Abstract
In Pseudomonas aeruginosa, cyclic AMP (cAMP) signaling regulates the transcription of hundreds of genes encoding diverse virulence factors, including the type II secretion system (T2SS) and type III secretion system (T3SS) and their associated toxins, type IV pili (TFP), and flagella. Vfr, a cAMP-dependent transcriptional regulator that is homologous to the Escherichia coli catabolite repressor protein, is thought to be the major cAMP-binding protein that regulates these important virulence determinants. Using a bioinformatic approach, we have identified a gene (PA4704) encoding an additional putative cAMP-binding protein in P. aeruginosa PAO1, which we herein refer to as CbpA, for cAMP-binding protein A. Structural modeling predicts that CbpA is composed of a C-terminal cAMP-binding (CAP) domain and an N-terminal degenerate CAP domain and is structurally similar to eukaryotic protein kinase A regulatory subunits. We show that CbpA binds to cAMP-conjugated agarose via its C-terminal CAP domain. Using in vitro trypsin protection assays, we demonstrate that CbpA undergoes a conformational change upon cAMP binding. Reporter gene assays and electrophoresis mobility shift assays defined the cbpA promoter and a Vfr-binding site that are necessary for Vfr-dependent transcription. Although CbpA is highly regulated by Vfr, deletion of cbpA did not affect known Vfr-dependent functions, including the T2SS, the T3SS, flagellum- or TFP-dependent motility, virulence in a mouse model of acute pneumonia, or protein expression profiles. Unexpectedly, CbpA-green fluorescent protein was found to be localized to the flagellated old cell pole in a cAMP-dependent manner. These results suggest that polar localization of CbpA may be important for its function.
Cyclic AMP (cAMP) is an evolutionarily conserved second messenger molecule that plays critical roles in signal transduction in many organisms, ranging from single-cell prokaryotes to multicellular higher eukaryotes (7, 8). cAMP signaling pathways are initiated by the activation of adenylate cyclases in response to extracellular or intracellular stimuli (39). In prokaryotes, the catabolite repressor protein (CRP) family of cAMP-binding proteins plays a central role in cAMP signaling (8, 26). These molecules form homodimers composed of an N-terminal cAMP-binding (CAP) domain and a C-terminal helix-turn-helix DNA-binding domain. Upon binding of cAMP to the CAP domain, CRP family regulators undergo a conformational change that allows them to bind to specific DNA sequences at the promoters of their target genes (57). Genome sequencing and functional studies have demonstrated that the CRP family transcriptional regulators are widely distributed in gram-negative and gram-positive bacteria, where they regulate diverse processes. These pathways include phototaxis and heterocyst formation in cyanobacteria (36), anaerobic respiration in Shewanella oneidensis (41), germination and morphological development in Streptomyces coelicolor (12), Mycobacterium tuberculosis virulence in mice (40), quorum sensing and motility in Vibrio cholerae (29), and phytopathogenicity and cell-cell communication in Xanthomonas campestris (11, 19).
Recent genome sequence studies have revealed that many bacterial species encode additional nucleotide cyclases and cyclic nucleotide (cNMP)-binding proteins. For example, M. tuberculosis strain H37Rv possesses 16 class III nucleotide cyclases and 10 putative cNMP-binding proteins (45). Eight of the 10 putative cNMP-binding proteins are structurally different from typical CRP family regulators, suggesting that cAMP-binding proteins are likely to regulate far-ranging processes in prokaryotes.
The gram-negative opportunistic human pathogen Pseudomonas aeruginosa is a leading cause of acute nosocomial infections in immunocompromised patients or hospitalized individuals with epithelial injury or extensive burns, as well as the cause of chronic infections in patients with cystic fibrosis (31). This pathogen possesses a large arsenal of cell-associated and secreted virulence factors, some of which are coordinately regulated by cAMP. The P. aeruginosa PAO1 genome encodes three adenylate cylases (48). One of these, ExoY, is a type III secreted effector that requires a host-encoded cofactor for its activity and is presumably inactive in the bacteria (59). In addition, a cytoplasmic adenylate cyclase, CyaA, and a membrane-localized class III adenylate cyclase, CyaB, have been identified (48, 56). In strain PAK, the cAMP signaling pathway is initiated mainly by upregulation and activation of CyaB in response to calcium depletion or host cell contact (56). cAMP binds and activates Vfr, an Escherichia coli CRP ortholog, leading to the expression of virulence factors important in acute infections. These include the type II secretion system (T2SS) and type III secretion system (T3SS) and their secreted toxins, as well as the flagellum- and type IV pilus (TFP)-associated motility systems. P. aeruginosa mutants defective in cyaB or vfr are significantly attenuated for virulence in a mouse model of acute infection (47).
In this study, we have characterized an additional cAMP-binding protein, PA4704, in P. aeruginosa, which we refer to as CbpA, for cAMP-binding protein A. Structural modeling and functional studies demonstrate that CbpA is comprised of a C-terminal functional CAP domain and an N-terminal highly degenerate CAP domain and binds to cAMP via the C-terminal CAP domain. We show that transcription of cbpA is directly regulated by Vfr, although CbpA does not appear to regulate known Vfr-dependent processes. Unexpectedly, CbpA localizes to a flagellated old cell pole in a cAMP-dependent manner, which may be critical to its function.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are described in Table 1. Bacteria were grown on Luria-Bertani (LB) agar. To isolate P. aeruginosa strains from the E. coli-P. aeruginosa mating mixture, 4.5% Difco pseudomonas isolation agar (PIA) (Becton Dickinson) was used. Antibiotics were used at the following concentrations: 15 μg/ml of gentamicin for E. coli and P. aeruginosa; 15 μg/ml and 500 μg/ml of tetracycline for E. coli and P. aeruginosa, respectively; 50 μg/ml of kanamycin for E. coli; 100 μg/ml of ampicillin for E. coli; and 500 μg/ml of carbenicillin for P. aeruginosa.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype and/or relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | 48 |
| PAO1Δvfr | In-frame deletion of vfr | 54 |
| PAO1ΔcbpA | In-frame deletion of cbpA | This study |
| PAO1ΔcbpA-mini-CTX-Pii-gfp | In-frame deletion of cbpA, mini-CTX-Pii-gfp integrated into attB site, Tetr | This study |
| PAO1ΔcbpA-mini-CTX-Pii-cbpA-gfp | In-frame deletion of cbpA, mini-CTX-Pii-cbpA-gfp integrated into attB site, Tetr | This study |
| PAO1ΔcyaAB | In-frame deletion of cyaA cyaB, exoTHA in attB site | Gift from J. West |
| PAO1ΔcyaABΔcbpA | PAO1 derivative, cyaA cyaB cbpA deletion, exoTHA in attB site | This study |
| PA103 | Wild type, exoU+ | 30 |
| PA103-mini-CTX-exoTHA-lacZ | Mini-CTX-exoTHA-lacZ integrated into attB site, Tetr | This study |
| PA103ΔcbpA | In-frame deletion of cbpA | This study |
| PA103ΔcbpA-mini-CTX-exoTHA-lacZ | In-frame deletion of cbpA, mini-CTX-exoTHA-lacZ integrated into attB site, Tetr | This study |
| PA103pscJ::Tn5 | Tn5 insertion in pscJ (T3SS mutant), Gmr | 23 |
| PA103pscJ::Tn5-mini-CTX- exoTHA-lacZ | Tn5 insertion in pscJ (T3SS mutant), mini-CTX-exoTHA-lacZ integrated into attB site, Gmr Tetr | This study |
| E. coli | ||
| DH5α | hsdR rec lacZYA φ80dlacZΔM15 | Invitrogen |
| BL21(DE3) | F−dcm ompT hsdS(rB− mB−) gal λ(DE3) | Stratagene |
| S17-1 λpir | thi pro hsdR recA RP4-2 (Tc::Mu) (Km::Tn7) | 46 |
| Plasmids | ||
| mini-CTX-lacZ | Containing attP site for integration at the attB site of P. aeruginosa chromosome, lacZ, Tetr | 20 |
| mini-CTX-gfp | Containing attP site for integration at the attB site of P. aeruginosa chromosome, gfp, Tetr | 20 |
| mini-CTX-exoTHA-lacZ | mini-CTX-lacZ derivative, expression of C-terminal HA-tagged ExoT under the control of exoT promoter, Tetr | This study |
| mini-CTX-Pii-gfp | mini-CTX-gfp derivative, expression of GFP under the control of cbpA promoter, Tetr | This study |
| mini-CTX-Pii-cbpA-GFP | mini-CTX-lacZ derivative, expression of C-terminal GFP-fused CbpA from cbpA promoter, Tetr | This study |
| pCR2.1-TOPO | TA cloning vector, Kmr | Invitrogen |
| pET24+ | T7 promoter-induced expression vector, Kmr | |
| pETPA0275His | pET24+ derivative, C-terminal Gly3-His6-tagged PA0275 expression, Kmr | This study |
| pETcbpAHis | pET24+ derivative, C-terminal Gly3-His6-tagged CbpA expression, Kmr | This study |
| pETcbpAMTHis | pET24+ derivative, CAP domain mutant (R207A S208A) of C-terminal Gly3-His6-tagged CbpA, Kmr | This study |
| pETvfrHis | pET24+ derivative, C-terminal Gly3-His6-tagged Vfr expression, Kmr | This study |
| pEX100T | Allelic-replacement suicide plasmid, Apr (Cbr) | 43 |
| pEXΔcbpA | pEX100T derivative containing cbpA flanking region, Tetr | This study |
| pMBAD18G | Broad-host-range expression vector, PBAD promoter, Gmr | This study |
| pMBAD-GFP | pMBAD18G derivative, C-terminal GFP fusion vector, Gmr | This study |
| pMBAD-cbpA-GFP | pMBAD-GFP derivative, expression of C-terminal GFP-fused CbpA, Gmr | This study |
| pMBAD-cbpAMT-GFP | pMBAD-GFP derivative, expression of C-terminal GFP-fused CbpAMT, Gmr | This study |
| pMBAD-cbpAΔH-GFP | pMBAD-GFP derivative, expression of C-terminal GFP-fused CbpAΔH, Gmr | This study |
| pMBAD-cbpAΔB1-GFP | Expression of C-terminal GFP-fused CbpAΔB1, Gmr | This study |
| pMBAD-cbpAΔCAP-GFP | Expression of C-terminal GFP-fused CbpAΔCAP, Gmr | This study |
| pME-lacZ | lacZ reporter vector, promoterless lacZ, Gmr | 14 |
| pME-Pi-lacZ | pME-lacZ derivative, 104-bp cbpA upstream region, Gmr | This study |
| pME-Pii-lacZ | pME-lacZ derivative, 200-bp cbpA upstream region, Gmr | This study |
| pME-PiiMT-lacZ | pME-lacZ derivative, 200-bp cbpA upstream region with mutations in VBS, Gmr | This study |
| pME-Piii-lacZ | pME-lacZ derivative, 602-bp cbpA upstream region, Gmr | This study |
| pOK12 | E. coli cloning vector, Kmr | 50 |
| pOKΔcbpA | pOK12 derivative containing cbpA flanking region, Kmr | This study |
| pOKΔcbpA-dwn | pOK12 derivative containing cbpA downstream region, Kmr | This study |
Plasmid construction.
All plasmids were purified by using QIAprep spin miniprep columns (Qiagen). Standard recombinant DNA manipulation techniques were used (42). Enzymes were purchased from New England Biolabs, Inc., or Roche Applied Science. All PCR primers employed in this study were designed based on the PAO1 genome sequence (http://www.pseudomonas.com) (48) and were purchased from Invitrogen. PCR primers used in this study are listed in Table S1 in the supplemental material. PCR amplifications were carried out with Herculase Hotstart DNA polymerase (Stratagene). PCR products were subcloned into pCR2.1-TOPO (Invitrogen) and sequenced to confirm that no mutations were introduced during amplification. Plasmids were introduced into a chemically competent E. coli DH5α strain or into P. aeruginosa strains by electroporation. Detailed methods for construction of plasmids used in this study are described in the supplemental material.
Construction of an in-frame deletion in cbpA.
In-frame unmarked cbpA deletion mutants of PAO1 and PA103 were constructed by allelic exchange (43). For the cbpA deletion mutant, a 2.8-kb region upstream of the cbpA open reading frame and a 1.3-kb region downstream of the cbpA open reading frame were amplified with two sets of primers, ΔcbpA-up-Fw and ΔcbpA-up-R and ΔcbpA-dwn-F and ΔcbpA-dwn-R, respectively (see Table S1 in the supplemental material). The subcloned 1.3-kb cbpA downstream region was digested with HindIII and XbaI and subcloned into HindIII-XbaI-digested pOK12, resulting in pOKΔcbpA-dwn. The subcloned 2.8-kb cbpA upstream region was digested with EcoRI and HindIII, and the resulting 1.9-kb EcoRI-HindIII fragment was subcloned into EcoRI-HindIII-digested pOKΔcbpA-dwn, resulting in pOKΔcbpA. The resultant 3.2-kb EcoRI-XbaI fragment containing the cbpA flanking region was blunted and cloned into SmaI-digested pEX100T, resulting in pEXΔcbpA. E. coli S17-1 λpir containing pEXΔcbpA was mated with P. aeruginosa PAO1 or PA103 on LB at 37°C overnight to induce homologous recombination between pEXΔcbpA and the corresponding chromosomal regions of P. aeruginosa strains. P. aeruginosa strains in which pEXΔcbpA was integrated into a cbpA flanking region were isolated on PIA containing carbenicillin. Finally, carbenicillin-resistant exconjugants (merodiploids) were then resolved by growth on PIA plus 5% sucrose as described previously (15). Candidate mutants were screened by PCR and confirmed by Southern blotting.
cAMP-binding assay.
E. coli BL21(DE3) containing pETcbpAHis, pETcbpAMTHis, or pETPA0275His was cultured in 5 ml of LB at 37°C overnight. A 1:100 aliquot of subculture was inoculated into 100 ml of LB and grown at 37°C to an optical density at 600 nm (OD600) of 0.8. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture (final concentration of 1 mM), and the culture was incubated at 20°C overnight with reciprocal shaking (300 rpm). E. coli cells were harvested by centrifugation, washed twice with chilled suspension buffer (20 mM Tris-HCl, pH 7.5, 0.5 M NaCl, and 10% glycerol), and resuspended in suspension buffer (10 ml final volume). All of the steps for preparation of the crude protein sample were carried out on ice or at 4°C. Cells were disrupted by sonication at 10 W for 15 min (Branson sonifier 150). After removal of E. coli cell debris by centrifugation at 14,000 rpm for 15 min at 4°C, the supernatant was filtered through a 0.2-μm disk filter (Millipore). The soluble fraction of the protein sample was applied to an open column (diameter = 15 mm) containing 1.5 ml of cAMP-conjugated agarose resin (Sigma-Aldrich). The column was washed with 100 ml of the suspension buffer and eluted with 50 ml of elution buffer (20 mM Tris-HCl, pH 7.5, 0.5 M NaCl, 10% glycerol, and 1 mM cAMP). The eluted protein was concentrated with an Amicon Ultra-4 filter unit (Millipore) and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis.
SDS-PAGE and immunoblot analysis.
The protein concentration was determined with a Bio-Rad protein assay (Bio-Rad Laboratories). Protein (20 μg) was separated on a 12% SDS-polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (Millipore) in transfer buffer (15 mM Tris, 120 mM glycine, 20% methanol). Membranes were incubated in blocking buffer (phosphate-buffered saline [PBS] containing 5% skim milk and 0.5% Tween 20). An anti-His5 tag antibody (Qiagen) was used as the primary antibody at a concentration of 0.4 μg/ml, while the secondary antibody, horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Jackson ImmunoResearch Laboratories), was used at a dilution of 1:2,000. Reactive bands were visualized using ECL Western blotting detection reagents (GE Healthcare) and were detected with an Amersham Hyperfilm ECL kit (GE Healthcare).
β-Galactosidase assays.
P. aeruginosa cultures were grown in LB broth with the appropriate antibiotics to an OD600 of ∼0.8. A β-galactosidase assay was performed as described previously (32).
Purification of C-terminally His6-tagged CbpA (CbpAHis) and VfrHis.
E. coli BL21(DE3) containing pETvfrHis or pETcbpAHis was grown in LB broth containing kanamycin at 37°C overnight. The overnight culture was diluted 1:10,000 into 100 ml of LB broth containing kanamycin and grown to an OD600 of 0.5 to 0.8. IPTG was added to a final concentration of 0.5 mM, and the culture was further grown with shaking at 20°C overnight. Cells were harvested by centrifugation, washed twice with 30 ml of a suspension buffer (20 mM Tris-HCl, pH 7.5, 10 mM imidazole, 0.5 M NaCl, 10% glycerol), and suspended in 20 ml of suspension buffer. Cells were disrupted by sonication at 10 W for 15 min (Branson sonifier 150), and cell debris was removed by centrifugation. The supernatant was filtered through a 0.45-μm filter disk and then applied to a column (diameter = 15 mm) containing 2 ml of HIS-Select nickel affinity gel (Sigma-Aldrich). After the column was washed with 200 ml of suspension buffer, proteins were eluted with 50 ml of His elution buffer (20 mM Tris-HCl, pH 7.5, 300 mM imidazole, 0.5 M NaCl, 10% glycerol). The eluted fraction was then applied to a column containing 2 ml of cAMP-conjugated agarose. After the column was washed with 300 ml of His elution buffer, proteins bound to cAMP-conjugated agarose were eluted with 100 ml of cAMP-BP elution buffer (20 mM Tris-HCl, pH 7.5, 300 mM imidazole, 0.5 M NaCl, 1 mM cAMP, 10% glycerol). Eluted proteins were concentrated with an Amicon Ultra-4 filter unit (Millipore) and then subjected to SDS-PAGE to confirm the purity of VfrHis or CbpAHis.
Trypsin protection assay.
The effects of cAMP on CbpAHis conformation were determined by treating 5.6 μg of CbpAHis with 0.2 μg trypsin for 5 min at 37°C (3, 27). Trypsin digestion was stopped by adding 2× SDS-PAGE buffer to and boiling the reaction mixture. CbpAHis digested by trypsin was analyzed by SDS-PAGE.
EMSA.
An electrophoresis mobility shift assay (EMSA) was performed with a digoxigenin gel shift kit (Roche Applied Science) according to the manufacturer's instruction. A 50-bp DNA fragment containing a wild-type Vfr-binding site (VBSWT) or a mutant version (VBSMT) was generated by PCR with sets of primers VBS-Fw and VBS-Rv or VBSMT-Rv, respectively (see Table S1 in the supplemental material). VBSWT (0.8 ng) and VBSMT (0.8 ng) were labeled with digoxigenin-11-ddUTP, and the labeled probes (0.08 ng) were mixed with purified VfrHis solution (18 or 50 ng) containing 1 mM cAMP in EMSA binding buffer containing 50 ng/μl of poly(dI-dC) and 5 ng/μl of poly-l-lysine. The reaction mixture was incubated for 15 min at 15 to 25°C and was subjected to electrophoresis (150 V, 10 min, 15 to 25°C) on 8% polyacrylamide gels with Tris-borate-EDTA buffer. The polyacrylamide gels were transferred to a Hybond-N+ (GE Healthcare) membrane and detected with a CSPD system (Roche Applied Science).
Motility and protease assays.
Twitching motility was measured by Coomassie brilliant blue staining of stab assays, as previously described (53), after 24 h of growth on LB plates poured on polystyrene dishes. For the swimming motility assay, P. aeruginosa strains were grown on LB containing 0.3% agar and appropriate antibiotics at 37°C (53). The diameter of the swimming zone was measured after 12 h. Protease assays were performed as previously described (13). P. aeruginosa strains were stabbed on the milk plate and grown at 37°C for 24 h. The diameter of clearing around colonies, in which milk proteins are degraded by secreted proteases from P. aeruginosa, was measured.
Cytotoxicity assays.
Cytotoxicity assays were performed as described previously (53). HeLa cells (ATCC) were plated in 24-well dishes at 5 × 104 cells per well 48 h prior to infection. P. aeruginosa strains (PA103 derivatives) were grown overnight at 37°C in LB broth and used at a multiplicity of infection of 50. Prior to infection, HeLa cells were washed and placed in minimal essential medium. At 3 h postinfection, the entire supernatant was transferred to a tube and centrifuged at 800 rpm for 5 min to remove cell debris. Lactate dehydrogenase was measured in an aliquot of the supernatant by use of a CytoTox96 nonradioactive cytotoxicity assay kit (Promega) according to the manufacturer's instructions.
Murine model of acute pneumonia.
Eight- to 12-week-old female C57BL/6 mice were housed under specific-pathogen-free conditions. Infections were carried out as described previously (4). Mice were infected intranasally with 50 μl of bacterial suspension (5 × 107 CFU) and sacrificed 18 h postinfection. The lungs, liver, and spleen were harvested and homogenized in PBS, and bacterial counts were determined by plating serial dilutions on LB agar plates. Statistical analyses were performed using a two-tailed Mann-Whitney U test (a P value of <0.05 was considered significant). All experiments were approved by the University of California, San Francisco, Committee on Animal Research.
ExoT secretion assay.
P. aeruginosa strains PA103-mini-CTX-exoTHA-lacZ, PA103ΔcbpA-mini-CTX-exoTHA-lacZ, and PA103pscJ::Tn5-mini-CTX-exoTHA-lacZ (Table 1) were grown in LB broth containing 5 mM CaCl2 or 5 mM EGTA at 37°C overnight. Cells were removed by centrifugation, and the supernatant was filtered through a 0.2-μm filter disk (Millipore). Secreted proteins in the supernatant were concentrated with an Amicon Ultra-4 filter unit, separated by SDS-PAGE, and immunoblotted using mouse anti-hemagglutinin (HA) IgG (Sigma-Aldrich).
Biofilm formation assay.
Biofilm formation was measured as previously described (37), with the following modifications. Strains PAO1, PAO1Δvfr, and PAO1ΔcbpA were grown in LB broth at 37°C overnight and diluted 1:1,000 into 2 ml of LB broth in 12-well polystyrene plates. After 24 h of incubation at 37°C, the culture supernatant was removed, each well was washed three times with 1 ml of distilled water, and the biofilm was stained with 0.05% crystal violet. After three washes with distilled water, the crystal violet-stained biofilm was dissolved by the addition of 1 ml of 99.8% ethanol, and the OD590 of the solution was measured.
2D-PAGE.
PAO1 containing pMBAD18G, PAO1ΔcbpA containing pMBAD18G, and PAO1ΔcbpA containing pMBAD-cbpA were grown in 2 ml of LB borth at 37°C overnight. The overnight culture was diluted 1:1,000 into 100 ml of LB broth containing 0.2% l-arabinose and 5 mM EGTA, and cells were grown at 37°C to an OD600 of 1.5. Cells were harvested by centrifugation at 4°C and washed once with 50 ml of PBS. The pellet was suspended in SDS boiling buffer (5% SDS, 5% β-mercaptoethanol, 10% glycerol, 60 mM Tris-HCl, pH 6.8). Two-dimensional PAGE (2D-PAGE) was performed by Kendrick Labs, Inc., using the carrier ampholine method of isoelectric focusing (34). Isoelectric focusing was carried out in glass tubes of an inner diameter of 3.5 mm, using 1% pH 4 to 6 (GE Healthcare) and 1% pH 5 to 8 (GE Healthcare) for 20 kV·h. After equilibrium in SDS sample buffer (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, and 62.5 mM Tris-HCl, pH 6.8), each tube gel was sealed to the top of a stacking gel that overlays a 10% acrylamide slab gel (1.0 mm thick). SDS-polyacrylamide slab gel electrophoresis was carried out for about 5 h at 25 mA per gel. Each sample was run on duplicate gels and was scanned with a laser densitometer, model PDSI (Molecular Dynamics, Inc.). The scanner was checked for linearity prior to scanning with a calibrated neutral density filter set (Melles Griot). The images were analyzed using Progenesis PG240 software with TT900 (Nonlinear Dynamics). The general method of computerized analysis for these pairs included image warping with TT900 software followed by automatic spot finding, background subtraction (average on border), matching, and quantification in conjunction with detailed manual checking. Student's t test values for fluctuation of each spot were generated by the software for two gels.
Fluorescent microscopy.
P. aeruginosa harboring green fluorescent protein (GFP)-expressing plasmids was grown at 37°C in LB broth containing 0.1% l-arabinose to an OD600 of 1.5 to 2.0. One hundred microliters of the culture was diluted with 900 μl of PBS buffer containing 2 μg/ml of FM4-64FX (Invitrogen) and was incubated at room temperature for 15 min. Cells were fixed with 4% paraformaldehyde on poly-l-lysine-coated glass slides. Coverslips were mounted in Vectashield mounting media containing DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories) to count the total number of bacterial cells in images. Images for all GFP fluorescence and immunofluorescence studies were acquired with a charge-coupled-device camera (Retiga) using a 100× lens objective mounted on a Nikon TE2000 inverted microscope driven by Simple PCI software (Compix, Inc.). A 10× ocular lens was used, making the total magnification ×1,000. For each set of experiments, the exposure times were identical for all images. Images were processed with Adobe Photoshop CS3. To confirm expression level and stability of CbpA-GFP and its variants, the culture used for immunofluorescent microscopy was diluted to an OD600 of 1.0 and cytoplasmic extracts were subjected to immunoblotting using an anti-GFP antibody (Invitrogen).
Indirect immunofluorescent staining.
Immunofluorescent staining for P. aeruginosa FliC was performed as previously described (33). Briefly, P. aeruginosa PAO1ΔcbpA containing pMBAD-cbpA-GFP was grown overnight at 37°C in LB broth containing 0.1% arabinose and 5 mM EGTA. The overnight culture was diluted 1:1,000 in fresh media containing 0.1% arabinose and 5 mM EGTA and then grown at 37°C to an OD600 of 1.5. Cells were pelleted by centrifugation and fixed in 4% paraformaldehyde for 30 min. After the cells were washed once with PBS, the cell pellets were suspended in 0.7% (wt/vol) fish scale gelatin (PBS-FSG), and the cell suspension was spotted on a poly-l-lysine-coated coverslip and incubated for 30 min at room temperature. Flagella were stained with rabbit anti-FliC polyclonal antiserum (1:100 in PBS-FSG) (kindly provided by D. Wozniak) and detected with Alexa Fluor 594-conjugated donkey anti-rabbit IgG (1:1,000 in PBS-FSG) (Invitrogen).
RESULTS
Identification of putative cAMP-binding proteins in P. aeruginosa.
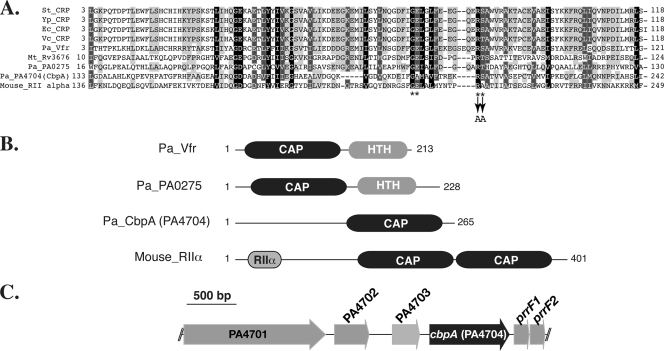
We carried out a BLAST-P search of the P. aeruginosa PAO1 genome for potential cAMP-binding proteins by using the CAP domain sequence (amino acids 1 to 144) of Vfr as a query. Three proteins with an E value of less than 1.0e−2 were identified, i.e., Vfr, PA4704, and PA0275, with E values (bit score) of 2.0e−79 (287), 2.0e−7 (48.5), and 5.0e−4 (37.4), respectively. BLAST-P search and alignment analysis revealed that the CAP domains of PA4704 and PA0275 showed more than 20% identity and 45% similarity to those of well-characterized cAMP-binding transcriptional regulators (Fig. 1A). Additionally, the amino acid residues involved in direct binding of cAMP are highly conserved (Fig. 1A).
FIG. 1.
Putative cAMP-binding proteins in P. aeruginosa. (A) Alignment of the CAP domains. Amino acid residues highlighted in black (100% similarity), dark gray (80 to 99% similarity), and light gray (60 to 79% similarity) are conserved among CAP domains of different proteins. Amino acid residues that are involved in direct binding to cAMP in CRP family regulators are indicated by asterisks. Two of these amino acids, Arg207 and Ser208 in CbpA (indicated by arrows), were changed to Ala to create a PA4704 CAP domain mutant (CbpAMT). Abbreviations are as follows (with NCBI database accession numbers in parentheses): Pa_Vfr, P. aeruginosa Vfr (NP_249343); Pa_PA0275, P. aeruginosa PA0275 (NP_248966); Pa_PA4704, P. aeruginosa PA4704 (CbpA) (NP_253392); St_CRP, Salmonella enterica serovar Typhimurium CRP (NP_462369); Ec_CRP, E. coli CRP (AP_004432); Vc_CRP, Vibrio cholerae CRP (YP_001218107); Yp_CRP, Yersinia pestis CRP (NP_671249); Mt_Rv3676, M. tuberculosis Rv3676 (NP_218193); and Mouse_RII alpha, Mus musculus PKA RIIα (P12367). (B) Domain organization of cAMP-binding proteins. HTH indicates a helix-turn-helix DNA-binding domain. RIIα represents the dimerization/docking domain of mouse PKA RIIα. (C) Loci that encode putative cAMP-binding protein PA4704 (CbpA).
PA0275 encodes a 228-amino-acid protein that is composed of an N-terminal CAP domain (amino acids 16 to 130) and a C-terminal, helix-turn-helix DNA-binding domain (amino acids 170 to 228) (Fig. 1B), suggesting that the PA0275 protein may function as a cAMP-dependent transcriptional regulator. The protein encoded by PA0275 shows 26% identity and 50% similarity to Rv3676, a cAMP-binding protein of M. tuberculosis (3).
The PA4704 gene (herein referred to as cbpA, for cAMP-binding protein A) encodes a 265-amino-acid protein composed of an N-terminal region (amino acids 1 to 132) and a C-terminal CAP domain (amino acids 133 to 265) (Fig. 1A and B). Pfam and PROSITE database searches failed to identify any conserved domain in the N-terminal half of CbpA; however, an iterative PSI-BLAST search revealed that the region may encode a highly degenerate CAP domain (see below for details). CbpA has no identifiable signal sequence for known secretion systems. It was detectable in cell lysates under calcium-rich and calcium-limiting conditions but not in culture supernatants (data not shown), suggesting that CbpA is not secreted and is a cytoplasmic protein. cbpA is located immediately (53 bp) upstream from two genes encoding iron-regulated small RNAs, prrF1 and prrF2 (Fig. 1C), which play a crucial role in iron homeostasis by posttranscriptionally regulating expression of their target genes, including bacterioferritin (PA4880), superoxide dismutase (sodB), and anthranilate dioxygenase (antABC) genes (35, 55). Transcription of these tandem small RNAs has been reported to be independently repressed by binding of the iron-bound form of the ferric uptake regulator (Fur) to their respective promoters (55). Notably, a Fur box was not detected in the intergenic region between PA4703 and cbpA, suggesting that cbpA transcription may not be regulated by an iron condition. A BLAST-P search revealed that CbpA structural orthologs are found in three pseudomonads, P. stutzeri, P. fluorescens, and P. mendocina (58 to 78% identity), and in five nonpseudomonad species, Reinekea species, Cellvibrio japonicus, Hahella chejuensis, Congregibacter litoralis, and Oceanobacter species (32 to 42% amino acid identity), suggesting a conserved function. Homologs of the small iron-regulated RNAs are not always found adjacent to cbpA.
CbpA binds cAMP.
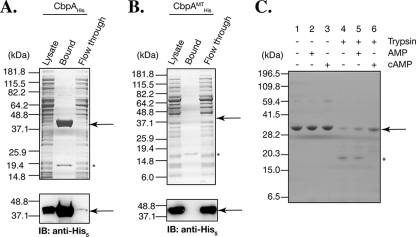
To examine whether CbpA and PA0275 are bona fide cAMP-binding proteins, we tested whether they bind to cAMP. C-terminally His6-tagged PA0275 (PA0275His) and CbpAHis were overexpressed in E. coli, and the soluble cell lysates were tested for binding to cAMP-conjugated agarose beads (see Materials and Methods for details). PA0275His did not appear to bind to the cAMP column, even though it was present in the soluble fraction of the E. coli cell lysates (data not shown). In contrast, CbpAHis bound to the cAMP-agarose resin and could be eluted with cAMP (Fig. 2A). Since PA0275His did not bind to cAMP-agarose beads under our experimental conditions, we focused on CbpA for further analysis.
FIG. 2.
CbpA (PA4704) binds cAMP and undergoes a conformational change upon cAMP binding. (A) CbpAHis binds to cAMP-conjugated agarose. The soluble lysate from E. coli overexpressing CbpAHis was applied to a cAMP-conjugated agarose column. The starting lysates, flowthrough fraction, and bound fraction were subjected to SDS-PAGE and visualized by Coomassie blue staining (top) or by immunoblot (IB) analysis (bottom) using anti-His5 antibody. CbpAHis is enriched in the bound fraction and depleted from the flowthrough fraction. The ∼19-kDa band (asterisk) present in the bound fraction likely represents E. coli CRP. (B) CbpAMTHis fails to bind to cAMP agarose. CbpAMTHis was overexpressed in E. coli, and lysates were applied to the cAMP agarose column. CbpAMTHis failed to bind to the cAMP agarose column and was not depleted from the flowthrough fraction. The ∼19-kDa band (asterisk) present in the bound fraction likely represents E. coli CRP, as observed for panel A. (C) Trypsin protection assay. Equal amounts of purified CbpAHis (see Materials and Methods) were digested with trypsin in the presence (+) or absence (−) of cAMP or AMP, separated by SDS-PAGE, and visualized by Coomassie blue staining. Undigested CbpAHis is shown by the arrow. The asterisk represents a major digested product in the absence of cAMP. Note that the molecular size markers are different from those in panels A and B. The spectrum of trypsin digestion products of CbpA is altered in the presence of cAMP but not in the presence of AMP.
To determine whether CbpAHis specifically binds to cAMP-agarose beads, we tested whether it could be eluted with AMP. As shown in Fig S1 in the supplemental material, CbpAHis was eluted with 1 mM cAMP but not with 1 mM AMP. To examine whether the C-terminal CAP domain of CbpA is required for binding, the Arg207 and Ser208 amino acid residues within the CAP domain, which are both predicted to be involved in direct binding to cAMP (18), were changed to Ala (Fig. 1A), and the resulting mutant protein (CbpAMTHis) was tested for binding to cAMP-agarose. As shown in Fig. 2B, CbpAMTHis no longer bound to cAMP-conjugated agarose beads, indicating that the two amino acid residues Arg207 and Ser208, conserved in the C-terminal CAP domain, are essential for binding to cAMP-agarose.
It has been reported that cAMP-binding proteins undergo an allosteric change in structure following the binding of cAMP (3, 27, 39). To test whether CbpA undergoes a conformational change upon binding to cAMP, we carried out an in vitro protease digestion assay on CbpA in the presence or absence of cAMP or AMP. Purified CbpAHis migrated as a 32-kDa band on SDS-polyacrylamide gels (Fig. 2C, lanes 1 to 3) and was degraded into smaller fragments after 5 min of trypsin treatment (Fig. 2C, lane 4). The pattern of trypsin digestion products was different in the presence of cAMP but was unaffected when digestion was carried out in the presence of AMP (Fig. 2C, lanes 4, 5, and 6). Together, these results confirm that CbpA is a cAMP-binding protein.
Vfr directly regulates cbpA expression.
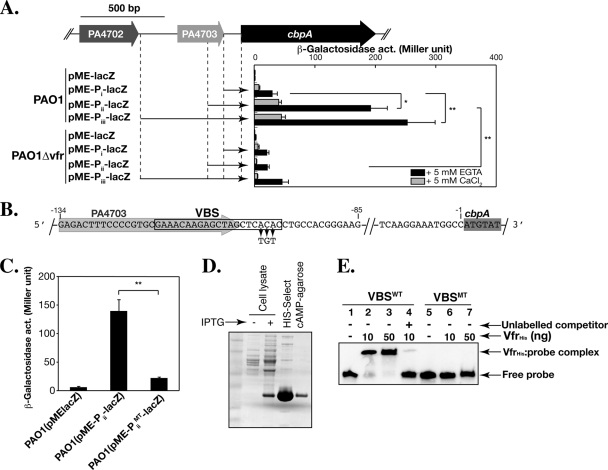
Previous microarray analysis has revealed that cbpA is highly downregulated in vfr or retS mutants (17, 56). We tested whether cbpA is a direct target of Vfr by examining its transcriptional regulation. Because cbpA is located within a cluster of six genes transcribed in the same direction (PA4701-prrF2) (Fig. 1C), we first defined the cbpA promoter. DNA fragments containing 104 bp, 200 bp, or 602 bp of DNA upstream of the cbpA coding region were cloned into a lacZ transcriptional fusion plasmid pME-lacZ, yielding pME-Pi-lacZ, pME-Pii-lacZ, and pME-Piii-lacZ, respectively (Fig. 3A). These reporter gene fusions were introduced into PAO1 and PAO1Δvfr, and β-galactosidase activity was measured. The transcription from all three promoter fragments increased when the bacteria were grown in the absence of calcium, but the β-galactosidase activity from regions Piii and Pii was approximately five- to sixfold higher (P < 0.05) than that from the Pi region (Fig. 3A). The transcription from the Pii and Piii regions in PAO1Δvfr was significantly reduced (P < 0.05) even under calcium-depleted conditions (Fig. 3A). These results indicate that Vfr is necessary for the regulation of cbpA transcription and that the Vfr-responsive element localizes between the Pi and Pii regions.
FIG. 3.
Transcription of cbpA is directly regulated by Vfr. (A) LacZ reporter gene assays. The schematic diagram shows cbpA and its upstream genes. The cbpA upstream regions cloned into a reporter plasmid, pME-lacZ, are indicated by the arrows. Plasmids pME-Pi-lacZ, pME-Pii-lacZ, pME-Piii-lacZ, and a control plasmid, pME-lacZ, were introduced into PAO1 and PAO1Δvfr, and β-galactosidase activity (act.) was measured in the presence of calcium (plus 5 mM CaCl2, gray bar) and in the absence of calcium (plus 5 mM EGTA, black bar). Error bars indicate standard deviation. *, P < 0.05; **, P < 0.01 (two-tailed Student's t test). (B) A putative VBS overlaps the 3′ end of the coding sequence of PA4703. The black box encloses the putative VBS. The nucleotides ACA at the 3′ end of the VBS were changed to TGT in the plasmid pME-PiiMT-lacZ and in the EMSA probe VBSMT. The sequences highlighted in gray and dark gray show the regions encoding PA4703 and cbpA, respectively. (C) The VBS sequence is required for expression of cbpA. The β-galactosidase activities of pME-Pii-lacZ and pME-PiiMT-lacZ expressed in PAO1 were measured. Error bars indicate standard deviation. **, P < 0.01 (two-tailed Student's t test). (D) VfrHis purification. VfrHis was expressed in E. coli BL21(DE3) and purified by two-step purification using a HIS-Select column followed by a cAMP-conjugated agarose column. (E) Vfr binds to the VBS. An EMSA was performed using purified VfrHis and DNA fragments containing VBS. VBSWT contains the DNA sequence from −85 to −134 relative to the translational start site of cbpA. An excess of unlabeled VBSWT was used as a competitor. The VfrHis gel shifts VBSWT in a dose-dependent manner but fails to shift VBSMT. The gel shift is inhibited by the addition of excess unlabeled VBSWT.
A 21-bp consensus Vfr-binding site, 5′-ANWWTGNGAWNYAGWTCACAT-3′, where W, Y, and N represent A or T, C or T, and any nucleotide, respectively, was recently defined (22). We identified a putative Vfr-binding sequence (VBS) (15/21 nucleotide identity) between −98 bp and −118 bp upstream of the translational start site of cbpA, which overlapped with the stop codon of PA4703 (Fig. 3B). Three highly conserved nucleotides, 5′-ACA-3′, in the putative VBS were replaced by 5′-TGT-3′ to yield pME-PiiMT-lacZ (Fig. 3B). The β-galactosidase activity from pME-PiiMT-lacZ was sixfold lower (P < 0.01) than that from pME-Pii-lacZ (Fig. 3C), indicating that the putative VBS plays an important role in the expression of cbpA. To confirm whether Vfr directly binds to the VBS, we performed an EMSA using purified VfrHis (Fig. 3D and E). VfrHis bound to a DNA probe (VBSWT) composed of nucleotides −84 to −134 relative to the cbpA start codon (Fig. 3B and E, lanes 1 to 3). Binding was inhibited upon addition of an excess of unlabeled VBSWT (Fig. 3E, lane 4), indicating that Vfr specifically binds to this probe. However, VfrHis was unable to bind to the VBSMT probe containing the three nucleotide mutations (Fig. 3E, lanes 5 to 7). Together, these results demonstrate that cbpA is a direct target of Vfr.
CbpA is not necessary for known Vfr-dependent functions.
Since transcription of cbpA is directly regulated by Vfr, we considered the possibility that CbpA may be involved in regulating a subset of cAMP- and Vfr-dependent events. To address the hypothesis, we constructed in-frame cbpA deletion mutants for PAO1 and PA103 as well as overexpression strains and investigated growth rate, quorum sensing-dependent protease secretion, flagellum-dependent motility, TFP-dependent twitching motility, T3SS-dependent effector secretion, host cell cytotoxicity, virulence in mice, and biofilm formation, all of which are associated with cAMP signaling through Vfr (2, 6, 10, 47, 52, 54, 56).
PAO1ΔcbpA and PA103ΔcbpA grew normally in nutrient-rich media (see Fig. S2 in the supplemental material) and formed rod-shaped cells, indicating that CbpA is dispensable for replication and is not involved in regulating cell morphology. PAO1ΔcbpA grown in rich media exhibited wild-type biofilm formation (see Fig. S3 in the supplemental material), quorum sensing-dependent and T2SS-dependent protease secretion (see Fig. S4 in the supplemental material), flagellum-dependent swimming motility (see Fig. S5 in the supplemental material), and TFP-dependent twitching motility (see Table S2 in the supplemental material). We also tested the role of CbpA in the virulence of P. aeruginosa PA103, a strain that produces and secretes high levels of the T3SS toxins ExoT and ExoU. We were unable to detect differences in ExoT secretion in calcium-depleted medium (see Fig. S6 in the supplemental material) or in ExoU-mediated cytotoxicity toward HeLa cells (see Fig. S7 in the supplemental material) in PA103ΔcbpA. Likewise, the virulence of PA103ΔcbpA in a mouse model of pneumonia was not decreased compared to the virulence of the parental strain; at 18 h postinfection, the bacterial burdens in the lungs, spleens, and livers were not statistically significantly different (see Fig. S8 in the supplemental material). To address whether CbpA affects the expression, degradation, or modification of proteins, we performed 2D-PAGE to compare the protein expression profiles of PAO1, a cbpA mutant, and a CbpA-overexpressing strain (see Materials and Methods for details). Except for some housekeeping proteins (RNA polymerase β subunit, 50S ribosomal protein, and PA2830 [HtpX], a heat shock-induced protease), no significant difference in the protein expression profiles was detected between any of these strains (see Fig. S9 in the supplemental material). These results suggest that CbpA may regulate as-yet-unknown functions in P. aeruginosa.
Structural modeling of CbpA.
As our results thus far failed to provide insights into the function of CbpA, we performed an additional bioinformatic analysis of the N-terminal region of CbpA. Iterative PSI-BLAST analysis demonstrated that amino acids 1 to 132 of CbpA are 47% (26%) and 35% (17%) similar (identical) to N-terminal regions (1 to 136 amino acid residues) of hypothetical proteins YP_433573 (NCBI accession no.; a 259-amino-acid protein) from H. chejuensis KCTC2396 and NP_214443 (NCBI accession no.; a 619-amino-acid protein) from “Aquifex aeolicus” VF5, respectively. The N-terminal regions (1 to 124 amino acid residues) of these proteins contain CAP domains, suggesting that the N-terminal region of CbpA may be composed of a degenerate CAP domain. We carried out structural modeling of CbpA using the Sequence Alignment and Modeling (SAM) system (version SAM-T06; http://compbio.soe.ucsc.edu/SAM_T06/T06-query.html) (28) and found that CbpA seems to consist of two β-barrel folding structures, which are typical structural folding patterns for CAP domains (see Fig. S10 in the supplemental material). We note that the predicted CbpA model is similar to the crystal structure of the protein kinase A (PKA) regulatory subunit RIα lacking the first 91 amino acids from Bos taurus (see Fig. S10 in the supplemental material) (49).
CbpA exhibits polar localization.
The regulatory subunits of eukaryotic PKA are anchored to specific subcellular compartments by binding to PKA-anchoring proteins and thereby play an important role in spatiotemporal regulation of the activity of PKA catalytic subunits (58). Based on the potential structural similarity of CbpA with eukaryotic PKA regulatory subunits, we tested whether CbpA exhibits spatial localization in P. aeruginosa.
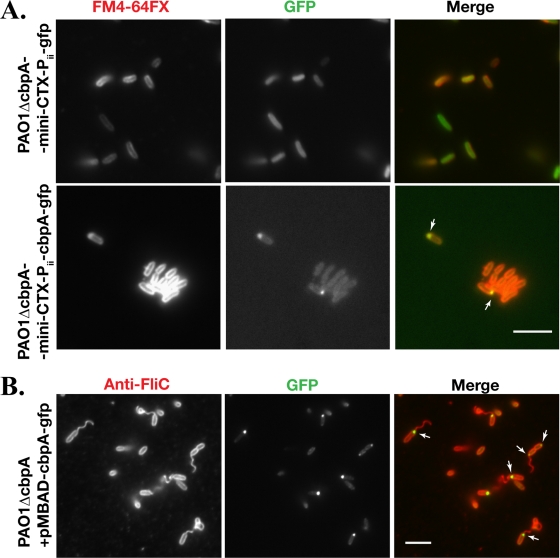
We constructed a CbpA-GFP fusion protein in which GFP was linked to the C terminus with a five-amino-acid flexible linker sequence (GGGKL). The fusion protein was placed under the control of the native cbpA promoter (the Pii region shown in Fig. 3A). The construct was integrated as a single copy into the attB site on the chromosome of PAO1ΔcbpA. We confirmed that the CbpA-GFP fusion protein binds to cAMP-conjugated agarose (data not shown), indicating that GFP did not disrupt the ability of CbpA to bind to cAMP. When PAO1ΔcbpA-mini-CTX-Pii-cbpA-gfp was grown to exponential phase, very little GFP signal was detected by fluorescence microscopy or by immunoblot analysis (data not shown). However, the CbpA-GFP fusion protein was detectable in stationary-phase-grown cells (OD600 of 3.0 to 3.5), suggesting that CbpA levels are growth phase regulated. Strikingly, CbpA exhibited polar localization (Fig. 4A, bottom). In the control strain PAO1ΔcbpA-mini-CTX-Pii-gfp, expressing GFP under the control of the Pii promoter from the attB site, only a diffuse cytoplasmic GFP signal was observed (Fig. 4A, top).
FIG. 4.
CbpA is polarly localized. (A) GFP or CbpA-GFP fusion protein was expressed from a single-copy gene integrated into the chromosome of PAO1ΔcbpA at the attB site. The bacteria were grown to an OD600 of ∼3.5, and the cell membrane was visualized by staining with FM4-64FX. Bar, 10 μm. The arrows indicate where polar localization of CbpA-GFP is observed. (B) PAO1ΔcbpA expressing CbpA-GFP from the plasmid pMBAD-cbpA-GFP under the control of an l-arabinose-inducible promoter (PBAD) was grown to an OD600 of ∼1.5. Cells were fixed and stained with rabbit anti-FliC antibody and Alexa Fluor 594-conjugated donkey anti-rabbit IgG. The arrows indicate where CbpA-GFP is localized. Bar, 10 μm. CbpA-GFP localizes to the old pole and to the flagellated pole.
Using an antibody to flagella, we next determined whether CbpA localized to a flagellated pole. As the expression level of CbpA-GFP from mini-CTX-Pii-cbpA-gfp was low even in stationary-phase cells, we constructed a multicopy plasmid, pMBAD-cbpA-gfp, that expresses the CbpA-GFP fusion protein under the control of an l-arabinose-inducible promoter (PBAD). Induction of CbpA-GFP fusion protein expression by the addition of up to 0.1% l-arabinose did not affect growth, cell morphology, twitching motility, or swimming motility (data not shown), and CbpA-GFP was found in the soluble fraction of the cell lysates (Fig. 5C). As shown in Fig. 4B, CbpA-GFP was concentrated mostly at the flagellated pole, though bipolar localization was observed in a small proportion of cells (data not shown). In dividing cells, CbpA-GFP foci were observed mostly at the pole distal to the division septum, suggesting that CbpA preferentially localizes to the old pole (Fig. 4B).
FIG. 5.
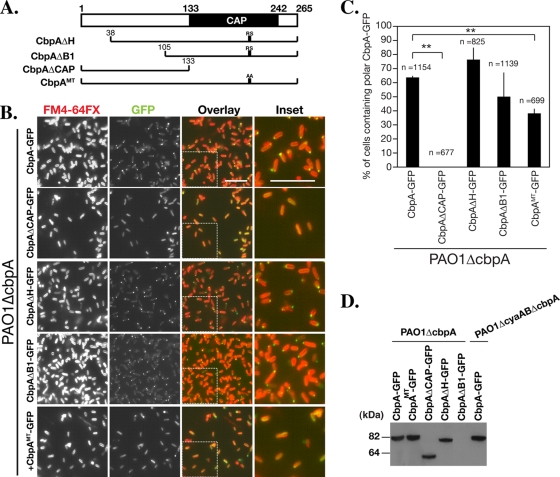
The CbpA CAP domain is necessary for polar localization. (A) Schematic diagram of the CbpA mutants used for the localization studies with results shown in panel B. RS indicates highly conserved two amino acid residues, Arg207 and Ser208, in the CAP domain of CbpA. AA indicates substitution of the two amino acid residues with Ala207 and Ala208 in CbpAMT. (B) Localization of CbpA mutants expressed from a plasmid under the control of an l-arabinose-inducible promoter in PAO1ΔcbpA. The cell membrane was stained with FM4-64FX. The rightmost set of panels shows enlargements of the sections indicated in the third set of panels. (C) Quantification of polar localization of CbpA-GFP mutants. The efficiency of polar localization was determined by the counting the number of DAPI-stained bacteria (the total cell number; see Materials and Methods) and the number of bacteria with polar CbpA-GFP, using ImageJ. Three images containing a total of at least 670 bacteria were counted for each sample. The CAP domain is required for CbpA-GFP localization. Error bars indicate standard deviation. **, P < 0.01 (two-tailed Student's t test). (D) Steady-state protein levels of CbpA-GFP mutants. The bacteria were grown to an OD600 of ∼3.5, and lysates were separated by SDS-PAGE and immunoblotted with an antibody to GFP. Each variant migrates at the expected molecular mass. The expression level of CbpAΔB1-GFP was lower than that of other CbpA-GFP variants but could be detected after longer exposures (data not shown).
We next examined whether Vfr is required for polar localization of CbpA, by expressing CbpA-GFP under an arabinose-inducible promoter in PAO1Δvfr. There was no significant difference in the efficiency of polar localization of CbpA-GFP in PAO1Δvfr (data not shown). These experiments suggest that while Vfr is required for CbpA transcription, it is not required for its polar localization.
The CbpA CAP domain is necessary for polar localization.
To investigate which regions of CbpA are essential for polar localization, we constructed truncated variants of CbpA, CbpAΔH, CbpAΔB1, and CbpAΔCAP (Fig. 5A), and examined the localization of the C-terminal GFP fusion proteins in PAO1ΔcbpA by fluorescent microscopy. With the exception of CbpAΔB1-GFP, the deletions did not affect the production and stability of the fusion proteins significantly (Fig. 5D). Although the levels of CbpAΔB1-GFP were reduced, sufficient protein was produced to allow detection by fluorescent microscopy. As shown in Fig. 5A and B, deletion of the N-terminal tandem helices (CbpAΔH-GFP) or the first putative β-barrel structure, consisting of amino acid residues 38 to 105 (CbpAΔB1-GFP), did not decrease polar localization of CbpA-GFP. Deletion of the C-terminal CAP domain (CbpAΔCAP) abolished polar localization (Fig. 5B and C). These studies indicate that the CAP domain of CbpA is necessary for polar localization.
cAMP binding is essential for polar localization of CbpA.
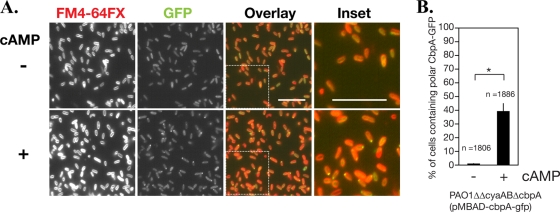
In order to distinguish whether structural characteristics of the CAP domain or cAMP binding is required for polar localization of CbpA, CbpAMT fused to GFP was expressed from an arabinose-inducible promoter in PAO1ΔcbpA, and the localization of CbpAMT-GFP was investigated. Although the CAP domain mutation did not abolish the polar localization completely, the frequency of polar localization of CbpAMT-GFP was significantly decreased (37.9% of cells, n = 699; P < 0.01) compared with that of the wild type (63.4%, n = 1,154) (Fig. 5); it is possible that the substitution of only two amino acid residues conserved in the CAP domain of CbpA may not abolish its ability to bind to cAMP completely in vivo. The expression level and stability of CbpAMT-GFP were comparable to those of CbpA-GFP (Fig. 5D). We further tested the role of cAMP by expressing CbpA-GFP in PAO1ΔcyaABΔcbpA, a strain in which no cAMP is synthesized because both adenylate cyclase genes cyaA and cyaB are deleted. Note that the stability and expression level of CbpA-GFP were not affected by the deletion of cyaA and cyaB (Fig. 5D). As shown in Fig. 6A and B, only 0.8% of cells (n = 1,806) harbored CbpA-GFP foci at the cell pole. Incubation of cells with cAMP restored CbpA-GFP polar localization in ∼40% of the cells (n = 1,886, P < 0.05) (Fig. 6A and B). These results indicate that binding of cAMP to the CAP domain is necessary and sufficient for polar localization of CbpA.
FIG. 6.
cAMP is necessary for CbpA polar localization. (A) CbpA-GFP was expressed from a plasmid under the control of an arabinose-inducible promoter in PAO1ΔcyaABΔcbpA, in which both adenylate cyclase genes cyaA and cyaB are deleted. The bacterial culture was grown to an OD600 of ∼2.0, diluted 10-fold with PBS buffer, and incubated with (+) or without (−) 10 mM cAMP at 4°C for 60 min. Cell membranes were visualized by staining with FM4-64FX. (B) The fractions of cells with polar localization of CbpA-GFP in the presence or absence of cAMP were determined from three different images (at least 1,800 cells), using ImageJ. Error bars indicate standard deviation. *, P < 0.05 (two-tailed Student's t test).
DISCUSSION
cAMP signaling in bacteria plays an important role in regulating the virulence of pathogenic microbes. Bioinformatics searches of over 1,000 bacterial species have demonstrated the presence of multiple cAMP-binding proteins with diverse domain organizations. However, no detailed studies of those cAMP-binding proteins, other than CRP family regulators, have been reported. In this study, we identified CbpA as a novel cAMP-binding protein in P. aeruginosa. Interestingly, CbpA seems to be more structurally similar to eukaryotic PKA regulatory subunits than to CRP family transcriptional regulators by virtue of the presence of an N-terminal CAP-like domain and a C-terminal CAP domain. We demonstrated that CbpA binds to cAMP via its C-terminal functional CAP domain and undergoes a conformational change upon cAMP binding. Although the N-terminal CAP-like region does not seem to be functional because of its low homology with functional CAP domains and because of the inability of CbpAMTHis to bind to cAMP-agarose (Fig. 2B), we cannot rule out the possibility that the N-terminal CAP-like domain may bind to cAMP cooperatively in conjunction with binding of cAMP to the C-terminal CAP domain. Transcription of cbpA is directly regulated by Vfr, suggesting the possibility that CbpA mediates Vfr-dependent phenotypes. However, exhaustive testing failed to reveal any differences between wild-type PAO1 or PA103 and in-frame deletions in cbpA in either of these strain backgrounds. Likewise, where examined, overexpression of CbpA did not have an apparent effect. Of particular interest is our finding that CbpA preferentially localizes to the old cell pole in a cAMP-dependent manner. We hypothesize that polar localization of CbpA may be important to its function.
CbpA-like proteins in other bacteria.
CbpA structural orthologs can be identified in three Pseudomonas species and five nonpseudomonad bacterial species. In addition, other proteins with a similar domain structure, comprising an N-terminal undefined region and one or two CAP domains at the C terminus, can be found in many other bacteria. One example, Rv0104 of M. tuberculosis H37Rv, encodes a 504-amino-acid protein with a C-terminal CAP domain whose transcription is predicted to be regulated by a cAMP-dependent transcriptional regulator, Rv3676 (1); its function in M. tuberculosis is still unknown. Likewise, Myxococcus xanthus possesses multiple cAMP-binding proteins (16). One of these cAMP-binding proteins, CbpB (YP_635154; NCBI database accession no.), is a 405-amino-acid protein with C-terminal tandem CAP domains, suggesting that CbpB has two β-barrel structures, similar to eukaryotic PKA regulatory subunits and CbpA in P. aeruginosa. Although the molecular function of CbpB is still unknown, CbpB seems to be involved in temperature and osmotic tolerance in M. xanthus (25). Of note, cbpA mutants of P. aeruginosa PAO1 did not exhibit temperature or osmotic sensitivity (T. Endoh and J. N. Engel, unpublished data). It will be interesting to determine whether these CbpA-like proteins exhibit distinct subcellular localization and/or change their distribution in a cAMP-dependent manner. Further studies of these CbpA-like proteins will provide clues that reveal the molecular function of those proteins and give important insights into the complexity of cAMP signaling systems in bacteria.
Regulation of cbpA.
Using previously identified Vfr-binding sites and mutational analysis, Kanack et al. defined a 21-bp Vfr-binding consensus sequence from which they identified 12 new potential Vfr-binding sites at upstream sites or in the intergenic regions of virulence-associated genes (22). The list of genes whose transcription is directly regulated by Vfr has expanded to include plcN, plcHR, pbpG, prpL, algD, vfr-PA0653, argH-fimS, and pilM-ponA in addition to previously reported Vfr-binding sites upstream of toxA, toxR, lasR, and fleQ (2, 10, 22, 52). Based on our comprehensive studies, we can now add cbpA to this growing list of Vfr-regulated genes. The VBS for cbpA is 71% (15 bp) identical to the 21-bp consensus sequence (Fig. 3), suggesting that the consensus sequence can be expanded (Fig. 3). Refining the Vfr-binding consensus sequence would help to predict direct Vfr targets out of more than 200 Vfr-regulated genes (56).
While we clearly showed that cbpA transcription in PAO1 is induced by a low-calcium condition (Fig. 3A), cbpA (PA4704) was not identified as a gene whose transcription is induced under low-calcium conditions in the extensive transcriptome analyses of P. aeruginosa strain PAK (56). This discrepancy may reflect strain differences. Alternatively, our transcriptional analysis was carried out with a multicopy reporter plasmid, which may have enhanced the sensitivity of the assay compared to that of microarray analysis.
Several of the genes upstream of cbpA are conserved in other pseudomonads. For example, homologs of PA4703, PA4702, and PA4701 are also found in P. fluorescens, P. mendocina, and P. stutzeri. Interestingly, P. putida, “P. entofila,” and P. syringae have maintained homologs of PA4701 and PA4702 but no longer encode PA4703 and/or cbpA (PA4704). The significance of the somewhat conserved nature of this locus may give clues to gene function.
CbpA is polarly localized by a cAMP-dependent mechanism.
It is increasingly recognized that many bacterial complexes are polarly localized, particularly gene products involved in bacterial cell cycle, motility, and secretion systems (44). Likewise, many bacterial signaling components are polarly localized, including proteins involved in chemosensory systems (5, 9, 51), other two-component signaling systems, and di-cyclic-GMP metabolism (21, 24, 38). Unexpectedly, we discovered that CbpA-GFP expressed under the control of its native promoter from the chromosome or under the control of an arabinose-inducible promoter from a multicopy plasmid is polarly localized and that cAMP binding to the CAP domain is essential for the polar localization (Fig. 5 and 6). Although we cannot eliminate the possibility that fusion to GFP may affect the molecular function of the native protein, we consider this unlikely because (i) CbpA-GFP bound to cAMP agarose and (ii) CbpA-GFP changes its localization in a cAMP-dependent manner. In conjunction with our results demonstrating that CbpA undergoes a conformational change upon binding cAMP, we speculate that a conformational change is required for polar localization. Whether this structural change allows the protein to bind to other proteins remains to be investigated. To the best of our knowledge, this is the first example of a bacterial protein whose subcellular localization is regulated by binding cAMP. Current studies are aimed at elucidating the physiologic role of CbpA by identifying factors required for CbpA polar localization and proteins that associate with CbpA.
Supplementary Material
Acknowledgments
We thank J. West and D. Wozniak for providing P. aeruginosa strains and the anti-FliC antibody, P. Balachandran and J. Pielage for technical help, and past and current Engel lab members for discussion and comments.
This work was supported by NIH grant R01 AI42806 (J.N.E.).
Footnotes
Published ahead of print on 2 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akhter, Y., S. Yellaboina, A. Farhana, A. Ranjan, N. Ahmed, and S. E. Hasnain. 2008. Genome scale portrait of cAMP-receptor protein (CRP) regulons in mycobacteria points to their role in pathogenesis. Gene 407:148-158. [DOI] [PubMed] [Google Scholar]
- 2.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai, G., L. A. McCue, and K. A. McDonough. 2005. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J. Bacteriol. 187:7795-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balachandran, P., L. Dragone, L. Garrity-Ryan, A. Lemus, A. Weiss, and J. Engel. 2007. The ubiquitin ligase Cbl-b limits Pseudomonas aeruginosa exotoxin T-mediated virulence. J. Clin. Investig. 117:419-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardy, S. L., and J. R. Maddock. 2005. Polar localization of a soluble methyl-accepting protein of Pseudomonas aeruginosa. J. Bacteriol. 187:7840-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beavo, J. A., and L. L. Brunton. 2002. Cyclic nucleotide research—still expanding after half a century. Nat. Rev. Mol. Cell Biol. 3:710-718. [DOI] [PubMed] [Google Scholar]
- 8.Botsford, J. L., and J. G. Harman. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang, P., M. Habash, and L. L. Burrows. 2005. Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta, N., E. P. Ferrell, K. J. Kanack, S. E. H. West, and R. Ramphal. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 184:5240-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Crecy-Lagard, V., P. Glaser, P. Lejeune, O. Sismeiro, C. E. Barber, M. J. Daniels, and A. Danchin. 1990. A Xanthomonas campestris pv. campestris protein similar to catabolite activation factor is involved in regulation of phytopathogenicity. J. Bacteriol. 172:5877-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derouaux, A., D. Dehareng, E. Lecocq, S. Halici, H. Nothaft, F. Giannotta, G. Moutzourelis, J. Dusart, B. Devreese, F. Titgemeyer, J. Van Beeumen, and S. Rigali. 2004. Crp of Streptomyces coelicolor is the third transcription factor of the large CRP-FNR superfamily able to bind cAMP. Biochem. Biophys. Res. Commun. 325:983-990. [DOI] [PubMed] [Google Scholar]
- 13.Dong, Y. H., X. F. Zhang, J. L. Xu, A. T. Tan, and L. H. Zhang. 2005. VqsM, a novel AraC-type global regulator of quorum-sensing signalling and virulence in Pseudomonas aeruginosa. Mol. Microbiol. 58:552-564. [DOI] [PubMed] [Google Scholar]
- 14.Endoh, T., H. Habe, T. Yoshida, H. Nojiri, and T. Omori. 2003. A CysB-regulated and sigma54-dependent regulator, SfnR, is essential for dimethyl sulfone metabolism of Pseudomonas putida strain DS1. Microbiology 149:991-1000. [DOI] [PubMed] [Google Scholar]
- 15.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. Hauser, and J. N. Engel. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, J. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745-754. [DOI] [PubMed] [Google Scholar]
- 18.Harman, J. G. 2001. Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta 1547:1-17. [DOI] [PubMed] [Google Scholar]
- 19.He, Y. W., A. Y. Ng, M. Xu, K. Lin, L. H. Wang, Y. H. Dong, and L. H. Zhang. 2007. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64:281-292. [DOI] [PubMed] [Google Scholar]
- 20.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 21.Huitema, E., S. Pritchard, D. Matteson, S. K. Radhakrishnan, and P. H. Viollier. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025-1037. [DOI] [PubMed] [Google Scholar]
- 22.Kanack, K. J., L. J. Runyen-Janecky, E. P. Ferrell, S. J. Suh, and S. E. H. West. 2006. Characterization of DNA-binding specificity and analysis of binding sites of the Pseudomonas aeruginosa global regulator, Vfr, a homologue of the Escherichia coli cAMP receptor protein. Microbiology 152:3485-3496. [DOI] [PubMed] [Google Scholar]
- 23.Kang, P. J., A. R. Hauser, G. Apodaca, S. Fleiszig, J. Wiener-Kronish, K. Mostov, and J. N. Engel. 1997. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 24:1249-1262. [DOI] [PubMed] [Google Scholar]
- 24.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60:1026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura, Y., H. Nakato, K. Ishibashi, and S. Kobayashi. 2005. A Myxococcus xanthus CbpB containing two cAMP-binding domains is involved in temperature and osmotic tolerances. FEMS Microbiol. Lett. 244:75-83. [DOI] [PubMed] [Google Scholar]
- 26.Kolb, A., S. Busby, H. Buc, S. Garges, and S. Adhya. 1993. Transcriptional regulation by cAMP and its receptor protein. Annu. Rev. Biochem. 62:749-795. [DOI] [PubMed] [Google Scholar]
- 27.Krakow, J. S., and I. Pastan. 1973. Cyclic adenosine monophosphate receptor: loss of cAMP-dependent DNA binding activity after proteolysis in the presence of cyclic adenosine monophosphate. Proc. Natl. Acad. Sci. USA 70:2529-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogh, A., M. Brown, I. S. Mian, K. Sjölander, and D. Haussler. 1994. Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235:1501-1531. [DOI] [PubMed] [Google Scholar]
- 29.Liang, W., A. Pascual-Montano, A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153:2964-2975. [DOI] [PubMed] [Google Scholar]
- 30.Liu, P. V. 1966. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis: identity of the lethal toxins produced in vitro and in vivo. J. Infect. Dis. 116:481-489. [DOI] [PubMed] [Google Scholar]
- 31.Mandell, G. L., J. E. Bennett, and R. Dolin. 2005. Principles and practice of infectious diseases, 6th ed. Churchill Livingstone Inc., New York, NY.
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Murray, T., and B. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 35.Oglesby, A., J. Farrow, J. Lee, A. Tomaras, E. Greenberg, E. Pesci, and M. Vasil. 2008. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 283:15558-15567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohmori, M., and S. Okamoto. 2004. Photoresponsive cAMP signal transduction in cyanobacteria. Photochem. Photobiol. Sci. 3:503-511. [DOI] [PubMed] [Google Scholar]
- 37.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 38.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rehmann, H., A. Wittinghofer, and J. L. Bos. 2007. Capturing cyclic nucleotides in action: snapshots from crystallographic studies. Nat. Rev. Mol. Cell Biol. 8:63-73. [DOI] [PubMed] [Google Scholar]
- 40.Rickman, L., C. Scott, D. M. Hunt, T. Hutchinson, M. C. Menéndez, R. Whalan, J. Hinds, M. J. Colston, J. Green, and R. S. Buxton. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56:1274-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saffarini, D. A., R. Schultz, and A. Beliaev. 2003. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 185:3668-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 44.Shapiro, L., H. H. McAdams, and R. Losick. 2002. Generating and exploiting polarity in bacteria. Science 298:1942-1946. [DOI] [PubMed] [Google Scholar]
- 45.Shenoy, A. R., and S. S. Visweswariah. 2006. New messages from old messengers: cAMP and mycobacteria. Trends Microbiol. 14:543-550. [DOI] [PubMed] [Google Scholar]
- 46.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 47.Smith, R. S., M. C. Wolfgang, and S. Lory. 2004. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 72:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 49.Su, Y., W. R. Dostmann, F. W. Herberg, K. Durick, N. H. Xuong, L. Ten Eyck, S. S. Taylor, and K. I. Varughese. 1995. Regulatory subunit of protein kinase A: structure of deletion mutant with cAMP binding domains. Science 269:807-813. [DOI] [PubMed] [Google Scholar]
- 50.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 51.Wadhams, G. H., A. C. Martin, A. V. Warren, and J. P. Armitage. 2005. Requirements for chemotaxis protein localization in Rhodobacter sphaeroides. Mol. Microbiol. 58:895-902. [DOI] [PubMed] [Google Scholar]
- 52.West, S. E. H., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. T. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52:873-893. [DOI] [PubMed] [Google Scholar]
- 54.Whitchurch, C. B., S. A. Beatson, J. C. Comolli, T. Jakobsen, J. L. Sargent, J. J. Bertrand, J. West, M. Klausen, L. L. Waite, P. J. Kang, T. Tolker-Nielsen, J. S. Mattick, and J. N. Engel. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 55:1357-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. USA 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 57.Won, H. S., Y. S. Lee, S. H. Lee, and B. J. Lee. 2009. Structural overview on the allosteric activation of cyclic AMP receptor protein. Biochim. Biophys. Acta 1794:1299-1308. [DOI] [PubMed] [Google Scholar]
- 58.Wong, W., and J. D. Scott. 2004. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5:959-970. [DOI] [PubMed] [Google Scholar]
- 59.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.