Abstract
Mycobacterium tuberculosis generally is assumed to depend on lipids as a major carbon and energy source when persisting within the host. The utilization of fatty acids requires a functional glyoxylate cycle with the key enzymes isocitrate lyase (Icl) and malate synthase. The open reading frame Rv0465c of M. tuberculosis H37Rv encodes a protein with significant sequence similarity to the transcriptional regulator RamB, which in Corynebacterium glutamicum controls the expression of several genes involved in acetate metabolism, i.e., those encoding enzymes of acetate activation and the glyoxylate cycle. We show here that the M. tuberculosis Rv0465c protein can functionally complement RamB in C. glutamicum and that it binds to the promoter regions of M. tuberculosis icl1 and Rv0465c. Construction and subsequent transcriptional and enzymatic analysis of a defined Rv0465c deletion mutant in M. tuberculosis revealed that the Rv0465c protein, now designated RamB, represses icl1 expression during growth with glucose and negatively autoregulates the expression of its own operon. Whole-genome microarray analysis of the M. tuberculosis ramB (ramBMT) mutant and the wild type furthermore showed that apart from icl1 and the ramBMT operon, the expression of all other M. tuberculosis genes involved in acetate metabolism remain unchanged in the mutant. Thus, RamBMT has a more specific regulatory function as RamB from C. glutamicum and is confined to expression control of icl1 and the ramBMT operon.
With nearly two million deaths each year and approximately one-third of the world's population harboring a latent infection, tuberculosis is a major contributor to the global burden of disease (43). Since Mycobacterium tuberculosis, the causative agent of tuberculosis, is able to persist within a host for decades, it must adapt its metabolism to tissue environments. Fatty acids are readily available within the macrophage phagosome, and M. tuberculosis might obtain fatty acids from lung surfactant (29), by hydrolysis of phagosomal membrane lipids (23), or from macrophage triacylglycerol stores (26). Genes encoding fatty acid β-oxidation enzymes are extensively duplicated in the mycobacterial genome (9) and, together with the glyoxylate cycle gene icl1, they are upregulated during infection of macrophages and mice (12, 37, 40). Therefore, it is generally assumed that M. tuberculosis subsists mainly on fatty acids rather than on carbohydrates within the host (6, 30).
When fatty acids are the sole source of carbon, a functional glyoxylate cycle with its key enzymes isocitrate lyase (Icl) and malate synthase (MS) is necessary for bacterial survival. The glyoxylate cycle is an anaplerotic pathway metabolizing acetyl coenzyme A (acetyl-CoA), the end product of β-oxidation. Icl cleaves isocitrate into succinate and glyoxylate, whereas MS subsequently condenses glyoxylate with another molecule, acetyl-CoA, to malate. Overall, the glyoxylate cycle ensures the bypass of two oxidative steps of the tricarboxylic acid cycle, thereby allowing the synthesis of carbohydrates from fatty acids and anaplerosis of tricarboxylic acid cycle intermediates. M. tuberculosis contains two Icl enzymes (Icl1, a prokaryotelike isoform, and Icl2, a eukaryotelike isoform) and one MS. Icl activity is necessary for virulence in mice and bacterial growth in macrophages (29), and Icl1-deficient M. tuberculosis are unable to sustain a chronic, persistent infection in mice (27). The facts that Icl enzymes are essential for M. tuberculosis virulence and that they are absent in mammals make them promising potential drug targets.
M. tuberculosis belongs to the group of the Corynebacterianeae (39), which includes also nonpathogenic bacteria, such as Corynebacterium glutamicum. This latter organism is widely used in the industrial production of amino acids, and its metabolism has been intensively studied (summarized in references 8 and 13). In the course of studies on the acetate metabolism of C. glutamicum, RamB (for the regulator of acetate metabolism B) has been identified to control the expression of genes encoding the glyoxylate cycle enzymes Icl and MS, as well as the genes for enzymes involved in acetate activation, i.e., acetate kinase (AK) and phosphotransacetylase (Pta) (2, 17). Although generally the specific activities of AK, Pta, Icl, and MS are high during growth on acetate and low during growth on glucose, deletion of the ramB gene led to high-level specific activities of all four enzymes during growth on glucose (17). Hence, C. glutamicum RamB functions as a repressor of the acetate activating enzymes, as well as the glyoxylate cycle enzymes under conditions when their activity is not needed. Furthermore, C. glutamicum RamB negatively regulates the adhA and ald genes, encoding alcohol and acetaldehyde dehydrogenases (1, 3), its own expression (10), and it acts as an activator of the pyruvate dehydrogenase complex subunit E1p gene aceE (5).
The open reading frame Rv0465c of M. tuberculosis H37Rv encodes a hypothetical protein which shares 72% similarity and 56% sequence identity with RamB from C. glutamicum. It is separated from icl1 by one open reading frame on the mycobacterial chromosome and, interestingly, a 13-bp DNA motif (CAAAATTTGCAAA) very similar to the RamB consensus binding motif in C. glutamicum (AA/GAACTTTGCAAA) could be identified within the promoter region of icl1 (17). This led to our hypothesis that Rv0465c from M. tuberculosis is an orthologue of ramB from C. glutamicum and might be involved in the regulation of icl1 expression and possibly also in the expression of other genes involved in acetate metabolism or the glyoxylate cycle.
In the present study, an M. tuberculosis ramB deletion mutant was constructed, and gene expression of the mutant was compared to that of the wild type (WT) on the whole genome and single gene level. These experiments, together with enzyme activity assays and DNA band-shift assays, provide clear evidence that M. tuberculosis RamB (RamBMT) acts as a transcriptional repressor that downregulates icl1 expression in M. tuberculosis when grown in the presence of glucose. Furthermore, we show that ramBMT forms an operon with the open reading frame Rv0464c and that this operon is subject to negative regulation by RamBMT which is independent of the available carbon source. We also show that of all enzymes involved in acetate activation and the glyoxylate cycle, only the expression of icl1 is induced in WT M. tuberculosis during growth in the presence of acetate and that RamBMT does not regulate the expression of any further genes.
MATERIALS AND METHODS
Bacteria, plasmids, oligonucleotides, and culture conditions.
Bacterial strains, plasmids, and oligonucleotides (primers), as well as their relevant characteristics and sources, are given in the supplemental material (see Tables S1 and S2 in the supplemental material, respectively). WT M. tuberculosis H37Rv and derivatives were cultured aerobically at 37°C in Middlebrook 7H9 broth or on Middlebrook 7H10 agar plates (Difco Laboratories, Detroit, MI) supplemented with 0.5% glycerol, 10% ADS (0.5% bovine serum albumin fraction V, 0.2% glucose, 140 mM NaCl) and 0.05% Tween 80 (broth only). For gene expression analysis, M. tuberculosis cells were incubated in M9 minimal salts medium (Serva, Heidelberg, Germany) containing 2 mM MgSO4, 0.1 mM CaCl2, and either d-glucose or sodium acetate at 1% (wt/vol). For Icl activity assays, M. tuberculosis was cultured in 7H9 broth containing 0.5% bovine serum albumin fraction V, 0.085% NaCl, 0.05% Tween 80, and either d-glucose or sodium acetate at 0.2% (wt/vol). TY medium was used as complex medium for C. glutamicum (36). The minimal medium used for C. glutamicum has been described (14) and contained glucose at a concentration of 1% (wt/vol). Cultivation was done at 30°C as 50 ml-cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm. Escherichia coli HB101 or E. coli DH5α were used as host for molecular cloning and cultured aerobically at 37°C in LB broth or on LB agar plates (28). Spectinomycin (100 μg/ml), hygromycin (50 μg/ml), or ampicillin (100 μg/ml) was added to the medium when necessary. Growth of the bacteria was monitored by measuring the optical density at 600 nm (OD600).
Construction of plasmid pEKEx5-Rv0465cx6His.
Rv0465c was amplified from chromosomal DNA of M. tuberculosis H37Rv by PCR with the primers 1 and 2. The PCR product was digested with BamHI and PstI, ligated into BamHI/PstI-restricted plasmid pEKEx5, (15; see Table 1 in the supplemental material), and transformed into E. coli DH5α. After transformation, the plasmid was isolated from E. coli DH5α by using the NucleoSpin extract II kit (Macherey-Nagel) and checked by restriction analysis and PCR. The original plasmid pEKEx5 and the newly constructed plasmid pEKEx5-Rv0465cx6His were transformed into C. glutamicum WT and C. glutamicum RG1 (ramB-negative) cells. C. glutamicum (pEKEx5), C. glutamicum RG1(pEKEx5), and C. glutamicum RG1(pEKEx5-Rv0465cx6His) were used for complementation experiments. In addition, C. glutamicum (pEKEx5-Rv0465cx6His) was used for synthesis and preparation of His6-tagged Rv0465c fusion protein.
Preparation of His-tagged Rv0465c fusion protein.
The synthesis of N-terminal His-tagged Rv0465c protein was induced in C. glutamicum (pEKEx5-Rv0465cx6His) by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when the culture had reached an OD600 of ∼2. The cells were grown to an OD600 of ∼8, harvested, washed twice with 50 mM phosphate buffer containing 300 mM NaCl and 10 mM imidazole (pH 8.0), and disrupted by preparation in a RiboLyser (Hybaid, Teddington, United Kingdom). Purification of the fusion protein was performed with nickel-nitrilotriacetic acid affinity chromatography according to the instructions of Qiagen (Hilden, Germany). For desalting, the Rv0465c fusion protein was loaded onto an equilibrated PD-10 column (Amersham Biosciences) and eluted with elution buffer (10 mM Tris-HCl [pH 7.6], 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]). The purified fusion protein was more than 90% pure and directly applied for promoter binding assays.
Promoter binding assays with His-tagged Rv0465c fusion protein.
The binding of His-tagged Rv0465c protein to the promoter regions of acs (Rv3667), ackA (Rv0409), pta (Rv0408), icl1 (Rv0467), icl2 (Rv1915/1916), glcB (Rv1837c), Rv0465c, and Rv0464c was tested by electrophoretic mobility shift assays (EMSAs) using the DNA probes indicated in Results. The respective DNA fragments were generated by PCR and purified with a NucleoSpin extract II kit. In the binding assays, 100- to 300-ng portions of the fragments were incubated with various amounts of Rv0465cx6His protein (0 to 1 μg) in 20 μl of 10 mM Tris-HCl reaction buffer (pH 7.6) containing 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% (wt/vol) glycerol, and 1 μg of poly(dI-dC) for 20 min at room temperature. Subsequently, the mixture was separated on a 2% agarose gel in 1 × TAE buffer (40 mM Tris-HCl [pH 7.5], 20 mM acetate, 1 mM EDTA) at 70 V and 80 mA and stained with ethidium bromide.
Generation and complementation of an Rv0465c deletion mutant in M. tuberculosis.
A 1,008-bp in-frame deletion in Rv0465c was generated by two-step homologous recombination, according to the method of Pavelka and Jacobs (32). The cosmid MAS15, containing the Rv0465c genomic region of M. tuberculosis H37Rv was obtained by colony blot hybridization from a genomic cosmid library (4). The DNA probe for hybridization was generated by PCR using primers 296 and 297. According to the protocol given by the manufacturer, the DNA probe was gel purified and labeled with a DIG DNA labeling and detection kit (Roche, Mannheim, Germany). A 6,273-bp SpeI-restricted fragment of cosmid pMAS15 containing the intact Rv0465c gene was subcloned into SpeI-restricted pBluescript SK(−), leading to pMAS9, and digestion with BglII and subsequent religation led to a plasmid carrying a 1,008-bp in-frame deletion within Rv0465c (pMAS10B). To eliminate one SpeI restriction site, pMAS10B was digested with HindIII and StuI, and subsequent 5′-overhang fill-in using Klenow enzyme and religation led to pMAS11. A 7,710-bp fragment containing hyg (hygromycin resistance) and sacB (sucrose sensitivity), which was obtained from pYUB657 by digesting with XbaI and SpeI, was cloned into the SpeI restriction site of pMAS11. The resulting plasmid pMAS14 was transformed into M. tuberculosis H37Rv by electroporation, and transformants were selected on 7H10 agar containing hygromycin. Integration of pMAS14 next to the Rv0465c locus was confirmed by PCR, using primers 296 and 345, and by Southern blot hybridization, using NheI-restricted genomic DNA and a probe generated by PCR with the primers 296 and 297. Cointegrants were cultured in 7H9 medium without selective pressure to allow for a second crossover. Sucrose-containing 7H10 agar plates were used for counterselection as described previously (32) to obtain clones in which double crossover had occurred. The genotype of Rv0465c deletion mutants was confirmed by PCR, using the primers 296 and 345, and Southern blot hybridization, using AgeI-restricted genomic DNA and a probe generated by PCR using the primers 343 and 344.
The complementing plasmid pMAS32 was constructed by cloning a 7,359-bp fragment carrying the intact Rv0465c gene, obtained by digesting pMAS9 with EcoRV, into the PvuII restriction site of the integrating vector pMV306.hyg. pMAS32 was transformed into MAS24 by electroporation, and transformants were selected on 7H10 agar containing hygromycin. Uptake of the plasmid was confirmed by two PCRs: one amplifying the DNA region containing the Rv0465c deletion (using primers 296 and 345) and one amplifying part of the oriE origin of replication (using primers 161 and 162).
Extraction of RNA for gene expression analysis.
RNA was extracted from cultures of M. tuberculosis incubated with M9 medium supplemented with either acetate or glucose. Since this minimal medium does not sustain robust growth of M. tuberculosis, bacteria were precultured in 7H9 broth to an OD600 of 0.5 to 0.7. The bacteria were then washed twice with M9 minimal medium (containing either acetate or glucose), resuspended in the respective minimal medium, and incubated in a shaking incubator for 3 or 24 h. The cultures were then incubated with an equal amount of 5 M GTC buffer (5 M guanidinium isothiocyanate, 0.5% n-laurylsarcosine, 0.7% sodium citrate, 0.7% β-mercaptoethanol) at room temperature for 15 min, followed by centrifugation at 4,500 × g for 15 min. The pellet was resuspended in 1 ml of TRIzol (Invitrogen, Karlsruhe, Germany) and incubated for 15 min at room temperature. Cells were lysed in a Lysing Matrix B tube (MP Biomedicals, Illkirch, France) using the Hybaid RiboLyser at setting 6.0 for 40 s. The tubes were centrifuged at 13,000 × g for 3 min, and the supernatant was mixed with chloroform. After centrifugation at 13,000 × g for 7 min, the upper phase was mixed with an equal volume of ethanol (70%), and RNA was purified by using an RNeasy kit (Qiagen) according to the manufacturer's instructions. The optional DNase I digest, using DNase I supplied by Qiagen, was extended to 1 h. A second DNase I (NEB) digest was carried out after eluting the RNA from the columns for 45 min at 37°C. Subsequent RNA cleanup was performed with the RNeasy kit. The RNA concentration and purity were determined photometrically at 260 and 280 nm.
Reverse transcription-PCR.
Total RNA was isolated from M. tuberculosis as described above and transcribed into cDNA, using the reverse transcriptase Superscript II and random primers (Invitrogen). First-strand synthesis was performed in a T3000 Thermocycler (Biometra, Göttingen, Germany) according to the Invitrogen protocol. For each RNA sample, two reactions were prepared—one as described in the protocol and one control that was treated in exactly the same way except that no Superscript II was added—to verify efficient DNase I treatment. The cDNA generated served as a template for subsequent PCRs by using primer pairs 391 to 398, as listed in Table S2 in the supplemental material. As controls, M. tuberculosis H37Rv genomic DNA and H2O were used as templates. M. tuberculosis H37Rv genomic DNA was obtained by boiling a broth culture for 10 min and removing the cell pellet by centrifugation. PCRs were performed in a T3000 Thermocycler (Biometra).
Quantitative real-time PCR.
TaqMan real-time PCR was used for the quantification of mRNA using a custom gene expression assay (Applied Biosystems, Foster City, CA) for each gene studied. For measuring ramBMT expression in the ramBMT deletion mutant, a TaqMan gene expression assay specific for the nondeleted part of ramBMT was used. Reactions were performed in a 96-well plate format according to the Applied Biosystems protocol. Samples were measured in duplicates and quantified using a standard curve obtained by serial dilution of a cDNA standard generated by pooling all cDNAs analyzed. TaqMan real-time PCR was performed in an ABI Prism 7000 sequence detection system (Applied Biosystems). As normalized results, the quotient of sample gene mRNA quantity and sigA mRNA quantity were calculated. The cutoff for significant regulation was arbitrarily set at 1.8-fold change.
Microarray analysis.
A custom-designed GeneChip M. tuberculosis array (Affymetrix, Santa Clara, CA) was used for DNA microarray analysis. RNA was extracted as described above. cDNA synthesis, fragmentation, labeling, target hybridization, staining, and scanning of the probe array were performed according to the Affymetrix protocol and as described previously (25). Analysis of microarray data was performed by using the Affymetrix GCOS 1.2 software. For normalization, all array experiments were scaled to a target intensity of 150, otherwise using the default values of GCOS 1.2. Further downstream analysis was performed using ArrayAssist (Stratagene, La Jolla, CA). The data were normalized by using the RMA (Robust Multi-Array Average) algorithm. The cutoff for significant regulation was arbitrarily set at 1.8-fold change.
Preparation of cell extracts of M. tuberculosis and of C. glutamicum.
M. tuberculosis cells were grown to an OD600 of 0.65 to 0.85, washed with 50 mM Tris-HCl buffer I (pH 7.6; containing 10 mM MgCl2, 1 mM EDTA, 1 mM DTT, and 30% glycerol), and resuspended in 500 μl of the same buffer. The cell suspension was transferred to a Lysing Matrix B tube (MP Biomedicals), and mechanical disruption was achieved by repeated shaking in a Silamat S5 Amalgamator (Ivoclar Vivadent GmbH, Ellwangen, Germany) five times for 25 s, with intermittent cooling on ice for 5 min. Silica spheres and cellular debris were removed by centrifugation at 13,000 × g for 10 min. The supernatant was used for the Icl activity assay.
For preparation of cell extracts of C. glutamicum, cells were grown in minimal medium to the exponential growth phase, washed twice in 50 mM Tris-HCl buffer I, and resuspended in 700 μl of the same buffer. The cell suspension was placed into 2-ml screwcap vials together with 250 mg of glass beads (Sigma-Aldrich) and subjected (five times for 25 s, setting 6.5) to mechanical disruption with the Hybaid RiboLyser at 4°C with intermittent cooling on ice for 5 min. After disruption, glass beads and cellular debris were removed by two consecutive centrifugation steps (20,000 × g, 4°C, 20 min, and 45,000 × g, 4°C, 120 min) and the supernatant was used for the assays.
The protein concentration in the cell extracts was determined by measuring absorbance at 595 nm after incubation with dye reagent concentrate (Bio-Rad Laboratories GmbH, Munich, Germany). Bovine serum albumin was used to prepare a standard curve of absorbance.
Icl activity assay.
Icl activity was assayed by measuring the conversion of glyoxylate to glycolate by lactate dehydrogenase I and monitoring the concomitant decrease of NADH at 340 nm (41). The assay was performed with cell extracts in 1 ml of a buffer containing 50 mM morpholinepropanesulfonic acid (pH 7.3), 5 mM DTT, 15 mM MgCl2, 0.2 mM NADH, 1 mM EDTA, and 24 U of lactate dehydrogenase (pig heart isoenzyme I). Reactions were started by adding 50 μl of 100 mM threo-Ds-isocitrate. One unit of Icl activity corresponds to 1 μmol of glyoxylate formed per min.
Computational analysis.
Searches for protein domains were performed by using the SUPERFAMILY server (19) and the PROSITE database (22).
Statistics.
Student unpaired t test (two-tailed) was used to assess statistical significance. P values of <0.05 were considered significant.
RESULTS
Analysis of the Rv0465c genomic locus.
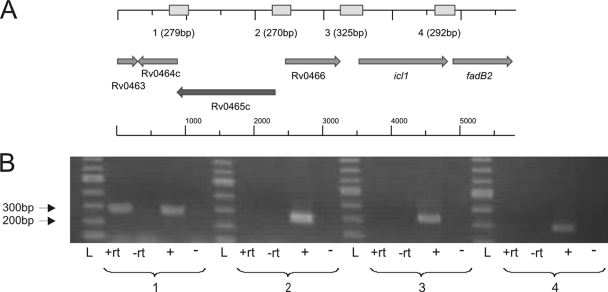
The open reading frame Rv0465c of M. tuberculosis H37Rv encodes a protein that has been annotated as a probable transcriptional regulator (9) and that shares significant identity with the RamB protein of C. glutamicum (see the introduction). It has a length of 475 amino acids (aa) and contains an N-terminal lambda repressor-like DNA-binding domain, which includes a helix-turn-helix domain at aa 21 to 40 (PROSITE database). The chromosomal region of M. tuberculosis containing Rv0465c and surrounding genes is depicted in Fig. 1A. Rv0465c is located ∼1.2 kb upstream of icl1 (Rv0467), which encodes one of the two mycobacterial Icl enzymes and which is preceded by a sequence motif with similarity to the RamB consensus motif in C. glutamicum (17) (see also the introduction). Rv0465c is separated from icl1 by the open reading frame Rv0466, which encodes an uncharacterized hypothetical protein. Rv0466 and icl1 are transcribed in the opposite direction to Rv0465c. Downstream of Rv0465c and overlapping with it by four base pairs, the open reading frame Rv0464c is located, which encodes a further, as-yet-uncharacterized hypothetical protein.
FIG. 1.
Rv0465c (ramBMT) region on the chromosome of M. tuberculosis. (A) Gene arrangement (arrows) and expected PCR products (boxes above); (B) transcriptional organization of the Rv0465c genomic region determined by reverse transcription-PCR amplification of the intergenic regions between individual open reading frames. Numbers: 1, Rv0464c and Rv0465c; 2, Rv0465c and Rv0466; 3, Rv0466 and icl1; 4, icl1 and fadB2. L, DNA ladder; +rt, cDNA synthesized by reverse transcriptase; −rt, negative control (no reverse transcriptase added); +, M. tuberculosis H37Rv genomic DNA; −, negative control (H2O).
To elucidate the transcriptional organization of the Rv0465c/icl1 genomic region, the intergenic regions between the genes located next to Rv0465c were subjected to reverse transcription-PCR, using mycobacterial cDNA as a template. As shown in Fig. 1B, a PCR product could only be obtained for the region between Rv0465c and Rv0464c. This observation suggests that these two genes are cotranscribed as a single polycistronic mRNA and form an operon. No PCR product was obtained for the intergenic regions between icl1 and the genes located directly up- or downstream (Fig. 1B), indicating that icl1 is transcribed as a single gene and is not part of an operon.
Substitution of ramB in C. glutamicum with Rv0465c.
Previously, it has been shown that glucose-grown C. glutamicum WT cells possess low specific Icl activity, whereas acetate-grown cells possess high specific Icl activity (42). In contrast, the ramB-deficient strain C. glutamicum RG1 showed high Icl activities also in glucose medium, and this deregulation was found to be due to the lack of transcriptional repression of the respective Icl gene by RamB (17). In order to test for a ramB-like function of Rv0465c in C. glutamicum, we tested for complementation of the Icl phenotype in C. glutamicum RG1. For this purpose, plasmid pEKEx5-Rv0465cx6His, containing Rv0465c from M. tuberculosis was transformed into the ramB-deficient mutant C. glutamicum RG1, and the specific Icl activities of the resulting strain C. glutamicum RG1(pEKEx5-Rv0465c) were compared to those of the WT and the ramB-deficient mutant C. glutamicum RG1 carrying the empty vector [C. glutamicum WT(pEKEx5) and C. glutamicum RG1(pEKEx5), respectively]. As previously observed with C. glutamicum WT (17), the specific Icl activities in C. glutamicum WT (pEKEx5) were relatively low (90 ± 10 mU mg of protein−1) when the cells were grown in glucose medium. As C. glutamicum RG1 (17), C. glutamicum RG1(pEKEx5) exhibited ∼5-fold higher specific Icl activities (450 ± 30 mU mg of protein−1) than the WT derivative. In contrast, C. glutamicum RG1 carrying plasmid pEKEx5-Rv0465cx6His showed the same low specific Icl activity of 90 ± 20 mU mg of protein−1 as C. glutamicum WT (pEKEx5). These results indicate that the M. tuberculosis Rv0465c gene functionally substitutes ramB in C. glutamicum and suggest similar roles of the respective proteins in both organisms. In accordance with the similarity of the Rv0465c protein with RamB and due to the ability of functional substitution, we designate Rv0465c in the following discussion as ramBMT.
Binding of purified RamBMT to the promoter regions of genes involved in acetate metabolism of M. tuberculosis.
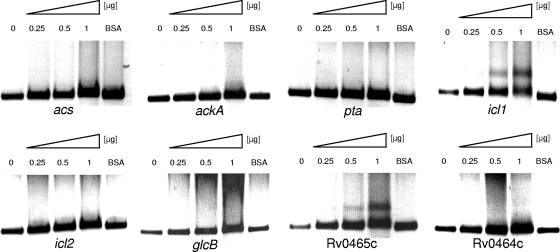
In the genome of M. tuberculosis, several genes have been annotated as proteins possibly involved in acetate activation and in glyoxylate cycle: Rv3667 as acs, encoding an acetyl-CoA synthetase; Rv0409 as ackA, encoding an AK; Rv0408 as pta, encoding a Pta; Rv0467 as icl1 and Rv1915/1916 as icl2, encoding the two mycobacterial Icl enzymes; and Rv1837c as glcB, encoding MS. To test for a direct interaction of RamBMT and the promoter regions of acs, ackA, pta, icl1, icl2, and glcB, we performed EMSA experiments with the respective promoter fragments and RamBMT. In addition, we tested the promoter regions of ramBMT itself and of the subsequent Rv0464c gene for binding to RamBMT. For these purposes, RamBMT was synthesized as a His-tagged fusion protein in C. glutamicum WT(pEKEx5-Rv0465cx6His) and purified by affinity chromatography. Different amounts of purified RamBMT were incubated separately with the promoter fragments and separated on agarose gels. As shown in Fig. 2, no retardation was observed with the acs, ackA, pta, icl2, glcB, and Rv0464c promoter fragments. In contrast, the icl1 and ramBMT promoter fragments clearly were retarded by RamBMT. It is, however, noteworthy to mention that the retardation of both fragments was not complete with 1 μg of RamBMT, and also with 2 μg of RamBMT no complete shift was observed (data not shown), suggesting that the binding of RamBMT to both regions is rather weak. These results indicate (i) binding of RamBMT to the promoter regions of icl1 and of its own gene and (ii) no binding to the other promoter regions tested.
FIG. 2.
Representative EMSAs using His-tagged Rv0465c protein (RamBMT). The genes of the tested promoter fragments are indicated below the different parts of the gels. The fragment sizes were 468 bp (acs), 505 bp (ackA), 509 bp (pta), 534 bp (icl1), 615 bp (icl2), 509 bp (glcB), 515 bp (Rv0465c), and 510 bp (Rv0464c). The “BSA” lanes represent negative controls with 1 μg of bovine serum albumin used instead of Rv0465c protein.
Effect of ramBMT inactivation on icl1, ramBMT, and icl2 expression.
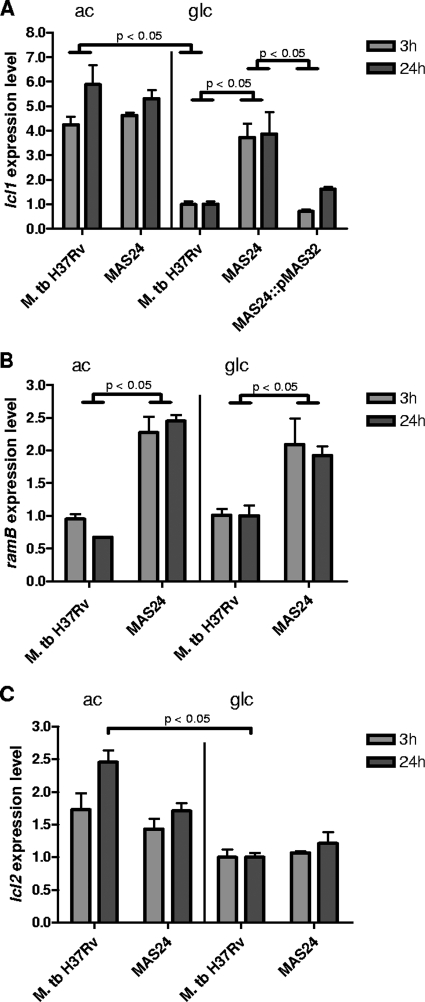
To determine whether RamBMT in fact functions as a transcriptional regulator in M. tuberculosis, the mutant MAS24, carrying a 1,008-bp in-frame deletion within ramBMT, was constructed and analyzed. The deletion was confirmed by PCR and Southern blot analysis (data not shown). Having gained evidence that RamBMT binds to the promoter regions of icl1 and the ramBMT/Rv0464c operon, we determined the mRNA levels (expression levels) of icl1 and ramBMT in the ramBMT-deficient mutant M. tuberculosis MAS24 in comparison to the WT by quantitative real-time PCR (Fig. 3A and B). In the latter strain, the expression level of icl1 was high during incubation with acetate and significantly lower during incubation with glucose (4.2-fold after 3 h and 5.9-fold after 24 h; Fig. 3A). However, in the mutant MAS24, icl1 expression remained high during incubation with glucose and reached approximately the same level observed in the WT grown in the presence of acetate. This derepression pattern of icl1 in glucose-medium could be reversed by reintroduction ramBMT on an integrating plasmid (strain MAS24::pMAS32). M. tuberculosis MAS24::pMAS32 showed a similar icl1 expression level as the WT. No difference in icl1 expression between the WT and MAS24 was observed during incubation with acetate (Fig. 3A). The ramBMT expression levels did not differ significantly in M. tuberculosis H37Rv when incubated with acetate or glucose (Fig. 3B). However, ramBMT expression levels were found to be 2.4- to 3.6-fold higher in MAS24 than in the WT during incubation with acetate and 1.8- to 2.1-fold higher during incubation with glucose (Fig. 3B). Taken together, these results indicate (i) that icl1 expression in M. tuberculosis is downregulated when glucose is used as carbon source, i.e., under conditions when Icl activity is not needed; (ii) that Rv0465c is responsible for repression of icl1 in the presence of glucose as carbon source; (iii) that ramBMT expression is similar in cells grown on glucose or acetate; and (iv) that ramBMT is repressed by its own gene product, i.e., is negatively autoregulated independent of the carbon source.
FIG. 3.
Expression level of icl1, ramBMT, and icl2 in M. tuberculosis (M. tb.) H37Rv, the ramBMT deletion mutant MAS24, and the complemented mutant MAS24::pMAS32 (where applicable) during incubation with acetate (ac) or glucose (glc) as sole carbon source. (A) icl1; (B) ramBMT; (C) icl2. For normalization, the expression level of the WT during growth on glucose was set to 1. P values are indicated for fold changes of >1.8. Means were calculated from three independent experiments and two determinations per experiment. Error bars represent standard errors of the mean.
We tested also for icl2 expression (determined as Rv1915 mRNA levels) in M. tuberculosis H37Rv and ramBMT-deficient mutant MAS24 in acetate and glucose media. As shown in Fig. 3C, icl2 expression was also higher in bacteria incubated with acetate as the sole carbon source compared to bacteria incubated with glucose, but to a lesser extent than icl1 and only after an incubation period of 24 h. Deletion of ramBMT had a minimal effect on icl2 expression, resulting in a 1.4-fold higher expression of icl2 in MAS24 after incubation with acetate for 24 h (Fig. 3C), compared to the WT. However, this effect is very small. Taking into consideration that icl2 of M. tuberculosis H37Rv is thought to be a nonfunctional pseudogene (21), it most likely has no biological significance.
Whole-genome expression analysis of the ramBMT deletion mutant MAS24.
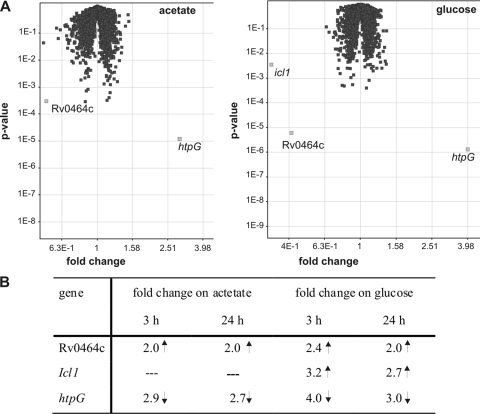
To identify potential further genes regulated by RamBMT, DNA microarray analysis was carried out, using a DNA chip covering all of the nearly 4,000 open reading frames of M. tuberculosis H37Rv. The gene expression pattern of M. tuberculosis H37Rv was compared to that of the ramBMT deletion mutant MAS24 after incubation in minimal medium containing either acetate or glucose. Surprisingly, only two genes (Rv0464c and htpG, encoding a putative heat shock protein) and three genes (Rv0464c, htpG, and icl1) were found to be differentially expressed in the WT and in strain MAS24 after 3 h of incubation with acetate or glucose, respectively (Fig. 4A). The same expression patterns were observed after 24 h of incubation with both carbon sources (Fig. 4B). In strain MAS24, Rv0464c was found to be upregulated 2.0- to 2.4-fold and htpG to be downregulated 2.7- to 4-fold in either medium. In accordance with our results obtained by quantitative real-time PCR (see above), the icl1 mRNA level was not influenced in the ramBMT deletion mutant MAS24 during incubation with acetate; however, it was upregulated 2.7- to 3.2-fold in strain MAS24 in glucose medium (Fig. 4B), supporting our finding that RamBMT negatively regulates icl1 expression in the presence of glucose.
FIG. 4.
Microarray analysis (combined data from four independent experiments) comparing gene expression of M. tuberculosis H37Rv and the ramBMT deletion mutant MAS24 during incubation with acetate or glucose. (A) Volcano plot of fold changes and P values after 3 h of incubation with acetate or glucose (WT versus mutant). Differentially expressed genes are in light gray. (B) Fold change of genes found to be differentially expressed in the WT and the mutant MAS24 during incubation with acetate or glucose for 3 h or 24 h (expressed as the fold change of gene expression in MAS24 compared to the WT under the same conditions).
The higher mRNA level of Rv0464c in the ramBMT deletion mutant MAS24 is in agreement with our results that Rv0464c forms an operon with ramBMT and that ramBMT expression is repressed by its own gene product (see above). Autoregulation of ramBMT itself could not be detected by DNA microarray analysis since the DNA probes cover the whole range of the ramBMT gene including the deleted part. Therefore, the signal for ramBMT expression was extremely low in the ramBMT deletion mutant (10.5- to 20.5-fold lower than in the WT [data not shown]), independent of regulation.
One additional gene, encoding the heat shock protein HtpG, was found to be downregulated in the ramBMT deletion mutant on both carbon sources. However, this protein has no known function in acetate metabolism. Further experiments using EMSAs and quantitative real-time PCR showed that RamBMT does not bind to the promoter region of htpG and downregulation of htpG in the ramBMT deletion mutant could not be complemented by introducing an intact copy of ramBMT into the ramBMT deletion mutant (data not shown).
Taken together, the DNA microarray experiments confirmed the results obtained by quantitative real-time PCR, showing that RamBMT downregulates icl1 expression when glucose is the sole carbon source and expression of the ramBMT/Rv0464c operon when glucose or acetate is the carbon source. Interestingly, and in contrast to the findings in C. glutamicum, the results of the DNA microarray analysis suggests that ramBMT does not regulate the expression of other genes involved in acetate activation or in the glyoxylate cycle.
RamBMT inactivation leads to increased Icl activity during growth on glucose.
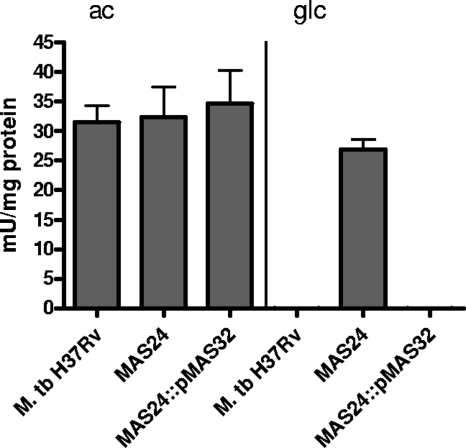
To prove that inactivation of ramBMT influences not only mRNA synthesis but also has an effect on enzyme activity, specific isocitrate lyase activities were determined in crude extracts of the M. tuberculosis WT, the ramBMT-deficient mutant MAS24, and the complemented mutant MAS24::pMAS32 during growth in medium containing either acetate or glucose (Fig. 5). In the WT strain and also in M. tuberculosis MAS24::pMAS32, Icl activity was below the detection limit when these strains were grown on glucose, whereas during growth on acetate, Icl activity increased to 32 to 35 mU mg of protein−1. In contrast, glucose- and acetate-grown cells of the ramBMT mutant MAS24 showed high specific Icl activities on either carbon source, with Icl activity in glucose-grown cells reaching nearly identical levels as in acetate-grown cells (27 mU mg of protein−1 in glucose-grown cells and 32 mU mg of protein−1 in acetate-grown cells). These data correspond to the increased levels of icl1 mRNA observed in MAS24 grown on glucose (Fig. 3 and 4) and indicate that downregulation of icl1 expression by RamBMT in glucose-incubated cells in fact leads to a decreased amount of enzyme and therefore to a lower specific Icl activity. The finding that Icl activity was abolished in the complemented mutant MAS24::pMAS32 during growth on glucose further emphasizes that deregulation of Icl activity in MAS24 is in fact due to ramBMT deletion.
FIG. 5.
Specific Icl activities in cell extracts of M. tuberculosis H37Rv, the ramBMT deletion mutant MAS24 and the complemented mutant MAS24::pMAS32 during growth on acetate (ac) or glucose (glc). Means were calculated from three independent experiments and two determinations per experiment.
Expression analysis of acetate metabolism genes during growth of M. tuberculosis on acetate or glucose.
As our results suggested that RamBMT regulates the expression of icl1, but not the expression of other genes involved in acetate metabolism (i.e., acs, ackA, pta, icl2, and glcB; see above), we compared the DNA microarray data to determine whether these genes were differentially expressed in the WT grown with acetate or with glucose (Table 1) The icl1 gene was the only gene whose expression level increased by a factor of 3 to 4 when incubated with acetate instead of glucose. The expression of Rv1915/Rv1916, encoding Icl2, increased only very slightly, whereas expression of glcB, encoding the mycobacterial MS, did not change at all. The expression of the three genes potentially encoding enzymes involved in acetate activation did not differ in acetate- and glucose-grown cells (Table 1). These results indicate that of all genes specific for acetate metabolism, only icl1 is upregulated in the presence of acetate, and therefore support our findings that only icl1 expression is regulated by RamBMT.
TABLE 1.
Microarray analysis dataa
| Gene | Fold changeb in expression at: |
|
|---|---|---|
| 3 h | 24 h | |
| icl1 | +4.2* | +3.0* |
| icl2a (Rv1915) | +1.1 | +1.1 |
| icl2b (Rv1916) | +1.4 | +1.5 |
| glcB | +1.1 | 1.0 |
| ackA | 1.0 | +1.2 |
| Pta | -1.1 | +1.1 |
| Acs | -1.2 | +1.1 |
The combined data from four independent experiments are presented.
That is the fold change in gene expression of genes encoding enzymes of the glyoxylate cycle and potential acetate-activating enzymes after incubation with acetate (compared to incubation with glucose). *, P < 0.05 (only indicated for fold changes of >1.8).
DISCUSSION
Together with enzymes of the citric acid cycle, the two mycobacterial Icl enzymes (Icl1 and Icl2) and MS constitute the glyoxylate cycle in M. tuberculosis, which metabolizes acetyl-CoA derived from carbon sources such as acetate or fatty acids. Icl activity has been shown to be essential not only for growth of M. tuberculosis on acetate and fatty acids, but also for growth and persistence of M. tuberculosis in mice, supporting the hypothesis that within the host, M. tuberculosis subsists on fatty acids rather than on carbohydrates (6, 29, 30). Interestingly, M. tuberculosis Icl1 and Icl2 have been shown to be dual-specificity enzymes that are able to function as Icl in the glyoxylate cycle (acetyl-CoA metabolism), as well as methylisocitrate lyases in the methylcitrate cycle (propionyl-CoA metabolism) (20, 31). In earlier studies, Icl1 of M. tuberculosis has been shown to have a 10-fold-higher affinity to its substrate isocitrate and a 3-fold-higher specific activity than Icl2, leading the authors to speculate that Icl2 plays a subordinate role in M. tuberculosis (21). This is also supported by the finding that transcription of icl1 but not icl2 is upregulated in M. tuberculosis in the human lung (16). The results obtained by Smith and coworkers (38) indicated regulation of Icl but not of MS in M. tuberculosis grown on glucose, acetate, and palmitate. Expression of the MS gene glcB remained also unaffected in infected mice compared to bacteria grown in vitro with glucose and glycerol (40). In contrast, in C. glutamicum, which is a close relative of M. tuberculosis, both enzymes, Icl and MS, are coordinately regulated. Interestingly, the genes encoding these enzymes are located next to each other on the chromosome, whereas in M. tuberculosis they are unlinked (17).
The regulatory mechanism or the regulatory proteins that control the expression of icl1 in M. tuberculosis have not been investigated thus far. In C. glutamicum, regulation of Icl and MS (genes annotated as aceA and aceB, respectively) has been studied extensively (reviewed in reference 18). The organism possesses only a single Icl (33), and both Icl and MS are essential for the growth of C. glutamicum on acetate (33, 34). Besides aceA and aceB, expression of the pta-ack-operon, encoding Pta and AK, is also upregulated on acetate (35). Expression of aceA, aceB, pta, and ackA is controlled by the two regulators RamA and RamB. The former represents a transcriptional activator, the latter a transcriptional repressor of the aforementioned genes (11, 17). Whereas RamA homologues and/or orthologues (with >40% sequence identity) were only found in some corynebacteria and in Streptomyces spp. (7, 11), genes encoding proteins with significant sequence similarity to RamB have been identified in other species of Corynebacterium (Corynebacterium efficiens, C. diphtheriae, C. urealyticum, and C. jeikeium; 91 to 58% identity) and also in related genera, such as Nocardia (Nocardia farcinica; 56%), Rhodococcus (55%), and Mycobacterium (Rv0465c protein of M. tuberculosis H37Rv, BCG_0505c protein of M. bovis BCG, MMAR_0790 and Mvan_0799 proteins of M. marinum and M. vanbaalenii; 72 to 55%). Thus, it can be concluded that RamB and its homologues or orthologues are specific for the suborder Corynebacterianeae. However, in none of these organisms except C. glutamicum has the function of RamB or RamB-like proteins been experimentally investigated thus far.
The results from the present study indicated functional similarity of C. glutamicum RamB and RamB from M. tuberculosis. As RamB in C. glutamicum, RamBMT represses icl1 expression during incubation with glucose, whereas this repression is released during incubation with acetate. These findings were further substantiated by Icl activity measurements, which showed a similar pattern as determined by the gene expression experiments. However, upregulation of icl1 mRNA by acetate was quite modest compared to the much more dramatic effect of upregulation of Icl activity by acetate. This difference might be due to the fact that different culture media were used for the two experiments (M9 for gene expression analysis and 7H9 for Icl activity measurements). The use of a richer medium supplemented with acetate, which allowed robust growth of M. tuberculosis, became necessary to measure enzyme activity. Furthermore, the fact that for gene expression experiments, bacteria were exposed to acetate or glucose for only 3 or 24 h, whereas for Icl activity measurements, bacteria were incubated with acetate or glucose for a period of approximately 7 days, might also account for the stronger Icl induction observed in the enzyme activity assays. Nonetheless, both experiments clearly show that RamBMT controls icl1 expression and Icl activity in M. tuberculosis. In addition, we found that RamBMT downregulates its own expression, together with the expression of the open reading frame Rv0464c, which we could show to be located within the same operon as ramBMT.
The finding of carbon source-dependent regulation of icl1 and carbon source-independent autoregulation by RamBMT is very interesting. The situation is the same as in C. glutamicum, i.e., the icl gene is repressed by RamB only during growth on glucose and derepressed on acetate, whereas the ramB gene is repressed by RamB on either carbon source (10). We do not have any (mechanistic) explanation based on experimental results. However, a possible explanation would be the involvement of an additional factor, which in the presence of acetate interferes with the binding of RamBMT to the icl1 promoter region. Such an acetate-induced interference might be due to specific motifs in the neighborhood of the RamBMT operator and/or to direct competition between RamBMT and the unknown factor specifically for binding at the icl1 promoter region. In any case such an interference would lead to a derepression of icl1 expression.
In contrast to RamB from C. glutamicum, RamBMT does not control the expression of other genes encoding enzymes involved in acetate or acetyl-CoA metabolism. Although for icl2 expression, a weak repression was observed in the ramBMT deletion mutant after 24 h of incubation with acetate, the effect was rather small and not supported by microarray results and Icl activity measurements. In contrast to other strains of M. tuberculosis, icl2 has been proposed to be a nonfunctional pseudogene in the strain H37Rv lineage (21). Therefore, carbon source regulation of icl2 expression might have been lost in H37Rv. However, our findings suggest that RamBMT represents a specific control factor of the carbon flux into the glyoxylate (and/or methylcitrate) cycle in M. tuberculosis. This very limited role of RamBMT is in striking contrast to the much wider control of acetate metabolism by RamB in C. glutamicum. It is noteworthy that C. glutamicum is a nonvirulent and ubiquitous organism, which actually might encounter acetate for utilization as a carbon source. M. tuberculosis is a pathogen, which is not found in the environment. Acetate is not likely to be a principal carbon source and so, rather than controlling acetate activating enzymes (Acs, Pta, and AckA), RamBMT has evolved to regulate only Icl activity, which is required for utilization of fatty acids as a carbon source.
The genome-wide microarray analysis revealed that except for icl1 and the ramBMT/Rv0464c operon, no other genes are regulated by RamBMT. Although a significantly reduced expression of htpG could be determined in the ramBMT deletion mutant on both carbon sources, it seems unlikely that this gene is subject to direct regulation by RamBMT, since this protein did not bind to the promoter region of htpG, and downregulation of htpG in the ramBMT deletion mutant could not be complemented by introducing an intact copy of ramBMT into the ramBMT deletion mutant (data not shown). At present, the lack of complementation of htpG downregulation cannot be explained. Since we were also unable to detect binding of RamB to the promoter region of htpG, it could be an effect unrelated to the regulator itself. Furthermore, htpG encodes a putative heat shock protein of the Hsp90 family that is involved in protein folding and that has no known function in acetate metabolism.
Based on a comparative bioinformatic approach, a much wider regulatory function of RamBMT in M. tuberculosis has recently been proposed by Krawczyk et al. (24). These authors transferred gene regulatory networks from C. glutamicum to M. tuberculosis and additionally predicted binding sites upstream of potential M. tuberculosis target genes and operons putatively regulated by transcription factor orthologues, such as RamBMT. Among more than 20 transferred/putative target genes for RamBMT were that for ramBMT itself; those for icl1, glcB, and pta-ackA; and also those for adhA, acn, and sdhA (24). However, in accordance with the results described here, it is evident that only the predicted RamBMT binding motifs in front of the icl1 gene (CAAAATTTGCAAA) and in front of the ramBMT gene (CTAACTCTGCGAA) share relatively high identity (11 and 9 bp, respectively, out of 13 bp) with the highly conserved 13-bp RamB consensus motif (AA/GAACTTTGCAAA) identified in C. glutamicum (17, 2). Taking into account the rather weak binding of RamBMT to the promoter regions of icl1 and of the ramBMT/Rv0464c operon, a binding of RamBMT to all other promoter regions is unlikely.
In summary, RamBMT binds to the promoter region of icl1 and to its own promoter region. It negatively regulates Icl1 in M. tuberculosis during growth on glucose and also controls its own expression. Thus, whereas RamB has been shown to act as a global regulator of acetate metabolism in C. glutamicum when cells were cultured on glucose, RamBMT acts more specifically under these conditions in M. tuberculosis. Since icl1 has been shown to be upregulated when M. tuberculosis resides inside activated macrophages or mice, RamBMT might play an important role during the adaptation of the pathogen to various conditions during infection of the host.
Supplementary Material
Acknowledgments
We thank S. Suerbaum for his support. We thank Lothar Eggeling for providing plasmid pEKEx5.
This study was supported by the Niedersächsische Verein zur Bekämpfung der Tuberkulose; by the German Research Foundation (DFG) through grant SFB 587 to J.C.M., M.S., and F.-C.B.; and by the state Lower Saxony through a Georg-Christoph-Lichtenberg Scholarship and a Wilhelm-Hirte-Foundation Scholarship to J.C.M. The support of the BMBF to B.J.E. (grants 0313704 [SysMAP] and 0313805G [GenoMik-Plus]) is gratefully acknowledged.
Footnotes
Published ahead of print on 18 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arndt, A., and B. J. Eikmanns. 2007. The alcohol dehydrogenase gene adhA in Corynebacterium glutamicum is subject to carbon catabolite repression. J. Bacteriol. 189:7408-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt, A., and B. J. Eikmanns. 2008. Regulation of carbon metabolism in Corynebacterium glutamicum, p. 155-182. In A. Burkovski (ed.), Corynebacteria: genomics and molecular biology. Horizon Scientific Press, Norwich, United Kingdom.
- 3.Auchter, M., A. Arndt, and B. J. Eikmanns. 2009. Dual transcriptional control of the acetaldehyde dehydrogenase gene ald of Corynebacterium glutamicum by RamA and RamB. J. Biotechnol. 140:84-91. [DOI] [PubMed] [Google Scholar]
- 4.Bange, F. C., F. M. Collins, and W. R. Jacobs, Jr. 1999. Survival of mice infected with Mycobacterium smegmatis containing large DNA fragments from Mycobacterium tuberculosis. Tuberc. Lung Dis. 79:171-180. [DOI] [PubMed] [Google Scholar]
- 5.Blombach, B., A. Cramer, B. J. Eikmanns, and M. Schreiner. 2009. RamB is an activator of the pyruvate dehydrogenase complex subunit E1p gene in Corynebacterium glutamicum. J. Mol. Microbiol. Biotechnol. 16:236-239. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff, H. I., and C. E. Barry III. 2005. Tuberculosis: metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3:70-80. [DOI] [PubMed] [Google Scholar]
- 7.Brinkrolf, K., I. Brune, and A. Tauch. 2007. The transcriptional regulatory network of the amino acid producer Corynebacterium glutamicum. J. Biotechnol. 129:191-211. [DOI] [PubMed] [Google Scholar]
- 8.Burkovski, A. 2008. Corynebacteria: genomics and molecular biology. Caiser Academic Press, Norfolk, United Kingdom.
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Cramer, A., M. Auchter, J. Frunzke, M. Bott, and B. J. Eikmanns. 2007. RamB, the transcriptional regulator of acetate metabolism in Corynebacterium glutamicum, is subject to regulation by RamA and RamB. J. Bacteriol. 189:1145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramer, A., R. Gerstmeir, S. Schaffer, M. Bott, and B. J. Eikmanns. 2006. Identification of RamA, a novel LuxR-type transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 188:2554-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubnau, E., J. Chan, V. P. Mohan, and I. Smith. 2005. Responses of Mycobacterium tuberculosis to growth in the mouse lung. Infect. Immun. 73:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eggeling, L., and M. Bott. 2005. Handbook of Corynebacterium glutamicum. CRC Press, Taylor & Friends Group, Boca Raton, FL.
- 14.Eikmanns, B. J., M. Metzger, D. Reinscheid, M. Kircher, and H. Sahm. 1991. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 34:617-622. [DOI] [PubMed] [Google Scholar]
- 15.Gande, R., L. G. Dover, K. Krumbach, G. S. Besra, H. Sahm, T. Oikawa, and L. Eggeling. 2007. The two carboxylases of Corynebacterium glutamicum essential for fatty acid and mycolic acid synthesis. J. Bacteriol. 189:5257-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garton, N. J., S. J. Waddell, A. L. Sherratt, S. M. Lee, R. J. Smith, C. Senner, J. Hinds, K. Rajakumar, R. A. Adegbola, G. S. Besra, P. D. Butcher, and M. R. Barer. 2008. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS. Med. 5:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstmeir, R., A. Cramer, P. Dangel, S. Schaffer, and B. J. Eikmanns. 2004. RamB, a novel transcriptional regulator of genes involved in acetate metabolism of Corynebacterium glutamicum. J. Bacteriol. 186:2798-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerstmeir, R., V. F. Wendisch, S. Schnicke, H. Ruan, M. Farwick, D. Reinscheid, and B. J. Eikmanns. 2003. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104:99-122. [DOI] [PubMed] [Google Scholar]
- 19.Gough, J., K. Karplus, R. Hughey, and C. Chothia. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313:903-919. [DOI] [PubMed] [Google Scholar]
- 20.Gould, T. A., L. H. van de, E. J. Munoz-Elias, J. D. McKinney, and J. C. Sacchettini. 2006. Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol. Microbiol. 61:940-947. [DOI] [PubMed] [Google Scholar]
- 21.Höner zu Bentrup, K., A. Miczak, D. L. Swenson, and D. G. Russell. 1999. Characterization of activity and expression of isocitrate lyase in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181:7161-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulo, N., A. Bairoch, V. Bulliard, L. Cerutti, E. de Castro, P. S. Langendijk-Genevaux, M. Pagni, and C. J. Sigrist. 2006. The PROSITE database. Nucleic Acids Res. 34:D227-D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo, E., K. Suzuki, K. Kanai, and T. Yasuda. 1985. Liposomes-mycobacteria incubation systems as a partial model of host-parasite interaction at cell membrane level. Jpn. J. Med. Sci. Biol. 38:169-180. [DOI] [PubMed] [Google Scholar]
- 24.Krawczyk, J., T. A. Kohl, A. Goesmann, J. Kalinowski, and J. Baumbach. 2009. From Corynebacterium glutamicum to Mycobacterium tuberculosis: towards transfers of gene regulatory networks and integrated data analyses with MycoRegNet. Nucleic Acids Res. 37:e97. doi: 10.1093/nar/gkp453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malm, S., Y. Tiffert, J. Micklinghoff, S. Schultze, I. Joost, I. Weber, S. Horst, B. Ackermann, M. Schmidt, W. Wohlleben, S. Ehlers, R. Geffers, J. Reuther, and F. C. Bange. 2009. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155:1332-1339. [DOI] [PubMed] [Google Scholar]
- 26.Mason, R. J., T. P. Stossel, and M. Vaughan. 1972. Lipids of alveolar macrophages, polymorphonuclear leukocytes, and their phagocytic vesicles. J. Clin. Investig. 51:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKinney, J. D., K. Höner zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics, p. 433. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Munoz-Elias, E. J., and J. D. McKinney. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz-Elias, E. J., and J. D. McKinney. 2006. Carbon metabolism of intracellular bacteria. Cell Microbiol. 8:10-22. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Elias, E. J., A. M. Upton, J. Cherian, and J. D. McKinney. 2006. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol. Microbiol. 60:1109-1122. [DOI] [PubMed] [Google Scholar]
- 32.Pavelka, M. S., and W. R. Jacobs. 1999. Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guerin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J. Bacteriol. 181:4780-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinscheid, D. J., B. J. Eikmanns, and H. Sahm. 1994. Characterization of the isocitrate lyase gene from Corynebacterium glutamicum and biochemical analysis of the enzyme. J. Bacteriol. 176:3474-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinscheid, D. J., B. J. Eikmanns, and H. Sahm. 1994. Malate synthase from Corynebacterium glutamicum: sequence analysis of the gene and biochemical characterization of the enzyme. Microbiology 140:3099-3108. [DOI] [PubMed] [Google Scholar]
- 35.Reinscheid, D. J., S. Schnicke, D. Rittmann, U. Zahnow, H. Sahm, and B. J. Eikmanns. 1999. Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology 145:503-513. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., D. W. Russell, N. Irwin, and U. A. Janssen. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, C. V., C. C. Huang, A. Miczak, D. G. Russell, J. C. Sacchettini, and K. Höner zu Bentrup. 2003. Biochemical and structural studies of malate synthase from Mycobacterium tuberculosis. J. Biol. Chem. 278:1735-1743. [DOI] [PubMed] [Google Scholar]
- 39.Stackebrandt, E., F. A. Rainey, and N. L. Ward-Rainey. 1997. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 47:479-491. [Google Scholar]
- 40.Timm, J., F. A. Post, L. G. Bekker, G. B. Walther, H. C. Wainwright, R. Manganelli, W. T. Chan, L. Tsenova, B. Gold, I. Smith, G. Kaplan, and J. D. McKinney. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. USA 100:14321-14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren, W. A. 1970. Catalysis of both oxidation and reduction of glyoxylate by pig heart lactate dehydrogenase isozyme 1. J. Biol. Chem. 245:1675-1681. [PubMed] [Google Scholar]
- 42.Wendisch, V. F., M. Spies, D. J. Reinscheid, S. Schnicke, H. Sahm, and B. J. Eikmanns. 1997. Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes. Arch. Microbiol. 168:262-269. [DOI] [PubMed] [Google Scholar]
- 43.World Health Organization. 2007. Fact sheet no. 104: tuberculosis. World Health Organization, Geneva, Switzerland.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.