ompA (which encodes MOMP) polymorphism: serovars A to C and Ba are responsible for trachoma; serovars D to K, Da, Ia, and Ja are normally associated with infection of the urogenital tract; and serovars L1 to L3 cause lymphogranuloma venereum (22). This preference for particular cell types is not exclusive, and therefore ocular strains can occasionally be found in the urogenital tract and vice versa. However, it is thought that only L1 to L3 strains possess the ability to invade the inguinal lymph nodes.
MOMP has been implicated in the mechanisms of attachment, infection, and/or pathogenesis due to its variability, surface exposure, and antigenic properties. Previous studies have shown that MOMP may act as a putative cytoadhesin by promoting nonspecific interactions with host cells (64). This major chlamydial membrane component, which constitutes about 60% of the membrane dry weight (9), is also thought to play a role in maintaining structural integrity of the organism (9, 10) by forming a trimeric structure (66). Also, during chlamydial replication, MOMP may act as a porin (6) that is folded into a β-barrel structure containing five constant domains (CDI to CDV) of transmembrane β-strands and periplasmic turns and four highly variable surface-exposed domains (VDI to VDIV) (34, 59, 69). Furthermore, MOMP possesses species- and serovar-specific epitopes (2, 48, 73, 74) that are able to elicit both the humoral (B-cell mediated through the production of antibodies) and cellular (T-cell mediated and also influencing the B-cell response) immune responses, making this dominant chlamydial antigen a potential candidate for the development of vaccines and therapeutic strategies (8, 17, 23, 61). Indeed, although no efficacious chlamydial vaccine has been developed so far, the use of inactivated or live-attenuated pathogens has been replaced by peptide or subunit vaccines, and MOMP is definitely one of the leading candidates (19).
To improve our knowledge of the effects of the host pressure on MOMP and also of the molecular epidemiology of the circulating C. trachomatis strains, it is imperative to investigate genetic variability in ompA. Here, we performed a sequence-based analysis of the ompA mutational trends in clinical isolates that were collected from patients with sexually transmitted C. trachomatis infections. So far, most studies have been limited to a small number of strains with variations in ompA (16, 33, 36, 42, 44, 47, 49, 52, 57, 62) or were restricted to the analysis of VDs (7, 14, 15, 32, 63), discarding the CDs, which contain numerous cytotoxic T lymphocyte (CTL) and T helper (Th) cell epitopes (30, 38, 39, 55, 56). We performed a detailed bioinformatic and statistical analysis of the mutational dispersion on both VDs and CDs, based on MOMP structure and on the mapping of all the B- and T-cell epitopes reported in the literature. We present statistically validated genomic evidence of the adaptation of this pathogen's key antigen to the host pressure, which strongly indicates a strategy to evade the human immune system.
MATERIALS AND METHODS
Study population and ompA genotyping.
From 2001 to 2007, a total of 15,135 first-void urine samples as well as cervical, urethral, and rectal swab samples were collected from patients attending general practice, family planning, and sexually transmitted disease clinics in the Lisbon area, and these samples were analyzed for the presence of C. trachomatis at the Portuguese National Institute of Health. This assessment was performed by DNA amplification methods (Amplicor PCR and Cobas-Amplicor) (Roche Molecular Systems) as per the manufacturer's instructions. For the 1,103 clinical specimens (7.3%) that were positive for C. trachomatis, DNA was extracted from 3 ml of first-void urine samples or 500 μl of exudate transport medium (Cobas-Amplicor collection kit or sucrose phosphate buffer [2SP]) using the QIAamp DNA mini kit (Qiagen) according to the manufacturer's instructions. Identification of clinical specimens was performed using ompA sequencing-based genotyping with subsequent BLAST comparison with the available GenBank sequences from C. trachomatis prototype strains, as previously described (28). Primers ompA-S (5′-TGCTGAACCAAGCCTTATGA-3′) and sero2A (5′-TTTCTAGATTTCATTTTGTT-3′) were used for the automated sequencing in the present survey. To avoid any epidemiological linkage that may lead to biased results, specimens collected from sexual partners during the same year were excluded from this study if they presented the same ompA sequence. All specimens presenting ompA variant sequences were confirmed by resequencing newly extracted DNA in order to overcome any amplification or sequencing artifact. The 18 prototype strains (see Table S1 in the supplemental material) that were used as the baseline for genotype comparisons were A/Har13, B/TW5, Ba/Apache2, C/TW3, D/UW3, Da/TW448, E/Bour, F/IC-Cal3, G/UW57, H/UW4, I/UW12, Ia/UW202, J/UW36, Ja/IUA795, K/UW31, L1/440, L2/434, and L3/404. Except for Ja/IUA795, the designation of these strains as prototype strains is consensual. Nevertheless, Ja/IUA795, which was isolated in 1986, has been the Ja strain most used as a reference. Additionally, other strains also referred to in the literature as reference strains (see Table S1 in the supplemental material) were used for comparative purposes in the present study.
Genomic and phylogenetic analyses.
For each ompA genotype, nucleotide and protein alignments of all clinical and reference strains were generated using the ClustalW method of the LaserGene (DNASTAR) and MEGA 4.0.2 (http://www.megasoftware.net) software. MEGA 4.0.2 was also used to create matrices of pairwise comparisons and to estimate the number of variable sites. For all clinical specimens, the existence of any recombination event in the ompA gene was evaluated using the SimPlot/BootScan software (http://sray.med.som.jhmi.edu/SCRoftware/), as previously described (27). Phylogenies were generated using the neighbor-joining method (60) with both nucleotide differences and Kimura two-parameter (40) or gamma (50) models to estimate evolutionary distances at the nucleotide or protein level, respectively. For all genomic and phylogenetic analyses, the pairwise-deletion option was chosen to remove all sites containing missing data or alignment gaps from all distance estimations, only when the need arose and not prior to the analyses.
Analysis of molecular evolution.
We used the Nei-Gojobori method (51) of MEGA 4.0.2 to calculate the overall mean of synonymous (dS) and nonsynonymous (dN) substitution rates for each ompA genotype, as previously described (25). Given the degeneracy of the genetic code, the p-distance model was used to normalize the computed differences against the number of potential synonymous and nonsynonymous sites.
Specific evaluation of the selective pressure (neutral [dN/dS = 1], purifying [dN/dS < 1], or positive [dN/dS > 1] selection) acting on the ompA gene was performed for the genotypes Da, G, and Ia, as these genotypes presented the highest number of variant clinical specimens (Fig. 1). As the selective pressures vary among different regions of the protein, this analysis was performed using a maximum-likelihood model of codon substitution along the phylogeny of ompA (53, 71) implemented by the codeml program in the PAML v4 software (http://abacus.gene.ucl.ac.uk/software/paml.html), which allows dN/dS to differ across different codons. Briefly, the distribution of the dN/dS ratio (ω) across sites was estimated using the following codon substitution models (71): M0 (one ratio), M1a (nearly neutral), M2a (positive selection), M3 (discrete), M7 (beta distribution), and M8 (beta distribution + ω). The existence of amino acid sites under positive selection was tested with a likelihood ratio test by comparing the nested models that differ by only one parameter (the class of sites with ω > 1), i.e., M2a versus M1a and M8 versus M7. The null distribution of nonexistence of sites under positive selection was evaluated by the likelihood ratio test statistic (2Δl, where Δl is the difference between the log-likelihood scores of the two methods), assuming that under null hypothesis the 2Δl follows a χ2 distribution where the degrees of freedom are the difference in the number of free parameters between the two methods. The empirical Bayes method was used to calculate the posterior probabilities of each site falling into the class of positively selected sites (i.e., class with ω > 1) (72) as well as the posterior distribution of the ω parameter describing that class. Thus, sites likely under positive selection are the ones with higher probabilities of belonging to this class.
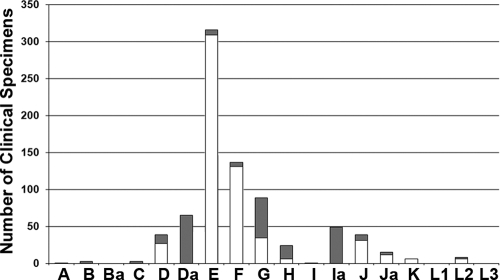
FIG. 1.
Distribution of the 795 C. trachomatis clinical specimens by ompA genotype. The bars represent the absolute numbers of nonvariant (in white) and variant (in gray) specimens.
Analysis of protein features.
To shed some light on the putative impact of each mutation on the MOMP sequence, the Protean program of LaserGene (DNASTAR) was used to perform a comparative analysis of the protein sequences of all variant and reference strains for the following features: charge density (70), secondary structure (method of Garnier et al. [24]), hydropathy (methods of Kyte and Doolittle [41], Hopp and Woods [31], and Engleman et al. [21]), antigenicity (Jameson-Wolf [35] and AMPHI [46] methods), amphiphilicity (method of Eisenberg et al. [18]), surface probability (method of Emini et al. [20]), and flexibility (Karplus-Schultz method [37]). Basically, for all these protein features, with the exceptions of the secondary structure and antigenicity, values were assigned for all residues and were then averaged over a sliding window of a specified range of amino acids for each method. For these analyses, the default parameters of each method were used, except for the Kyte-Doolittle and Hopp-Woods methods, where a window size of seven amino acids was used.
Statistical analysis.
For each ompA genotype, only the variable sites where clinical specimens presented nucleotide changes to the corresponding prototype strain (Table 1) were analyzed. Globally, a total of 80 variable sites were considered, as follows: genotype A, 1 site; B, 15; C, 5; D, 2; Da, 6; E, 5; F, 6; G, 7; H, 4; I, 1; Ia, 14; J, 10; Ja, 2; and L2, 2. To investigate the variability inside both the VDs and CDs of ompA, we evaluated whether the 31 nonsynonymous mutations found in VDs are overrepresented relative to the ones that occurred in CDs. We found that there were 23 nonsynonomous substitutions in CDs, 2 synonymous substitutions in VDs, and 24 synonomous substitutions in CDs.
TABLE 1.
Nucleotide sequence variation in ompA genotype variants compared with the respective prototype strain
| Genotype (no. of variants) | No. of variable sites | MOMP region | Nucleotide positiona | Amino acid changeb | Type of changec |
No. of isolates (x/y)d | dN/dS (SE) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| New | B cell | T cell | Loop | |||||||
| A (1) | 1 | CDIII | 736 | Val → Ile | x | x | 1/1 | —e | ||
| B (3) | 15 | CDI | 154 | Thr → Ala | 2/3 | 0.35 (0.19) | ||||
| 163/164 | Val → Cys | E | 3/3 | |||||||
| 184/186 | Met → Val | 2/3 | ||||||||
| 195 | 2/3 | |||||||||
| 198 | 2/3 | |||||||||
| 216 | 3/3 | |||||||||
| 228 | 2/3 | |||||||||
| 246 | 2/3 | |||||||||
| 249 | 2/3 | |||||||||
| VDI | 268 | Thr → Ala | 2/3 | |||||||
| 286/287 | Val → Thr | 2/3 | ||||||||
| VDII | 514 | Ala → Thr | x | 3/3 | ||||||
| C (3) | 5 | CDI | 155 | Thr → Ile | x | 1/3 | 0.59 (0.51) | |||
| 183 | x | 1/3 | ||||||||
| VDI | 284 | Gly → Asp | x | x | E | 1/3 | ||||
| CDIII | 693 | x | C | 1/3 | ||||||
| VDIV | 1003 | Ala → Ser | x | 3/3 | ||||||
| D (12) | 2 | VDII | 488 | Glu → Gly | x | E | 1/12 | — | ||
| VDIV | 976 | Ala → Thr | x | 12/12 | ||||||
| Da (65) | 6 | VDII | 485 | Asn → Ser | 11/65 | 0.29 (0.25) | ||||
| CDIII | 636 | 46/65 | ||||||||
| VDIV | 939 | x | 61/61 | |||||||
| 976 | Ala → Thr | x | 11/58 | |||||||
| 997 | Thr → Ala | x | 57/57 | |||||||
| CDV | 1050 | x | 1/1 | |||||||
| E (7) | 5 | VDII | 512 | Asn → Ser | x | x | 1/7 | — | ||
| 514 | Ser → Ala | x | x | C | 1/7 | |||||
| CDIII | 568 | Ala → Thr | x | C | 2/7 | |||||
| VDIV | 995 | Ser → Asn | x | x | 1/7 | |||||
| 997 | Ala → Thr | x | 2/7 | |||||||
| F (6) | 6 | CDI | 167 | Asp → Gly | x | E | 1/6 | 1.53 (1.60) | ||
| 180 | Met → Ile | x | 1/6 | |||||||
| CDII | 388 | Ile → Val | x | 1/6 | ||||||
| CDIII | 703 | Phe → Leu | x | 1/6 | ||||||
| CDIV | 780 | 1/6 | ||||||||
| 802 | Asp → Asn | x | P | 1/6 | ||||||
| G (54) | 7 | CDI | 228 | 15/54 | 2.48 (2.55) | |||||
| CDII | 374 | Cys → Tyr | x | P | 1/54 | |||||
| VDII | 487 | Gly → Ser | 24/54 | |||||||
| CDIII | 700 | Glu → Gln | P | 15/54 | ||||||
| CDIV | 857 | Thr → Ile | x | W | 1/54 | |||||
| VDIV | 1003 | Ser → Ala | 29/49 | |||||||
| Ser → Thr | 12/49 | |||||||||
| 1015 | Ile → Val | x | 1/45 | |||||||
| H (18) | 4 | VDI | 272 | Asn → Ser | x | 18/18 | 0.30 (0.29) | |||
| CDII | 327 | x | 1/18 | |||||||
| 383 | Ala → Val | x | 1/18 | |||||||
| CDIV | 850 | W | 18/18 | |||||||
| I (1) | 1 | VDIII | 764 | Ile → Thr | x | x | 1/1 | — | ||
| Ia (49) | 14 | CDI | 129 | 1/1 | 0.84 (0.45) | |||||
| 184/186 | Val → Met | 47/47 | ||||||||
| 228 | 47/47 | |||||||||
| VDI | 277 | Ile → Val | 2/49 | |||||||
| VDII | 526 | Ile → Val | 22/49 | |||||||
| CDIII | 571 | Asn → Asp | P | 49/49 | ||||||
| 577 | Thr → Ala | x | 1/1 | |||||||
| VDIII | 764 | Ile → Thr | x | 47/47 | ||||||
| CDIV | 837 | W | 46/46 | |||||||
| 840 | W | 46/46 | ||||||||
| 932 | Lys → Arg | x | 1/44 | |||||||
| CDV | 1141 | Ala → Thr | 1/1 | |||||||
| 1188 | 1/1 | |||||||||
| J (8) | 10 | CDI | 213 | x | 1/6 | 0.22 (0.13) | ||||
| CDII | 369 | 7/8 | ||||||||
| CDIII | 684 | x | 1/8 | |||||||
| 720 | x | 1/8 | ||||||||
| VDIII | 742 | Ala → Thr | x | x | 1/8 | |||||
| 753 | x | x | 1/8 | |||||||
| 782 | Ala → Glu | x | E | 1/7 | ||||||
| VDIV | 991 | Thr → Ala | 1/7 | |||||||
| 1000 | Ala → Ser | x | 1/7 | |||||||
| 1020 | Asp → Glu | x | E | 1/7 | ||||||
| Ja (3) | 2 | VDII | 544 | Asn → Asp | x | E | 2/3 | 0.50 (0.25) | ||
| CDIII | 634 | x | 1/3 | |||||||
| L2 (2) | 2 | VDII | 485 | Asn → Ser | x | 2/2 | — | |||
| 517 | Leu → Phe | x | x | 1/2 | ||||||
Based on the alignment of the strains belonging to the same genotype.
Amino acid change presented by the variant clinical specimen compared with the prototype strain of the respective ompA genotype.
Mutations that were observed only for the Portuguese clinical specimens or that occurred within well-defined B-cell epitopes (x), mutations that occurred within (W) or in close proximity (C) to T-cell (CTL and Th) epitopes, and mutations found in MOMP external loops (E) or periplasmic turns (P). For this last type of mutation, only the residues known to be involved in MOMP structural constrains were considered for this analysis.
x represents the total number of clinical specimens sharing the same variable site; y represents the total number of clinical specimens for which the nucleotide sequence involving the variable site is available.
—, calculation of the dN/dS ratio was not feasible for these genotypes as they presented solely nonsynonymous mutations.
We also investigated the mutational trend in MOMP B-cell antigenic regions (see Fig. S1 in the supplemental material) for all genotypes containing variant clinical specimens. To achieve this, we estimated the total length of the MOMP B-cell antigenic regions for each genotype, where all the epitope sequences described for each VD were overlapped and the lengths of the resulting regions were summed. Thereafter, we calculated the mean of the final value for all genotypes, which yielded an antigenic region encompassing a length of 105 bp per ompA (35 amino acids per MOMP). Since it was experimentally demonstrated (75) that a change in the amino acid immediately adjacent to a defined epitope may alter its immunoreactivity, we also considered all variable sites occurring one amino acid before and after each epitope. We then evaluated whether 20 variable sites restricted to the 105 bp coding for the B-cell antigenic regions are overrepresented relative to the 60 variable sites found in the rest of the ompA gene. With a restricted mean gene sequence of 900 bp (the ompA sequence length available for all clinical specimens) for each strain, we found 85 nonvariable sites for the B-cell antigenic regions and 735 nonvariable sites for the nonantigenic regions.
Also, given that 18 of the above 20 variable sites occurring in B-cell antigenic regions yielded amino acid replacement, we also evaluated whether these sites restricted to the superimposed antigenic regions are overrepresented relative to the 36 sites found outside that region. We found 2 synonymous sites in the B-cell antigenic regions and 24 synonymous sites in the nonantigenic regions.
For all these statistical analyses, the SPSS Base version 15.0 (SPSS Inc. Chicago, IL) was used to estimate the P values by Fisher's exact test as well as the odds ratios with a 95% confidence interval.
Nucleotide sequence accession numbers.
The sequence data for all types of genetic variants (see Table S1 in the supplemental material) were submitted to GenBank under accession numbers DQ116393, DQ116396 to DQ116398, DQ116400, DQ116402, EU676180, and FJ943511 to FJ943546.
RESULTS
Analysis of ompA genotypes and genetic variants.
We initially characterized C. trachomatis isolates by analyzing ompA nucleotide variation. Of the 15,135 urogenital samples that were received at the Portuguese NIH, 1,103 were positive for C. trachomatis, with 795 specimens (493 from women, 294 from men, and 8 for which the gender of the donor was unknown) successfully typed. These represented 15 ompA genotypes; Ba, L1, and L3 were not found (Fig. 1). Genotype E was the most prevalent type (39.8%), followed by genotypes F (17.3%), G (11.2%), Da (8.2%), Ia (6.2%), D and J (4.9%), H (3.0%), Ja (1.9%), L2 (1.0%), K (0.8%), B and C (0.4%), and finally A and I (0.1%). Globally, 232 clinical specimens (29.2%) of all identified genotypes (with the exception of genotype K) were found to present ompA nucleotide changes compared with the respective prototype strain (Fig. 1). The latter were isolated up to 50 years ago (see Table S1 in the supplemental material) and have been used worldwide as references for comparative purposes. The highest number of clinical variants was presented by genotypes Da, G, and Ia, with 65, 54, and 49 strains, respectively. Moreover, all specimens typed as A, B, C, Da, I, and Ia presented ompA sequences that varied from that of the respective prototype strain. Interestingly, for the two most prevalent genotypes, E and F, which together represent almost 57% of all typed specimens, only 7 out of 317 E strains (2.2%) and 6 out of 138 F strains (4.4%) had ompA variant sequences. This is intriguing as it suggests that conservation of the major antigen favors ecological success. In support of this divergent ompA molecular trend for these genotypes, we have previously shown a mutational dynamic involving the entire chromosome that separated E and F from the remaining genotypes (54).
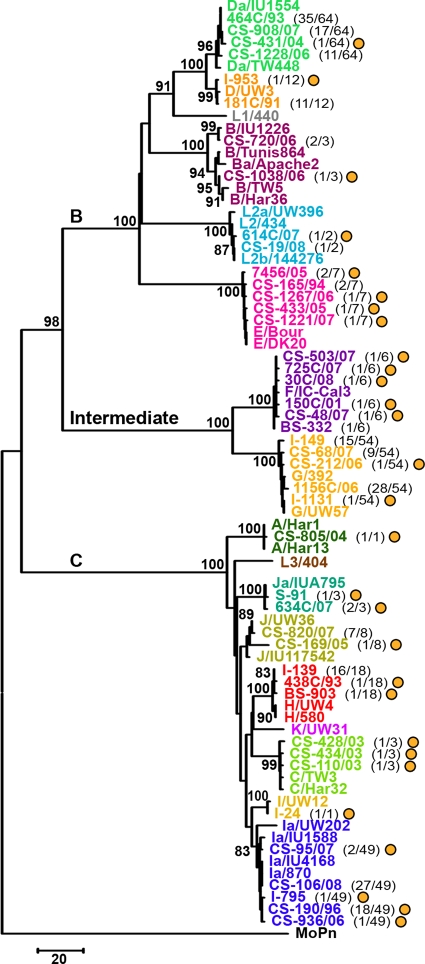
Overall, 43 types of genetic variants (i.e., strains with a distinct ompA mutational pattern) were identified among our 232 variant clinical specimens, involving 14 ompA genotypes (Fig. 2). Twenty-nine out of the 43 genetic variants have not been reported before, and these represent a total of 49 strains. Curiously, 44.9% of these belong to genotype Ia. Also of note is that all genetic variants of genotypes A, C, I, and Ja found for the Portuguese population were never described for any other country. No insertion or deletion (indel) event was found in the ompA gene for any of our 43 genetic variants compared to the respective prototype strain (data not shown). Furthermore, based on SimPlot/BootScan analyses, none of them revealed any trace of recombination within ompA, presenting only slight dissimilarities to the corresponding prototype and/or other reference strains (data not shown). Supporting this, both the nucleotide (Fig. 2) and amino acid (data not shown) phylogenetic analyses showed strain segregation according to three main clusters corresponding to the traditional B, C, and intermediate ompA serogroups with maximum bootstrap values (98 to 100%). In fact, within each serogroup, closely related clusters were found for all variant genotypes, where genetic distances among strains varied from 0.1% (standard error [SE], 0.1%) for genotypes A, I, and Ja to 0.9% (SE, 0.2%) for genotype B (data not shown). Interestingly, all the Da and Ia variants showed higher similarity to reference strains Da/IU1554 and Ia/IU4168, respectively, than to the corresponding traditional prototype strains (Da/TW448 and Ia/UW202). Altogether, this indicates that in the population studied, ompA diversity is strictly a consequence of the occurrence of point mutations rather than recombination or indel events.
FIG. 2.
Evolutionary relationship between all C. trachomatis prototype strains and the 43 types of genetic variants. The neighbor-joining phylogenetic tree is based on the number of nucleotide differences observed among the ompA sequences to better illustrate the genetic distances between genetic variants. Kimura two-parameter and gamma models yielded similar topologies. The tree was rooted using the ompA sequence from C. muridarum strain MoPn. Bootstrap values (1,000 replicates) are shown next to the branch nodes. The traditional ompA serogroups (B, C, and Intermediate) are shown above the three major branches of the tree. All types of genetic variants and reference strains belonging to the same genotype are represented by the same color together with the respective prototype strain. Values in parentheses indicate the number of variant specimens represented by that type of genetic variant and the total number of variant specimens found for the same ompA genotype. Types of genetic variants that had never been described before are represented by yellow circles.
Analysis of ompA variable sites per genotype.
We proceeded with a detailed analysis of ompA variability within same-genotype strains. Table 1 describes the nucleotide sequence variation in ompA for each genotype, showing all mutations relative to the corresponding prototype strain. A total of 80 nucleotide-variable sites were found, where B, Ia, and J presented the highest number of sites (with 15, 14, and 10 changes, respectively), while genotypes A and I were the least variable ones (both with 1 change). Each variable site was represented by no more than two types of nucleotides, with exception of genotype G, which presented a parsimony-informative site, where two different nonsynonymous changes were found for the same variable site at position 1003. Interestingly, for the vast majority of genotypes, variable sites were dispersed throughout ompA, reflecting a general mutational trend on both VDs and CDs. One notable exception occurred for B strains, where 11 of the 15 mutations were located at CDI (P < 10−7), suggesting a highly restricted mutational clustering for genotype B. Also interestingly, all six mutations displayed by F genetic variants were found solely in CDs.
Overall, from the 80 variable sites found for all types of genetic variants (Table 1), 33 occurred in VDs and 47 occurred in CDs, with CDI encompassing almost half (44.7%) of them. Moreover, 93.9% (31/33) of the mutations that occurred in VDs yielded amino acid changes, while the reverse was seen for CDs, with 51.1% (24/47) of the mutations resulting in silent substitutions. In fact, we found a global nonsynonymous mutation rate that was 16.2-fold higher for VDs than for CDs (P < 10−4), indicating a nonsynonymous variability targeted mainly on MOMP surface-exposed domains, which are involved in the interaction with the host.
Impact of mutations on MOMP antigenic regions.
Considering that well-defined B- and T-cell epitopes were already described in MOMP VDs and CDs (see Fig. S1 in the supplemental material), we also investigated the specific mutational trend in these antigenic regions for all genotypes presenting variant strains in order to evaluate the host immune pressure on the chlamydial major antigen. Therefore, we constructed a comprehensive “map” of C. trachomatis B- and T-cell (CTL and Th) epitopes described in the literature for all 18 prototype strains (3, 4, 12, 29, 30, 38, 39, 55, 56, 65, 75, 76) (see Fig. S1 in the supplemental material). B-cell epitopes encompassing full VDs were not considered, as immunoreactivity is generally due to small epitopes inside those large regions (75). Also, although the Th-stimulatory peptides reported in the literature are known to contain multiple core Th epitopes, only a minority were already mapped and only these were considered for the present study. This conservative approach was used to minimize any bias that could result from considering an overrepresentation of antigenic regions in MOMP, which would substantially favor the statistical analysis.
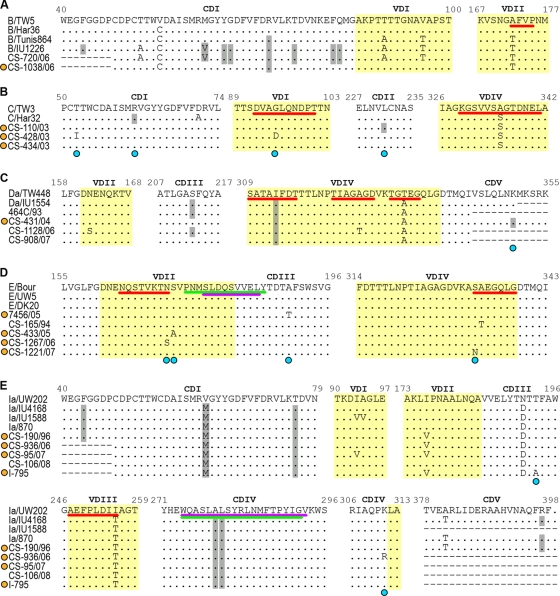
We found that 20 out of the 80 mutations (25%) occurred within B-cell epitopes with species, B-serogroup, and serovar specificities (P < 10−3) (Table 1; Fig. 3). This phenomenon was observed for all genotypes except F and Ja, where mutations were found solely outside these antigenic regions. Additionally, we also found a global nonsynonymous mutation rate sixfold higher for the B-cell antigenic regions than for the rest of the ompA gene (P = 0.027). Indeed, 90% (18/20) of the B-cell epitope mutations resulted in amino acid replacements. Furthermore, for six genotypes (C, E, G, H, Ia, and J), mutations also occurred within or close to the few core CTL and/or Th cell epitopes that were already mapped on MOMP (Table 1; Fig. 3). An interesting example occurred for genotype E, where two nonsynonymous mutations at positions 514 and 568 were found to flank a CTL epitope that spans VDII and CDIII and is specifically recognized by subjects infected with C. trachomatis serovar E (38). Curiously, this epitope is within a serovar E-specific Th epitope-containing peptide (55, 56) and is adjacent to three well-defined B-cell core epitopes (3) (Fig. 3). In addition, some silent mutations found among all H (at position 850) and Ia (at positions 837 and 840) strains as well as one nonsynonymous mutation found for one G variant clinical specimen (at position 857) occurred within a cluster of five HLA class I-restricted minimal CTL epitopes, which are located at CDIV and are C. trachomatis species specific (38). In addition, for all genotypes (except G), these CTL epitopes fully overlap at least six known HLA class II-restricted core Th cell epitopes (55). Thus, this remarkable concentration of amino acid alterations in MOMP antigenic regions points to a pathogen's strategy for host immune evasion, where these changes may lead to disruption of functional/structural epitopes.
FIG. 3.
Variability of C. trachomatis MOMP. Panels A to E correspond to genotypes B, C, Da, E, and Ia, respectively. Within each panel, the upper row represents the prototype strain for the respective genotype. Each row in the alignment consists of a single type of genetic variants that may represent several specimens. For comparative proposes, sequences from other reference strains are shown below the prototype sequence. Types of genetic variants (yellow circles) and variable sites (blue circles) that had never been described before are represented. Silent mutations are highlighted in gray. Positions 62 in panels A and E constitute a simultaneous occurrence of one silent and one nonsilent mutation on the respective codon. Protein VDs are highlighted in yellow. B-cell, CTL, and Th cell antigenic regions (which may contain various overlapping epitopes) are underlined in red, purple, and green, respectively.
Global and codon-based evaluation of selective pressure.
Considering the targeted occurrence of nonsilent mutations in MOMP antigenic regions, we used bioinformatics to examine the existence of codons under positive selection. The analysis of the molecular evolution of the whole ompA gene showed dN/dS values of <1 for genotypes B, C, Da, H, Ia, J, and Ja, while genotypes F and G showed dN/dS values of >1 (1.53 [SE, 1.60] and 2.48 [SE, 2.55], respectively) (Table 1), suggesting that the ompA genes of the latter may be subject to positive selection. In fact, 83.3% and 85.7% of the mutations displayed by all F and G strains, respectively, yielded amino acid alterations. In addition, the dN/dS ratio for genotypes A, D, E, I, and L2 is expectedly high (>1), as all the mutations displayed by their genetic variants were nonsynonymous. However, the nonexistence of silent mutations for these genotypes rules out the estimation of dN/dS. Still, a calculation of dN/dS for the whole gene may be biased by both the existence of a nonrepresentative number of variants for each genotype and the nondiscrimination of codons with distinct evolutionary signatures. In fact, it is known that when very few amino acids are under positive selection, the global dN/dS statistics are not very sensitive to that selection. If synonymous substitutions accumulate at a higher rate than nonsynonymous substitutions, the few amino acid replacements that occur due to positive selection will not influence dN enough to cause it to be significantly greater than dS.
Thus, we applied the codon substitution model to evaluate positive selection. Considering software constraints, we studied genotypes Da, G, and Ia, as they presented the highest number of variant clinical specimens. A proportion of 0.5% of sites under positive selection and 99.5% under neutral selection (ω = 1.00) was estimated for genotype G (including all variant and nonvariant G strains), considering models M2a and M8 (see Table S2 in the supplemental material). The proportion of sites under positive selection was not statistically significant (M2a versus M1a, P = 0.064; M8 versus M7, P = 0.063) (Table 2). Both models (M2a and M8) identified six sites (125, 163, 234, 286, 335, and 339) with a posterior probability of belonging to a class of sites under positive selection higher than 0.5. From these, only position 335 presented a probability of higher than 0.87, showing a posterior ω of 5.6 (SE = 3.1). Interestingly, site 286 is within the above-cited cluster of five HLA class I-restricted CTL species-specific epitopes overlapping six known HLA class II-restricted Th cell epitopes (38) (see Fig. S1 in the supplemental material).
TABLE 2.
Likelihood ratio test statistics for testing positive selection
| Genotype and models | 2Δla | dfb | Pc |
|---|---|---|---|
| G | |||
| M2a vs M1a | 5.509764 | 2 | 0.064 |
| M8 vs M7 | 5.526772 | 2 | 0.063 |
| Ia | |||
| M2a vs M1a | 3.347866 | 2 | 0.188 |
| M8 vs M7 | 6.345914 | 2 | 0.042 |
Log-likelihood difference between compared models.
Degrees of freedom between compared models.
p of the log-likelihood test.
For genotype Ia (including all variant and nonvariant Ia strains), the hypothesis of the nonexistence of sites under positive selection was rejected from the comparison of models M8 versus M7 (P = 0.042) (Table 2). Estimates from model M8 showed that 7.3% of the sites were under positive selection and 92.7% of sites were under purifying selection, with ω = 0.0052 (see Table S3 in the supplemental material). Eight sites (62, 93, 94, 176, 191, 193, 255, and 311) were identified as belonging to the positive selection class (ω of >1, posterior probability of >0.5), although only positions 62 (ω = 5.7; SE = 3.2) and 176 (ω = 5.9; SE = 3.1) presented posterior probabilities higher than 0.89. Curiously, sites 93, 94, and 255 are within or close to B-cell epitopes (Fig. 3E; see Fig. S1 in the supplemental material).
With respect to genotype Da (including all variant and nonvariant Da strains), although the M8 model identified three sites (162, 326, and 333) as being positively selected (ω of >1, posterior probability of >0.5) (see Table S3 in the supplemental material), the likelihood ratio test of a positive selection was not statistical significant (data not shown). Once more, sites 326 and 333 are within B-cell epitopes with specificity for serogroup B and genotype Da, respectively (Fig. 3C; see Fig. S1 in the supplemental material).
Collectively, despite the lack of statistical significance for a positive selection, the six described sites (one for G, three for Ia, and two for Da) within MOMP antigenic regions were also found to be variable in C. trachomatis strains collected in other regions of the world (see Discussion). Thus, this strongly suggests that they are under positive selection, and that the lack of statistical significance may be a matter of population size.
Types of amino acid substitutions.
In order to evaluate the existence of mutations that are likely becoming fixed in the population, we investigated the frequency of each amino acid substitution per nonsynonymous variable site, involving all 232 variant clinical specimens (Table 1). The change of Thr to Ala was the most frequent, occurring 63 times (15.5%), 94.5% of which were for genotype Da. The Asn-to-Asp, Ile-to-Thr, and Val-to-Met changes were also highly common, with 51, 48, and 47 occurrences, respectively, and the vast majority involved genotype Ia. These high frequencies observed for both genotypes Da and Ia indicate a fixation of these mutations among the corresponding strains, where they likely play an important structural or functional role.
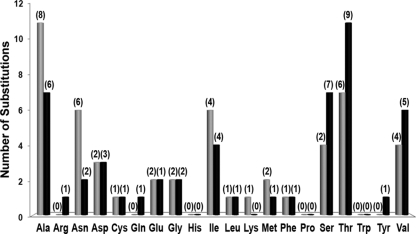
We also evaluated the frequency of each amino acid substitution per nonsynonymous variable site but considering only the 43 types of genetic variants, in order to avoid any bias due to the number of same-genotype variant specimens presenting that substitution. Overall, from the 30 distinct types of amino acid substitutions that took place, the change of Ala to Thr was the most frequent, occurring seven times for seven different genotypes (B, D, Da, E, Ia, J, and L2), followed by the Thr-to-Ala change, with five occurrences (Table 1). Moreover, among all types of genetic variants, Ala and Thr were also the most frequently replaced amino acids throughout MOMP (11 times for eight genotypes and 7 times for six genotypes, respectively), as well as the two most acquired ones (7 times for six genotypes and 11 times for nine genotypes, respectively) (Fig. 4). This is not surprising considering that these two amino acids are the most common among all chlamydial reference strains and the 43 genetic variants under study (data not shown).
FIG. 4.
Number and type of substitutions involving a specific amino acid. Comparative analysis of all types of genetic variants for all genotypes revealed 52 nonsynonymous variable sites in MOMP, representing 30 distinct types of amino acid substitutions. The size of each bar represents the number of times that a residue is replaced (in gray) or acquired (in black) among solely the 43 types of genetic variants (this excludes any bias due to the heterogeneous number of same-genotype variant specimens presenting that substitution). The number of ompA genotypes presenting that specific amino acid substitution is shown in parentheses above each bar.
Analysis of protein features.
To shed some light on the putative impact of each mutation on MOMP, a protein sequence-based comparative analysis was also performed for the 43 types of genetic variants and all reference strains. In general, although 80% of the 30 types of amino acid substitutions occurred between residues with dissimilar properties, no significant differences were observed for charge density, secondary structure, hydropathy, antigenicity, amphiphilicity, surface probability, or flexibility (data not shown). Interestingly, although only 23.3% of all substitutions are indicated to cause a gain or loss of charge density, a consequent reversal of charge density was frequently observed for their neighbor amino acids (data not shown).
Considering the porin role of MOMP, we also analyzed the loss or gain of amino acids that were shown to influence porin structure (59). Six of the nine changes involving residues Asp and Glu (Table 1) were on the MOMP external loops, leading to an alteration of the negative charge in these regions. Moreover, these substitutions occurred for specimens of several genotypes (C, D, F, J, and Ja). Similarly, a targeted addition of Asp, Glu, and Tyr residues in the MOMP periplasmic turns was also observed for all Ia (Asp) as well as for 15 G (Glu) and one G (Tyr) variant specimens (Table 1). Therefore, although the impact of the loss or gain of these specific amino acids is not known, the nonrandomness of these occurrences suggests some plasticity to deal with environmental changes/pressure.
DISCUSSION
C. trachomatis MOMP has been implicated as a porin, adhesin, and key antigen. In the present study, we performed a mutational trend analysis of this major membrane component from 795 clinical isolates from urogenital infections. In common with previous surveys for other countries (36, 43, 45, 49, 52), E and F were the most prevalent genotypes among the Portuguese population (Fig. 1), representing almost 57% of all C. trachomatis infections (39.8% E and 17.3% F), which indicates little fluctuation in genotypic prevalence worldwide. Although the secret of this apparent worldwide success still remains to be elucidated, it has been speculated that the biological advantage of E and F may reside in a more effective host immune evasion (7) as well as in the existence of specific adhesins or virulence factors that may give them some functional or structural facility in terms of infection and transmission (26, 54). However, further experimental evidence will be needed to support these assumptions.
Globally, 232 clinical specimens (29.2%) were found to present ompA variant sequences compared to the respective prototype strain (Fig. 1). Interestingly, the two most successful genotypes, E and F, were the least variable among the studied population, showing an ompA mutation rate 60.3-fold lower than that of the other genotypes (P < 10−8). In agreement with this, several studies from different geographic regions (36, 44, 45, 58) have reported a high level of conservation of both E and F strains. The apparent lack of ompA variability in these strains may indicate a better fitness for E and F, where the resultant MOMP may be less immunogenic, which could favor longer E/F “camouflage,” further dissemination, and, consequently, higher success. Supporting this, by testing serovar-specific lymphoproliferative responses to several MOMP synthetic peptides, Arno et al. (1) found that individuals infected with genotype E recognized a peptide containing a known CDIII T-cell epitope less frequently than did non-E-infected individuals. Moreover, although the lack of clinical data has hampered us from making any correlation with clinical symptoms, genotype E has been frequently associated with asymptomatic infections (14, 63, 68), reinforcing the speculation that the most prevalent genotypes may elicit a less vigorous host immune response. However, some weakly supported associations with clinical symptoms were previously found for genotype F (5, 14, 67, 68), which makes any assumption merely controversial and inconclusive.
Overall, 43 distinct types of genetic variants (involving 14 genotypes) were found (Fig. 2), and 67.4% of them had not been reported before. It is known that genetic variability in MOMP may arise from point mutations, recombination events, or indels as a result of host selective pressure and bacterial adaptation. However, no visible trace of recombination or any indel event was found in ompA in our genetic variants compared to the respective prototype strain, suggesting that the genetic variability observed is strictly a consequence of the occurrence of point mutations. Interestingly, of the total 80 nucleotide-variable sites exhibited by the 43 genetic variants (Table 1), 65% resulted in amino acid alterations, which suggest that they are evolutionarily nonneutral. In support of this, dN/dS values of >1 were found, at least for variant specimens F and G, suggesting the existence of positive selection. More specifically, various codons (some of them located in MOMP antigenic regions) were likely positively selected in Da, G, and Ia variant specimens, and statistical significance was achieved for the latter (P = 0.042). Curiously, 46 out of the 80 nucleotide changes were shared by other same-genotype strains already described in other regions of the world. This suggests a fixation of these mutations, likely as a biological advantage in terms of infectivity and transmission. Although no visible mutational “hot spots” were found among genetic variants of different genotypes, we observed a few cases where the occurrence of mutations for different ompA genotypes was in exactly the same gene location (Fig. 3 and Table 1), which may suggest the existence of spots that are more prone to change. We speculate that this may be due either to fitness advantages or simply to MOMP structural constraints.
For most genotypes, mutations were found in both the ompA VDs and CDs, indicating a general mutational trend throughout the entire gene. A notable exception occurred for B strains, which exhibited a 20.4-fold-higher probability of undergoing changes in CDI than in the remainder of the gene (P < 10−7). Nevertheless, we have no reasonable explanation for this highly restricted mutational clustering for genotype B, considering that CDI is the protein region that is thought to be more conserved and one of the less immunogenic. In contrast to what was expected, a greater number of mutations (1.42-fold higher) was seen for the CDs than for the VDs. However, 51.1% of the mutations in CDs were silent, suggesting that the conservation of these domains may be important to maintain either the MOMP porin function related to its monomeric form (34, 59) or the chlamydial membrane structural integrity, which is associated with the trimeric form of MOMP (66). In contrast, VDs showed a global nonsynonymous mutation rate 16.2-fold higher than that of the CDs, where 93.9% of the changes resulted in amino acid replacements (P < 10−4). This seems to be remarkable evidence of the existence of positive selection almost exclusively targeted to the MOMP surface-exposed domains. Considering that MOMP is the major chamydial membrane component and unquestionably its dominant antigen, these findings point to an evolutionary dynamic used by C. trachomatis to deal with the environment and the host immune pressures. In support of a chlamydial host evasion strategy, 25% (20/80) of the mutations occurred within well-defined B-cell epitopes for most genotypes (Table 1), which is statistically significant as these epitopes encompass only about 12% of the total MOMP length (P < 10−3).
Furthermore, 90% of the mutations found within B-cell antigenic regions resulted in amino acid replacements. In fact, these regions showed a global nonsynonymous mutation rate sixfold higher than that for the rest of the ompA gene (P = 0.027), which suggests a targeted amino acid variability in MOMP B-cell antigenic regions. Curiously, some of the nonsynonymous mutations reported in the present study were previously shown to abrogate monoclonal antibody binding and neutralization of infectivity of several prototype and/or genovariant strains in vitro (3, 4). For example, the VDIV Ala → Thr change found in all D and 11 Da variant specimens (Table 1) was previously shown to prevent the antibody binding at both the serogroup B 322TIAGAGD328 and 323IAGAG327 epitopes for the known genovariant D− (4). Also, one Portuguese genovariant, C, showed a Gly → Asp alteration in VDI that was found to prevent the antibody binding to the 92DVAGL96 epitope (specific for serovars A, C, I, J, and Ja) in a previous complete-replacement analysis of this antigenic site (75). Although it would be interesting to extend this kind of in vitro experimental demonstration to all mutations occurring within B-cell epitopes, their reflection in vivo remains speculative, as the true epitope conformation cannot be ensured and potential differences among genotypes cannot be evaluated.
Interestingly, for some genotypes, mutations were also found to occur within or close to two MOMP T-cell antigenic clusters (Table 1 and Fig. 3) (3, 38, 55, 56). Although the impact of mutations in these epitopes remains unknown, they may indicate an evasion strategy where single mutations are used to disrupt or affect clusters of multiple epitopes that simultaneously elicit different host protective immune responses, especially when they are serovar specific (39). This strategy would promote the persistence of the infection and/or disease progression, which could significantly increase transmission and/or reinfection. However, Kim and DeMars (39) have suggested, based on a previous study (16), that the existence of mutations in cluster epitopes with species specificity (and, thus, highly conserved among serovars) would be deleterious and greatly reduce pathogen fitness. This negative selection theory finds some support in our data. In fact, of the three mutations occurring inside that region, only one yielded an amino acid replacement, and it was exhibited solely by one specimen out of the 795 strains under study. Nevertheless, 64 variant strains from different genotypes (all 18 H and 46 Ia) presented silent mutations within that cluster, which points to a fixation event and may influence the subsequent evolutionary landscape of ompA. In fact, it was recently demonstrated that the fixation of silent mutations in open reading frames may influence both the transcriptional pattern (due to the modification of the primary mRNA structure) and the type of amino acids that can be added in a second round of mutations (11). Although we cannot evaluate the impact of this mutational clustering in B- and T-cell epitopes of the chlamydial key antigen on vaccine success, new antigenic profiles may be expected. Accordingly, acquisition of knowledge about the ompA evolutionary mutational pattern and the identification of new types of variants that are arising emerge as important tasks.
Besides the remarkable evidences of a MOMP mutational dynamics strongly focused on a host evasion strategy, our data also suggest that the variability exhibited by some genetic variants may have a direct influence on the protein structure. In fact, some replacements involved amino acids that are thought to be important to the MOMP conformation. For instance, most of the changes involving the negatively charged residues Asp and Glu have occurred on the protein external loops (Table 1), where, according to the literature (59), they are normally abundant and participate in binding to the lipopolysaccharide (13). Since the substitution of charged amino acids has a direct mimic effect on the charge density of the neighbor amino acids (data not shown), it is expected that changes altering the number of these residues may affect the MOMP porin topology. Similarly, the additional presence of Asp, Glu, and Tyr residues in the MOMP periplasmic turns of some genetic variants (Table 1) may also contribute to a more stable and flexible porin conformation. In fact, it was previously reported that residues with a higher bend potential (such as Asp, Glu, and Tyr) are well represented in the small periplasmic turns of several bacterial porins (59), where they likely play a critical role during the membrane mechanical movements. However, an evaluation of the conformational impact of these alterations on MOMP is likely an unfeasible task, considering the extremely complex putative protein structure (59, 69).
Collectively, these results show a global mutational evolutionary dynamics of C. trachomatis ompA that is associated with MOMP functional/structural constraints and mainly with evading host immune surveillance. In the light of the continuous coevolution between this pathogen and its human host (54) that takes place during infection, this evolutionary scenario is expectedly beneficial in terms of infectivity, persistence, and transmission.
Supplementary Material
Acknowledgments
This work was supported by grants from Fundação para a Ciência e a Tecnologia (FCT) (PTDC/BIA-BCM/71117/2006) and Comissão de Fomento da Investigação em Cuidados de Saúde (no. 112/2007) (to J.P.G.). A.N. is the recipient of a Ph.D. grant (SFRH/BD/25651/2005) from FCT.
Footnotes
Published ahead of print on 25 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arno, J. N., C. Xie, R. B. Jones, and B. Van Der Pol. 1998. Identification of T cells that respond to serovar-specific regions of the Chlamydia trachomatis major outer membrane protein in persons with serovar E infection. J. Infect. Dis. 178:1713-1718. [DOI] [PubMed] [Google Scholar]
- 2.Baehr, W., Y. X. Zhang, T. Joseph, H. Su, F. E. Nano, K. D. Everett, and H. D. Caldwell. 1988. Mapping antigenic domains expressed by Chlamydia trachomatis major outer membrane protein genes. Proc. Natl. Acad. Sci. USA 85:4000-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batteiger, B. E. 1996. The major outer membrane protein of a single Chlamydia trachomatis serovar can possess more than one serovar-specific epitope. Infect. Immun. 64:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batteiger, B. E., P. M. Lin, R. B. Jones, and B. J. Van Der Pol. 1996. Species-, serogroup-, and serovar-specific epitopes are juxtaposed in variable sequence region 4 of the major outer membrane proteins of some Chlamydia trachomatis serovars. Infect. Immun. 64:2839-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batteiger, B. E., W. Lennington, W. J. Newhall, B. P. Katz, H. T. Morrison, and R. B. Jones. 1989. Correlation of infecting serovar and local inflammation in genital chlamydial infections. J. Infect. Dis. 160:332-336. [DOI] [PubMed] [Google Scholar]
- 6.Bavoil, P., A. Ohlin, and J. Schachter. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44:479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunham, R. C., C. Yang, I. Maclean, J. Kimani, G. Maitha, and F. Plummer. 1994. Chlamydia trachomatis from individuals in a sexually transmitted disease core group exhibit frequent sequence variation in the major outer membrane protein (omp1) gene. J. Clin. Investig. 94:458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunham, R. C., D. J. Zhang, X. Yang, and G. M. McClarty. 2000. The potential for vaccine development against chlamydial infection and disease. J. Infect. Dis. 181:S538-543. [DOI] [PubMed] [Google Scholar]
- 9.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldwell, H. D., and R. C. Judd. 1982. Structural analysis of chlamydial major outer membrane proteins. Infect. Immun. 38:960-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cambray, G., and D. Mazel. 2008. Synonymous genes explore different evolutionary landscapes. PLoS Genet. 4:e1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conlan, J. W., I. N. Clarke, and M. E. Ward. 1988. Epitope mapping with solid-phase peptides: identification of type-, subspecies-, species- and genus-reactive antibody binding domains on the major outer membrane protein of Chlamydia trachomatis. Mol. Microbiol. 2:673-679. [DOI] [PubMed] [Google Scholar]
- 13.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghosh, R. A. Pauptit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 14.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 15.Dean, D., J. Schachter, C. R. Dawson, and R. S. Stephens. 1992. Comparison of the major outer membrane protein variant sequence regions of B/Ba isolates: a molecular epidemiologic approach to Chlamydia trachomatis infections. J. Infect. Dis. 166:383-392. [DOI] [PubMed] [Google Scholar]
- 16.Dean, D., and K. Millman. 1997. Molecular and mutation trends analyses of omp1 alleles for serovar E of Chlamydia trachomatis. Implications for the immunopathogenesis of disease. J. Clin. Investig. 99:475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Maza, M. A., and L. M. de la Maza. 1995. A new computer model for estimating the impact of vaccination protocols and its application to the study of Chlamydia trachomatis genital infections. Vaccine 13:119-127. [DOI] [PubMed] [Google Scholar]
- 18.Eisenberg, D., R. M. Weiss, and T. C. Terwilliger. 1984. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA 81:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eko, F. O., Q. He, T. Brown, L. McMillan, G. O. Ifere, G. A. Ananaba, D. Lyn, W. Lubitz, K. L. Kellar, C. M. Black, and J. U. Igietseme. 2004. A novel recombinant multisubunit vaccine against Chlamydia. J. Immunol. 173:3375-3382. [DOI] [PubMed] [Google Scholar]
- 20.Emini, E. A., J. Hughes, D. Perlow, and J. Boger. 1985. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 55:836-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman, D. M., T. A. Steitz, and A. Goldman. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15:321-354. [DOI] [PubMed] [Google Scholar]
- 22.Fields, P. I., and R. C. Barnes. 1992. The genus Chlamydia, p. 3691-3709. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K. H. Schleifer, (ed.), The prokaryotes. Springer-Verlag, New York, NY.
- 23.Fitch, W. M., E. M. Peterson, and L. M. de la Maza. 1993. Phylogenetic analysis of the outer-membrane-protein genes of Chlamydiae, and its implication for vaccine development. Mol. Biol. Evol. 10:892-913. [DOI] [PubMed] [Google Scholar]
- 24.Garnier, J., D. J. Osguthorpe, and B. Robson. 1978. Analysis of the accuracy and implications of simple method for predicting the secondary structure of globular proteins. J. Mol. Biol. 120:97-120. [DOI] [PubMed] [Google Scholar]
- 25.Gomes, J. P., A. Nunes, W. J. Bruno, M. J. Borrego, C. Florindo, and D. Dean. 2006. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 188:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomes, J. P., M. J. Borrego, B. Atik, I. Santo, J. Azevedo, A. Brito de Sá, P. Nogueira, and D. Dean. 2006. Correlating Chlamydia trachomatis infectious load with urogenital ecological success and disease pathogenesis. Microbes Infect. 8:16-26. [DOI] [PubMed] [Google Scholar]
- 27.Gomes, J. P., W. J. Bruno, A. Nunes, N. Santos, C. Florindo, M. J. Borrego, and D. Dean. 2007. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 17:50-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gomes, J. P., W. J. Bruno, M. J. Borrego, and D. Dean. 2004. Recombination in the genome of Chlamydia trachomatis involving the polymorphic membrane protein C gene relative to ompA and evidence for horizontal gene transfer. J. Bacteriol. 186:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes, L. J., M. A. Pickett, J. W. Conlan, S. Ferris, J. S. Everson, M. E. Ward, and I. N. Clarke. 1990. The major outer-membrane proteins of Chlamydia trachomatis serovars A and B: intra-serovar amino acid changes do not alter specificities of serovar- and C subspecies-reactive antibody-binding domains. J. Gen. Microbiol. 136:1559-1566. [DOI] [PubMed] [Google Scholar]
- 30.Holland, M. J., D. J. Conway, T. J. Blanchard, O. M. Mahdi, R. L. Bailey, H. C. Whittle, and D. C. Mabey. 1997. Synthetic peptides based on Chlamydia trachomatis antigens identify cytotoxic T lymphocyte responses in subjects from a trachoma-endemic population. Clin. Exp. Immunol. 107:44-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopp, T. P., and K. R. Woods. 1981. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA 78:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh, Y. H., L. D. Bobo, T. C. Quinn, and S. K. West. 2001. Determinants of trachoma endemicity using Chlamydia trachomatis ompA DNA sequencing. Microbes Infect. 3:447-458. [DOI] [PubMed] [Google Scholar]
- 33.Hsu, M. C., P. Y. Tsai, K. T. Chen, L. H. Li, C. C. Chiang, J. J. Tsai, L. Y. Ke, H. Y. Chen, and S. Y. Li. 2006. Genotyping of Chlamydia trachomatis from clinical specimens in Taiwan. J. Med. Microbiol. 55:301-308. [DOI] [PubMed] [Google Scholar]
- 34.Hughes, E. S., K. M. Shaw, and R. H. Ashley. 2001. Mutagenesis and functional reconstitution of chlamydial major outer membrane proteins: VS4 domains are not required for pore formation but modify channel function. Infect. Immun. 69:1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jameson, B. A., and H. Wolf. 1988. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput. Appl. Biol. Sci. 4:181-186. [DOI] [PubMed] [Google Scholar]
- 36.Jurstrand, M., L. Falk, H. Fredlund, M. Lindberg, P. Olcén, S. Andersson, K. Persson, J. Albert, and A. Backman. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karplus, P. A., and G. E. Schultz. 1985. Prediction of chain flexibility in proteins. Naturwissenschaft 72:212-213. [Google Scholar]
- 38.Kim, S. K., M. Angevine, K. Demick, L. Ortiz, R. Rudersdorf, D. Watkins, and R. DeMars. 1999. Induction of HLA class I-restricted CD8+ CTLs specific for the major outer membrane protein of Chlamydia trachomatis in human genital tract infections. J. Immunol. 162:6855-6866. [PubMed] [Google Scholar]
- 39.Kim, S. K., and R. DeMars. 2001. Epitope clusters in the major outer membrane protein of Chlamydia trachomatis. Curr. Opin. Immunol. 13:429-436. [DOI] [PubMed] [Google Scholar]
- 40.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 41.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 42.Lee, G., J. Park, B. Kim, S. A. Kim, C. K. Yoo, and W. K. Seong. 2006. OmpA genotyping of Chlamydia trachomatis from Korean female sex workers. J. Infect. 52:451-454. [DOI] [PubMed] [Google Scholar]
- 43.Lister, N. A., C. K. Fairley, S. N. Tabrizi, S. Garland, and A. Smith. 2005. Chlamydia trachomatis serovars causing urogenital infections in women in Melbourne, Australia. J. Clin. Microbiol. 43:2546-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lister, N. A., S. N. Tabrizi, C. K. Fairley, A. Smith, P. H. Janssen, and S. Garland. 2004. Variability of the Chlamydia trachomatis omp1 gene detected in samples from men tested in male-only saunas in Melbourne, Australia. J. Clin. Microbiol. 42:2596-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lysén, M., A. Osterlund, C. J. Rubin, T. Persson, I. Persson, and B. Herrmann. 2004. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J. Clin. Microbiol. 42:1641-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Margalit, H., J. L. Spouge, J. L. Cornette, K. B. Cease, C. Delisi, and J. A. Berzofsky. 1987. Prediction of immunodominant helper T cell antigenic sites from the primary sequence. J. Immunol. 138:2213-2229. [PubMed] [Google Scholar]
- 47.Millman, K., C. M. Black, R. E. Johnson, W. E. Stamm, R. B. Jones, E. W. Hook, D. H. Martin, G. Bolan, S. Tavaré, and D. Dean. 2004. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 186:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison, R. P., D. D. Manning, and H. D. Caldwell. 1992. Immunology of Chlamydia trachomatis infections: immunoprotective and immunopathogenic responses, p. 57-84. In T. C. Quinn (ed.), Sexually transmitted diseases. Raven Press, New York, NY.
- 49.Mossman, D., K. W. Beagley, A. L. Landay, M. Loewenthal, C. Ooi, P. Timms, and M. Boyle. 2008. Genotyping of urogenital Chlamydia trachomatis in Regional New South Wales, Australia. Sex. Transm. Dis. 35:614-616. [DOI] [PubMed] [Google Scholar]
- 50.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics, Oxford University Press, New York, NY.
- 51.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 52.Ngandjio, A., M. Clerc, M. C. Fonkoua, J. Thonnon, F. Njock, R. Pouillot, F. Lunel, C. Bebear, B. De Barbeyrac, and A. Bianchi. 2003. Screening of volunteer students in Yaounde (Cameroon, Central Africa) for Chlamydia trachomatis infection and genotyping of isolated C. trachomatis strains. J. Clin. Microbiol. 41:4404-4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen, R., and Z. Yang. 1998. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148:929-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nunes, A., P. J. Nogueira, M. J. Borrego, and J. P. Gomes. 2008. Chlamydia trachomatis diversity viewed as a tissue-specific coevolutionary arms race. Genome Biol. 9:R153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ortiz, L., K. P. Demick, J. W. Petersen, M. Polka, R. A. Rudersdorf, B. Van der Pol, R. Jones, M. Angevine, and R. DeMars. 1996. Chlamydia trachomatis major outer membrane protein (MOMP) epitopes that activate HLA class II-restricted T cells from infected humans. J. Immunol. 157:4554-4567. [PubMed] [Google Scholar]
- 56.Ortiz, L., M. Angevine, S. K. Kim, D. Watkins, and R. DeMars. 2000. T-cell epitopes in variable segments of Chlamydia trachomatis major outer membrane protein elicit serovar-specific immune responses in infected humans. Infect. Immun. 68:1719-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pedersen, L. N., H. O. Kjaer, J. K. Møller, T. F. Orntoft, and L. Ostergaard. 2000. High-resolution genotyping of Chlamydia trachomatis from recurrent urogenital infections. J. Clin. Microbiol. 38:3068-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez, P., B. de Barbeyrac, K. Persson, B. Dutilh, and C. Bebear. 1993. Evaluation of molecular typing for epidemiological study of Chlamydia trachomatis genital infections. J. Clin. Microbiol. 31:2238-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodríguez-Marañón, M. J., R. M. Bush, E. M. Peterson, T. Schirmer, and L. M. de la Maza. 2002. Prediction of the membrane-spanning beta-strands of the major outer membrane protein of Chlamydia. Protein Sci. 11:1854-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 61.Stephens, R. S. 2000. Chlamydial genomics and vaccine antigen discovery. J. Infect. Dis. 181:S521-S523. [DOI] [PubMed] [Google Scholar]
- 62.Stothard, D. R., G. Boguslawski, and R. B. Jones. 1998. Phylogenetic analysis of the Chlamydia trachomatis major outer membrane protein and examination of potential pathogenic determinants. Infect. Immun. 66:3618-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sturm-Ramirez, K., H. Brumblay, K. Diop, A. Guèye-Ndiaye, J. L. Sankalé, I. Thior, I. N′Doye, C. C. Hsieh, S. Mboup, and P. J. Kanki. 2000. Molecular epidemiology of genital Chlamydia trachomatis infection in high-risk women in Senegal, West Africa. J. Clin. Microbiol. 38:138-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su, H., N. G. Watkins, Y. X. Zhang, and H. D. Caldwell. 1990. Chlamydia trachomatis-host cell interactions: role of the chlamydial major outer membrane protein as an adhesin. Infect. Immun. 58:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su, H., R. P. Morrison, N. G. Watkins, and H. D. Caldwell. 1990. Identification and characterization of T helper cell epitopes of the major outer membrane protein of Chlamydia trachomatis. J. Exp. Med. 172:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun, G., S. Pal, A. K. Sarcon, S. Kim, E. Sugawara, H. Nikaido, M. J. Cocco, E. M. Peterson, and L. M. de la Maza. 2007. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 189:6222-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van de Laar, M. J., Y. T. van Duynhoven, J. S. Fennema, J. M. Ossewaarde, A. J. van den Brule, G. J. van Doornum, R. A. Coutinho, and J. A. van der Hoek. 1996. Differences in clinical manifestations of genital chlamydial infections related to serovars. Genitourin. Med. 72:261-265. [PMC free article] [PubMed] [Google Scholar]
- 68.van Duynhoven, Y. T., J. M. Ossewaarde, R. P. Derksen-Nawrocki, W. I. van der Meijden, and M. J. van de Laar. 1998. Chlamydia trachomatis genotypes: correlation with clinical manifestations of infection and patients' characteristics. Clin. Infect. Dis. 26:314-322. [DOI] [PubMed] [Google Scholar]
- 69.Wang, Y., E. A. Berg, X. Feng, L. Shen, T. Smith, C. E. Costello, and Y. X. Zhang. 2006. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 15:122-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White, A., P. Handler, and E. Smith. 1964. Principles of biochemistry, McGraw-Hill, New York, NY.
- 71.Yang, Z., R. Nielsen, N. Goldman, and A. M. Pedersen. 2000. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155:431-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang, Z., W. S. Wong, and R. Nielsen. 2005. Empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22:1107-1118. [DOI] [PubMed] [Google Scholar]
- 73.Zhang, Y. X., S. Stewart, T. Joseph, H. R. Taylor, and H. D. Caldwell. 1987. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J. Immunol. 138:575-581. [PubMed] [Google Scholar]
- 74.Zhang, Y. X., S. J. Stewart, and H. D. Caldwell. 1989. Protective monoclonal antibodies to Chlamydia trachomatis serovar- and serogroup-specific major outer membrane protein determinants. Infect. Immun. 57:636-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhong, G. M., and R. C. Brunham. 1991. Antigenic determinants of the chlamydial major outer membrane protein resolved at a single amino acid level. Infect. Immun. 59:1141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong, G. M., R. E. Reid, and R. C. Brunham. 1990. Mapping antigenic sites on the major outer membrane protein of Chlamydia trachomatis with synthetic peptides. Infect. Immun. 58:1450-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.