Abstract
Plants and fungi respond to environmental light stimuli via the action of different photoreceptor modules. One such class, responding to the blue region of light, is constituted by photoreceptors containing so-called light-oxygen-voltage (LOV) domains as sensor modules. Four major LOV families are currently identified in eukaryotes: (i) the plant phototropins, regulating various physiological effects such as phototropism, chloroplast relocation, and stomatal opening; (ii) the aureochromes, mediating photomorphogenesis in photosynthetic stramenopile algae; (iii) the plant circadian photoreceptors of the zeitlupe (ZTL)/adagio (ADO)/flavin-binding Kelch repeat F-box protein 1 (FKF1) family; and (iv) the fungal circadian photoreceptors white-collar 1 (WC-1). Blue-light-sensitive LOV signaling modules are also widespread throughout the prokaryotic world, and physiological responses mediated by bacterial LOV photoreceptors were recently reported. Thus, the question arises as to the evolutionary relationship between the pro- and eukaryotic LOV photoreceptor systems. We used Bayesian and maximum-likelihood tree reconstruction methods to infer evolutionary scenarios that might have led to the widespread appearance of LOV domains among the pro- and eukaryotes. The phylogenetic study presented here suggests a bacterial origin for the LOV domains of the four major eukaryotic LOV photoreceptor families, whereas the LOV sensor domains were most likely recruited from the bacteria in the course of plastid and mitochondrial endosymbiosis.
A plant family of photoreceptors, the phototropins, containing so-called light-oxygen-voltage (LOV) domains as the blue-light-sensitive signaling switches (12), were previously identified as the key modulators of a variety of plant blue-light responses, including plant phototropism (23), chloroplast relocation, and stomatal and leaf opening (25, 39). A second family (ZTL/ADO/FKF1 family) of eukaryotic LOV domain-containing proteins is constituted by the zeitlupe (ZTL) and the flavin-binding Kelch repeat F-box (FKF1) proteins. This family was found to play a primary role in the photocontrol of flowering and in the light-dependent regulation of the circadian period in plants (37, 53). Phototropins and ZTLs together with the fungal white-collar 1 (WC-1) proteins of, e.g., Neurospora crassa (3), which are involved in the blue-light dependent control of fungal circadian responses (19), constitute the three major eukaryotic LOV families in plants and fungi. A more recently discovered fourth family of LOV domain-containing blue-light receptors, the so-called aureochromes (58), seem to be restricted to stramenopile algae such as Vaucheria frigida and some marine diatoms (e.g., Thalassiosira pseudonana). Recently, the presence of LOV domain signaling modules was also predicted for animals, including humans. This functional assignment was solely based on sequence similarity to known LOV systems in plants and bacteria (62); however, experimental evidence is still missing. LOV domains are small (110 amino acids), bind flavin mononucleotide as chromophores, and fold independently to adopt a Per-Arndt-Sim (PAS) fold (40), which they share with other ligand-binding sensor modules (59). The best-characterized LOV domains are LOV1 and LOV2, found in plant phototropins (11), which probably regulate the physiological response via a blue-light-dependent phosphorylation of a serine/threonine kinase located C terminal to the LOV sensor domains in plant phototropins. (10). The light-sensitive function of the LOV proteins is based on the presence of a strictly conserved cysteine residue located at a distance of about 4 Å from the isoalloxazine ring of the flavin chromophore. In all LOV domains, this cysteine residue is found as the fourth residue in a highly conserved sequence motif GXNCRFLQG defined previously based on plant phototropin sequences (45). Irradiation with blue light results in the formation of a covalent bond between this cysteine and the carbon atom in position 4a of the flavin isoalloxazine ring. In the dark, this bond reopens within minutes to hours, depending on the respective LOV protein (32). The increasing number of completely sequenced microbial genomes revealed the presence of a variety of putative LOV photosensory proteins in bacteria and archaea (31) whose biological roles remain in most cases elusive. Only recently, experiments showed that LOV domain-containing proteins found in several bacterial taxa, namely, a LOV sulfate transporter anti-sigma factor antagonist protein from the common soil bacterium Bacillus subtilis and LOV histidine kinases of the mammalian pathogen Brucella abortus (57) and the freshwater-dwelling microbe Caulobacter crescentus (44), mediate physiological responses toward environmental blue-light stimuli. The latter LOV domain-containing histidine kinases are blue-light sensitive, displaying a photochemistry similar to that of the plant phototropins with the blue-light stimulus inducing autophosphorylation in the kinase which in turn triggers the respective physiological response (8, 44, 57). Furthermore, a number of biochemical and photochemical studies demonstrated a conserved primary LOV photochemistry among several bacteria (8, 28, 30, 44, 57). Finally, only a few years after the identification of plant phototropin serine/threonine kinase systems with their blue-light-sensitive LOV sensory modules, experimental evidence has accumulated for the presence of similar systems in bacteria, namely, LOV histidine kinases regulating blue-light-dependent responses via phototropin-like autophosphorylation systems. These findings prompted us to investigate the evolutionary history as well as the distribution of the LOV sensor domain in the pro- and eukaryotic kingdoms. To this end, we have performed a phylogenetic analysis comprising 223 putative LOV sequences obtained from completely sequenced genomes of 22 eukaryotes, 115 bacteria, and 3 archaea.
MATERIALS AND METHODS
Sequence retrieval and domain content analysis.
Protein sequences with significant sequence similarity to LOV domains were obtained using the PSI-BLAST (2) utility. Numerous eukaryotic and bacterial LOV photoreceptor proteins with demonstrated function or photochemistry were used as query sequences in independent BLAST searches at the NCBI GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Query sequences included Arabidopsis thaliana phototropin 1 (NM_114447) and ZTL/ADO-FKF1 LOV (NP_849983), Adiantum capillus-veneris neochrome (BAA36192), Neurospora crassa WC-1 (NCU02356), and Vaucheria frigida aureochome (BAF91488) on the eukaryotic side. Prokaryotic LOV sequences used for BLAST searches were Bacillus subtilis YtvA (O34627), the Pseudomonas putida sensory box proteins (Q88E39 and Q88JB0), and Brucella abortus (Q577Y7), Pseudomonas syringae (Q881J7), and Caulobacter crescentus (Q9ABE3) LOV histidine kinases. The resulting hits were manually scanned for the presence of the canonical LOV sequence motif GXNCRFLQG and, most importantly, for the presence of the photoreactive cysteine residue. This cysteine residue is strictly conserved and indispensable for functioning of all LOV proteins described so far. Independent genome mining in various fungal genome projects (accessible at the Munich Information Center for Protein Sequences, http://mips.gsf.de) employing protein sequence-seeded BLAST searches (1) was performed to obtain additional LOV domain-containing protein sequences from several fungal taxa. The resulting sequences (see Table S1 in the supplemental material for details) covered representatives of different taxonomic groups and kingdoms, including eukaryotic phototropins (6), ZTL/ADO/FKF1 sequences (53), fungal WC-1 sequences (3), stramenopile algal aureochromes (58), putative archaeal LOV domain-containing proteins, and, most importantly, a comprehensive list of putative LOV sequences from a broad variety of bacterial taxa. Phototropin sequences were divided into LOV1 and LOV2 domains and separately included in the alignment. Furthermore, phototropin LOV1 and LOV2 sequences of the chimeric phototropin-phytochrome photoreceptor neochrome (56) were included. Moreover, a representative list of animal sequences with similarity to plant and bacterial LOV domains was additionally included in the alignment. Those putative animal LOV sequences were Q9ULD8 (Homo sapiens), A8WHX9 (Danio rerio), Q7YW98 (Manduca sexta), and Q9WVJ0 (Mus musculus). The retrieved full-length protein sequences were subjected to a functional domain content analysis using the Simple Modular Architecture Research Tool (SMART) (41, 48) at the European Molecular Biology Laboratory (EMBL) (http://smart.embl-heidelberg.de/).
The small-subunit (SSU) rRNA gene sequence alignment that was used to reconstruct the phylogenetic species tree of the LOV photoreceptor protein-containing taxa was obtained from the SILVA database (http://www.arb-silva.de/) (43). Sequences not found in SILVA were retrieved from the European rRNA database (64) (http://bioinformatics.psb.ugent.be/webtools/rRNA/), from the GreenGenes SSU rRNA gene database (http://greengenes.lbl.gov) (13), or from the NCBI GenBank (www.ncbi.nlm.nih.gov). Sequences for species not present in any of these databases are considered missing. For the plant species in our analysis, the nucleus-, mitochondrion-, and chloroplast-encoded SSU rRNA sequences were included. For the fungal and animal taxa, the nuclear and mitochondrial SSU rRNA sequences were added correspondingly. Finally, since some SSU rRNA sequences are very diverged from the remaining, a few more taxa that help to bridge this gap were additionally included in the analysis. Those taxa were Schizosaccharomyces octosporus, Cryphonectria parasitica, and Prototheca wickerhamii (see Table S1 in the supplemental material for details).
Sequence alignment.
Protein sequences of the isolated LOV core domain, and not full-length photoreceptor protein sequences, were aligned with M-Coffee (63). M-Coffee is a meta-alignment method which combines five different aligning strategies, i.e., T-Coffee (38), ClustalW (60), Muscle (15), Mafft (26), and Probcons (14). Independent alignments were also constructed using these five methods to assess the quality of the final alignment. The DNA alignment of the LOV domain was subsequently obtained from the protein alignment based on the corresponding codon frames. Visual inspection and judgment based on alignment scores showed that M-Coffee produced the best result, although T-Coffee and ClustalW produced similar alignments. The alignment from M-Coffee was edited manually and used in subsequent phylogenetic analysis. For the SSU rRNA alignment (see Fig. S4 in the supplemental material), most of the sequences were already aligned in the SILVA database. rRNA sequences which were absent in SILVA but present in the above-mentioned databases were aligned into the existing alignment using the SILVA web aligner (http://www.arb-silva.de/aligner/). Sequences that the web aligner failed to align were excluded. In particular, mitochondrial rRNA sequences from the animal taxa were excluded as they represent the 12S rRNA regions and the web aligner did not align them correctly. Moreover, the Plant3_mt sequence (mitochondrial rRNA sequence of Chlamydomonas reinhardtii) was also excluded due to scrambled rRNA gene regions over the mitochondrial DNA (5). Finally, all columns consisting of more than 80% of gaps were discarded from the rRNA alignment. For the purpose of comparison, Gblocks (9) was applied with reduced stringency options to filter out divergent regions. It should be noted that gap columns were removed only from the rRNA alignments and not from the LOV alignments in order to retain as much phylogenetic information as possible from the comparatively short LOV sequences. The resulting LOV and SSU rRNA alignments are presented in the supplemental material.
Phylogenetic tree reconstruction. (i) Bayesian analysis.
Six different runs of MrBayes (24) were conducted to reduce the risk of getting stuck in local optima. Due to the large number of sequences in both alignments (rRNA and LOV) we used one Markov chain Monte Carlo chain per run, we set the total number of generations to 50 million, and we sampled trees every 1,000 generations. Thus, each run produced a collection of 50,000 trees. The convergence of each run was evaluated with Tracer v1.4 (A. Rambaut and A. J. Drummond, 2007; http://beast.bio.ed.ac.uk/Tracer/). The Tracer analysis showed that the LOV chain became trapped in a suboptimal area in three runs. This result suggests that a reconstruction of the LOV phylogeny is difficult but nevertheless feasible. For the remaining three runs, MrBayes converged to the same likelihood plateau. The best run that reached this plateau after 11 million generations was used for the subsequent tree analysis. The burn-in value was therefore set to 11,000. In contrast, the SSU rRNA data converged rapidly to the same likelihood plateau in all six runs. Thus, the SSU rRNA tree was summarized from all runs with a burn-in of 10,000.
(ii) ML and bootstrap analyses.
In order to find the best maximum-likelihood (ML) tree, 10 independent runs of RAxML 7.0.4 (54) were conducted. The tree with the highest likelihood among all runs was designated the ML tree. Interestingly, all RAxML runs on the LOV sequences inferred 10 different tree topologies, whereas the SSU rRNA runs provided the same tree 10 times.
Moreover, we applied nonparametric bootstraps using RAxML 7.0.4 and IQPNNI 3.3.1 (62a) with 1,000 bootstrap replicates to evaluate the branch support of the tree topology. For RAxML, we applied the rapid bootstrap method (55). For the IQPNNI heuristic, at least 200 iterations were executed and the stopping rule was turned on to automatically determine how many iterations are needed to reach an ML tree with 95% confidence. Subsequently, an extended majority-rule consensus tree was constructed using PHYLIP's “consense” program (17).
For RAxML we used the GTR+G model (66) as the only nucleotide substitution model implemented in RAxML. For IQPNNI and MrBayes, the HKY85+G model of substitution was used for the LOV domain data set, whereas the TN93+G model was used for the SSU rRNA gene data, both with four discrete gamma rate categories. Note that the Modeltest program (42) suggested TVM+I+G for the LOV data and TN93+I+G for the SSU rRNA data. However, we decided to use a simpler evolution model for the LOV domain and the SSU rRNA gene data because of the short sequence lengths of the LOV domain and the estimated proportion of invariable sites, which is not sufficiently high, for both data sets.
(iii) Evaluation of tree topologies.
To determine whether the ML, MrBayes, and bootstrap consensus trees are equally good explanations of the LOV data, we employed the Shimodaira-Hasegawa test (50) and the approximately unbiased test (52). In addition, we used the one-sided Kishino-Hasegawa test (20, 27) to compare the best tree against each of the other trees to test if they were significantly worse than the best tree. All tests assumed a significance of 0.05. The tests were performed with Tree-puzzle (46) and CONSEL (51) as described previously (47). For the SSU rRNA gene, all three trees were very similar and thus no tests were performed.
RESULTS
Taxonomic distribution of the LOV signaling module in the prokaryotes.
Previous studies indicated that the LOV signaling module is present in all three kingdoms of life (31). Our BLAST (2) analysis revealed that currently the LOV signaling module can be found in about 3.5% (115 species/3,254 sequenced genomes) of the sequenced bacterial genomes and in approximately 2.3% (3 species/126 sequenced genomes) of the sequenced archaeal species. Among the Archaea, LOV domains seem to be restricted to Euryarchaeota, whereas they are more widely distributed in the bacterial kingdom, being widely dispersed throughout the Proteobacteria and Cyanobacteria, represented among the Firmicutes, and are present in a few Chloroflexi and Actinobacteria. It should be noted that the analysis of LOV domain distribution across the tree of life may be biased due to the limited or uneven availability of completely sequenced genomes. Therefore, Tables 1 and 2 display the number of species that contain putative LOV domain sequences and the number of completely sequenced genomes for the respective phyla. Furthermore, we have calculated an upper limit (f*) for the probability that a taxon from a systematic group where no LOV module was found in n genomes may contain a LOV domain. The resulting f* values are summarized in Tables 1 and 2. We computed f* only if at least one sequenced genome was available for a taxonomic group.
TABLE 1.
General overview over the taxonomic distribution of LOV homologues in Bacteria and Archaea
| Phylum | f*a | No. of LOV homologues/no. of sequenced genomesb (%) |
|---|---|---|
| Bacteria | ||
| Aquificae | 0.451 | 0/5 |
| Thermotogae | 0.221 | 0/12 |
| Thermodesulfobacteria | 0/0 | |
| Chrysiogenetes | 0/0 | |
| Nitrospira | 0/0 | |
| Deferribacteres | 0/0 | |
| Chloroflexi | 3/14 (21.4) | |
| Thermomicrobia | 0.776 | 0/2 |
| Fibrobacteres | 0/0 | |
| Proteobacteria | 84/1637 (5.1) | |
| Plactomycetes | 0.527 | 0/4 |
| Chlamydiae | 0.221 | 0/12 |
| Spirochaetes | 0.015 | 0/201 |
| Bacteroidetes | 0.036 | 0/81 |
| Chlorobi | 0.193 | 0/14 |
| Actinobacteria | 3/270 (1.1) | |
| Deinococcus-Thermus | 0.153 | 0/18 |
| Cyanobacteria | 14/118 (11.7) | |
| Firmicutes | 11/734 (1.5) | |
| Fusobacteria | 0.181 | 0/15 |
| Verrumicrobia | 0.312 | 0/8 |
| Acidobacteria | 0.776 | 0/2 |
| Dictyoglomi | 0.776 | 0/2 |
| Gennatimoadetes | 0/0 | |
| Archaea | ||
| Crenarchaeota | 0.076 | 0/38 |
| Euryarchaeota | 3/86 (3.5) | |
| Korarchaeota | 0.950 | 0/1 |
| Nanoarchaetoa | 0.950 | 0/1 |
f* is the largest admissible frequency of the LOV domain in the corresponding systematic group such that the probability of not finding any LOV domain in a sample of size n (number of sequenced genomes) is above 5%, where a geometric distribution is assumed.
Number of sequenced genomes as of April 2009 (NCBI-TaxBrowser, http://www.ncbi.nlm.nih.gov).
TABLE 2.
Overview over the distribution of LOV homologs in Bacteria and Archaea
| Taxon | No. of LOV homologues/no. of sequenced genomes (%)a |
|---|---|
| Proteobacteriab | 84/1,637 (5.1) |
| Alphaproteobacteria | 42/371 (11.3) |
| Caulobacterales | 2/6 (33) |
| Kiloniellales | 0/0 |
| Kopriimonadales | 0/0 |
| Kordiimonadales | 0/0 |
| Parvularculales | 1/1 (100) |
| Rhizobiales | 21/178 (11.8) |
| Rhodobacterales | 10/81 (12.3) |
| Rhodospirillales | 2/34 (5.9) |
| Rickettsiales | 0/42 [0.067] |
| Sneathiellales | 0/0 |
| Sphingomonadales | 6/25 (24) |
| Betaproteobacteria | 11/250 (4.4) |
| Burkholderiales | 10/184 (5.4) |
| Hydrogenophilales | 0/1 [0.95] |
| Methylophilales | 0/2 [0.776] |
| Neisseriales | 0/46 [0.063] |
| Nitrosomonadales | 1/10 (10) |
| Procabacteriales | 0/0 |
| Rhodocyclales | 0/6 [0.393] |
| Gammaproteobacteria | 31/886 (3.5) |
| Acidithiobacillales | 0/6 [0.393] |
| Aeromonadales | 0/18 [0.153] |
| Alteromonadales | 11/58 (19) |
| Cardiobacteriales | 0/2 [0.776] |
| Chromatiales | 1/7 |
| Enterobacteriales | 0/439 [0.007] |
| Legionellales | 0/22 [0.127] |
| Methylococcales | 0/1 [0.95] |
| Oceanospirillales | 2/8 (25) |
| Pasteurellales | 0/68 [0.043] |
| Pseudomonadales | 10/95 (10.5) |
| Thiotrichales | 1/28 (3.6) |
| Vibrionales | 0/84 [0.035] |
| Xanthomonadales | 6/36 (16.6) |
| Cyanobacteria | 14/118 (11.7) |
| Chroococcales | 9/65 (13.8) |
| Oscillatorales | 1/8 (12.5) |
| Nostocales | 3/21 (14.3) |
| Gloeobacteria | 0/1 [0.95] |
| Acaryochloris (uncla) | 1/10 (10) |
| Pleurocapsales | 0/0 |
| Prochlorales | 0/13 [0.206] |
| Stigomatales | 0/0 |
| Firmicutes | 11/734 (1.5) |
| Bacilli | 11/589 (1.9) |
| Clostridia | 0/138 [0.021] |
| Erysipelotrichi | 0/7 [0.348] |
| Thermolithobacteria | 0/0 |
| Chloroflexi | 3/14 (21.4) |
| Chloroflexi | 3/8 (37.5) |
| Caldilineae | 0/0 |
| Dehalococcoidetes | 0/4 [0.527] |
| Caldilineae | 0/2 [0.776] |
| Anaerolineae | 0/0 |
| Actinobacteria | 3/270 (1.1) |
| Acidimicrobidae | 0/0 |
| Actinobacteridae | 2/263 (0.8) |
| Coriobacteridae | 0/0 |
| Rubrobacteridae | 1/1 (100) |
| Archaea | 3/126 (2.3) |
| Crenarchaeota | 0/38 [0.076] |
| Euryarchaeota | 3/86 (3.5) |
| Korarchaeota | 0/1 [0.95] |
| Nanoarchaetoa | 0/1 [0.95] |
Number of sequenced genomes as of April 2009 (NCBI-TaxBrowser, http://www.ncbi.nlm.nih.gov). The values represent the number of genera possessing a LOV homologue versus the number of completely sequenced genomes within the class, order, or phylum. f* (shown in square brackets) is the largest admissible frequency of the LOV domain in the corresponding systematic group such that the probability of not finding any LOV domain in a sample of size n (number of sequenced genomes) is above 5%, where a geometric distribution is assumed.
No LOV homologues were detectable for Delta- and Epsilonproteobacteria.
The only taxa with a sufficiently high number of sequenced genomes that apparently lack LOV homologues are the Spirochaetes (201 sequenced genomes) (f* = 0.015), the Bacteroidetes (81 sequenced genomes) (f* = 0.036), and the Crenarchaeota (38 sequenced genomes) (f* = 0.076).
Table 2 provides an overview over the presence/absence of LOV homologues within the different phyla that contain LOV domains. The highest frequency of occurrence of the LOV signaling module was detected in the Chloroflexi (21.4%). Presently, however, the total number of 14 completely sequenced Chloroflexi genomes is relatively small, and more Chloroflexi genome sequences would be required to decide whether or not the observed LOV occurrence frequency is significant.
The LOV signaling module is widely dispersed among the Proteobacteria throughout the Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria classes, with the highest frequency within alphaproteobacterial genera (11.3%). The Rickettsiales represent the only alphaproteobacterial order that apparently lacks the LOV signaling module, with no LOV homologues detectable within the 42 completely sequenced genomes (f* = 0.067). For the remaining alphaproteobacterial orders (e.g., Kiloniellales) that apparently lack LOV domain homologues, no completely sequenced genomes are available. Thus, those phyla are most probably underrepresented in our analysis. This underrepresentation of taxonomic groups also occurs in other proteobacterial classes that possess LOV homologues; i.e., orders with a small number of completely sequenced genomes contain fewer or no LOV homologues. Interestingly, among the Gammaproteobacteria, the Enterobacteriales (f* = 0.007) (e.g., Escherichia coli and Salmonella spp.) and Vibrionales (f* = 0.035) clearly lack a LOV homologue, with no LOV domain detectable within 439 or 84 sequenced genomes, respectively. In the Cyanobacteria, the LOV signaling module is widely distributed (11.7%), with representatives among the Chroococcales, Oscillatorales, Nostocales, and Acaryochloris. In contrast, among the Firmicutes LOV homologues seem to be restricted to the Bacilli (Bacillus spp. and Listeria spp.). Thus, in 138 completely sequenced genomes from Clostridia we did not detect LOV homologues (f* = 0.021). Erysipelotrichi and Thermolithobacteria are underrepresented in our analysis, and thus the data do not allow unequivocal conclusions as to the presence of LOV homologues. In the Actinobacteria the frequency of occurrence is low (1.1%), with representatives in a few Actinobacteridae and Rubrobacteridae.
Tree reconstruction and evaluation.
Our phylogenetic analysis was performed only on the conserved LOV domains of putative and known blue-light photoreceptors. This choice is dictated by the overwhelming diversity of the functional domains found fused to the LOV sensor domain (see Table S1 in the supplemental material for details). This holds true for the J-alpha helix containing extension to the LOV core (22), which is found well conserved (in sequence) only C terminally to the plant phototropin LOV2 domains. Although this LOV photoreceptor structural feature apparently plays an important role in the LOV-mediated light-signaling mechanisms, sequence similarity in this region of the full-length photoreceptor proteins in most eukaryotic and prokaryotic proteins is low. Therefore, no meaningful alignment can be generated using either full-length photoreceptor sequence information or LOV domain sequences extended in the LOV C-terminal region. Prior to tree reconstruction, we performed several analyses to determine whether DNA or amino acid sequence alignments are better suited to infer the LOV phylogeny. To this end, we performed ML mapping analyses. Furthermore, we analyzed the DNA alignment at all three codon positions for molecular saturation by computing the number of observed transitions relative to the number of transversions, which were then plotted against genetic distance values (65) (see the supplemental material for details). In conclusion, according to ML mapping analysis, the LOV nucleotide alignment contains the most phylogenetic information, and thus we decided to use the nucleotide alignment and to employ ML and Bayesian methods for tree reconstruction due to the saturation of pairwise genetic distances observed for this alignment. The MrBayes, bootstrap consensus, and ML trees were compared with each other as described in Materials and Methods. All tree topology tests rejected the IQPNNI bootstrap consensus and the MrBayes tree with a P value of <0.01. In contrast, the best RAxML tree and the RAxML bootstrap consensus tree are equally good explanations of the data. Therefore, the phylogenetic tree generated by RAxML is used for the illustration throughout this study.
The three-kingdom phylogeny.
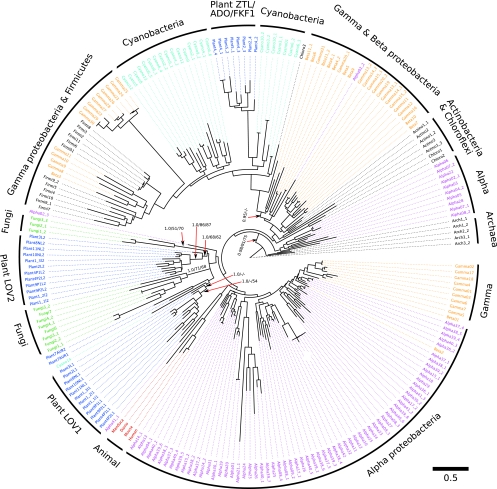
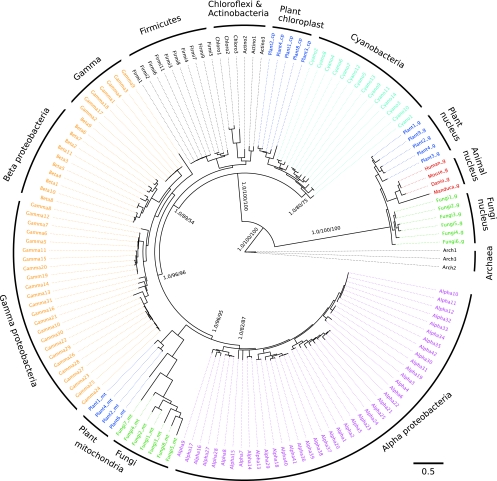
The tree shown in Fig. 1 shows a separation of bacterial and archaeal LOV sequences with strong support (BPP, 98%; RAxML BP, 91%; IQPNNI BP, 75%). This prokaryotic dichotomy is well in accordance with the SSU rRNA gene tree (Fig. 2), which comprises the same species as the LOV domain tree. Therefore, we rooted the LOV tree with the archaeal LOV sequences. However, we note that this rooting does not imply that the archaeal LOV sequences necessarily represent the evolutionarily oldest sequences. In fact, the “patchy” distribution of LOV sequences in the Archaea (restricted to a few mesophilic Euryarchaeota) might suggest acquisition through horizontal gene transfer from Bacteria. In the nonarchaeal part of the LOV tree we discern two subtrees, and this dichotomy is retained even if the archaeal outgroup is excluded from the tree construction (not shown). The “upper” subtree consists of cyanobacterial, actinobacterial, and proteobacterial LOV sequences as well as sequences from the Firmicutes and Chloroflexi. Moreover, the LOV sequences of the plant ZTL/ADO/FKF1 family are found in the upper subtree. Here, the ZTL/ADO/FKF1 LOVs form a sister group to the Cyanobacteria LOVs (BPP support, 95%). This grouping is congruent with the placement of chloroplast SSU rRNAs as a sister group to the Cyanobacteria (BPP, 100%; RAxML BP, 80%; IQPNNI BP, 75%) in the SSU rRNA tree. The “lower” subtree is a mixture of eukaryotic sequences from fungi (WC-1 LOVs), plants (phototropin LOVs, neochrome-LOVs, and aureochrome LOVs) and of sequences predominately from Proteobacteria. Here all eukaryotic LOV sequences cluster as a sister group together with an alphaproteobacterial clade. This grouping is, moreover, in accordance with the SSU rRNA tree (Fig. 2), where the respective mitochondrial SSU rRNAs group with alphaproteobacteria (BPP, 100%; RAxML BP, 96%; IQPNNI BP, 95%). Interestingly, the putative animal LOV domains group within the lower part of the LOV tree as a sister group to alphaproteobacterial lineages. Although support for this grouping is low, it nevertheless suggests that the putative animal LOV domains may actually be LOV homologues, related to the plant and bacterial LOV sequences. More detailed analyses, e.g., by including other PAS families, would be required to address this issue appropriately.
FIG. 1.
Phylogenetic tree determined by RAxML rapid bootstrap (55), reconstructed for the LOV gene sequences from divergent taxa. In addition, Bayesian analysis was performed using MrBayes (24) and bootstrap with ML by IQPNNI (62a) as described in Materials and Methods. Bayesian posterior probabilities (BPP) and RAxML and IQPNNI (in this order) bootstrap support values (BP) are shown at the relevant branches of the RAxML bootstrap consensus tree. The respective LOV sequences of the different eukaryotic photoreceptor families as well as the LOV sequences of prokaryotic origin are color coded: the Archaea (outgroup), the Actinobacteria, Chloroflexi, and Firmicutes, are shown in black, Alphaproteobacteria are in pink, Betaproteobacteria and Gammaproteobacteria are in orange, cyanobacterial genera are in cyan, fungal WC-1 sequences are in green, plant phototropin and neochrome LOV domains (LOV1 and LOV2), algal aureochrome LOV sequences (Plant7AUR2 and Plant7AUR1), and the ZTL/ADO/FKF1-LOV family are in blue, and the putative animal LOV domains are in red. Detailed sequence information, including protein accession numbers, is summarized in Table S1 in the supplemental material.
FIG. 2.
SSU rRNA gene tree reconstructed to infer the species evolution of the LOV domain-containing taxa. Bayesian, bootstrap, and ML trees were constructed as described for the LOV gene tree. All trees that were generated using the different phylogeny inference methods showed the same overall topology. Here, the MrBayes tree is depicted. Bayesian posterior probabilities (BPP) and bootstrap support values (BP) with RAxML and IQPNNI are added at relevant branches. The color code is the same as in Fig. 1. The suffixes g, mt, and cp indicate SSU rRNA sequences from nuclear, mitochondrial, and chloroplast regions, respectively (see Table S1 in the supplemental material for details). The branch lengths of the subtree containing mitochondrial rRNA were scaled down three times to better fit in the figure.
DISCUSSION
LOV is (not) all around: the taxonomic distribution of the LOV signaling module.
From our analysis of the taxonomic distribution, it is apparent that LOV homologues are missing only in highly “specialized microbes,” including (i) extremophiles such as Crenarchaetoa, which dwell under extreme conditions, e.g., in hot hydrothermal vents and cold deep-sea environments (34), (ii) obligate anaerobic bacteria, e.g., Bacteroidetes (34) and anaerobic Clostridia (34) of the Firmicutes; and (iii) Rickettsiales as obligate intracellular parasites (18). The absence of photoreceptors in obligate anaerobes, obligate intracellular parasites, and extremophilic microbes might readily be explained by the fact that light does not represent a frequent environmental stimulus under their respective living conditions. However, the absence of LOV homologues in the Enterobacterales and the Vibrionales is more intriguing, since those organisms are probably exposed to light many times in their respective living environments. Interestingly, instead of LOV proteins, species of both taxonomic classes possess blue-light photoreceptors of another family. For example, organisms such as Escherichia coli and certain Vibrio spp. contain so-called BLUF (blue-light-sensing FAD) proteins (33) that possess a FAD-binding BLUF domain as the light-sensing module (21). It is thus tempting to speculate that in the Enterobacterales and Vibrionales, which lack LOV photoreceptor systems, BLUF domain proteins might take over the blue-light-sensing function. This notion finds further support in the fact that bacterial BLUF photoreceptors often possess effector domains similar to those of their bacterial LOV counterparts (33). The same might hold true for the other blue-light photoreceptor families. Photoreceptors of the photoactive yellow protein (PYP) family are restricted to the Proteobacteria (Alpha- to Deltaproteobacteria subclasses), and their presence was suggested for Salinibacter ruber from the phylum Bacteroidetes (29). PYPs share a general PAS fold with LOV domains but, in contrast, occur mostly as stand-alone light-sensing proteins (62). It appears that LOVs, BLUFs, and PYPs can be found together in the same anoxygenic phototrophic Alphaproteobacteria, such as, e.g., Rhodobacter sphaeroides, or even in soil-dwelling chemotrophs such as Burkholderia phytofirmans (29, 33, 62). Interestingly, it appears that LOVs and BLUFs are absent in halophilic proteobacteria such as Halorhodospira halophila and Halochromatium salexigens, whereas those organisms often contain a PYP protein instead (29, 33, 62). Cryptochrome proteins (CRYs) have so far not been included in such analyses because it is not possible to make a clear distinction between cryptochromes and photolyases on the basis of sequence analysis alone (62). In particular, the definition that CRYs lack DNA repair activity, in contrast to the homologous DNA photolyases, seems not to hold true when it comes to the recently identified bacterial cryptochromes of the CRY-DASH family (7). For those putative CRYs, single-stranded-DNA-specific photolyase activity recently was demonstrated (49), and thus it seems yet possible that CRYs (per definition) might not be present in prokaryotes.
Congruency and incongruency between gene (LOV) and species (SSU rRNA) trees illustrate the evolutionary history of the LOV domain.
Although the phylogenetic signal is in most cases not very strong, the phylogenetic tree in Fig. 1 points in all cases toward a direct sister group relationship between bacterial LOVs and the eukaryotic LOV sequences. First, the eukaryotic ZTL/ADO/FKF1-LOV family clusters as a monophyletic group within the cyanobacterial LOV sequences but does not form a monophyletic group together with the other eukaryotes. Since in the SSU rRNA tree, the chloroplast SSU rRNAs group with the Cyanobacteria with strong support, the most parsimonious explanation for the cyanobacterial affiliation of ZTL/ADO/FKF1 LOV domains is plastid endosymbiosis. Second, the remaining eukaryotic LOV photoreceptor families, namely, the plant phototropin LOVs (including neochrome LOVs), the aureochrome LOVs, the fungal WC-1 LOVs, and the animal LOV sequences, are related to alphaproteobacterial clades (Fig. 1), which comprise mainly (anoxygenic) phototrophic Alphaproteobacteria (lower part of the tree). Since this grouping is in accordance with the SSU rRNA tree (Fig. 2), here the most parsimonious evolutionary scenario is that the once-free-living mitochondrial ancestors transferred the respective LOV precursor into the eukaryotic genome during endosymbiosis.
In conclusion, our results regarding the appearance of LOV domains in the eukaryotes are thus in agreement with the general endosymbiotic theory (36). This well-established concept argues for an alphaproteobacterium (16) as the ancestor of eukaryotic mitochondria, whereas the chloroplasts, on the other hand, are thought to have originated from the endosymbiosis of an ancestral cyanobacterium (35). The exclusive localization of the eukaryotic LOV photoreceptors in the nuclear but not the chloroplast or mitochondrial genomes of the respective plants is not surprising and can readily be accounted for by invoking gene transfer from organelles to the nucleus after the endosymbiotic uptake event by a process known as endosymbiotic gene transfer (61).
Not surprisingly, the picture is less clear for the bacterial part of the LOV tree (Fig. 1). Here the topology for the Firmicutes, the Actinobacteria, the Chloroflexi, most Alphaproteobacteria, and certain Gammaproteobacteria is congruent with the generally accepted branching order (4) and with the topology of the rRNA tree (Fig. 2). One incongruence observed in the lower part of the LOV tree is the divergence of a group of phototrophic Alphaproteobacteria (including, e.g., Roseobacter and Erythrobacter) before the separation of the other Alphaproteobacteria from the Gammaproteobacteria. In the upper part of the LOV tree, the position of the Actinobacteria and the Firmicutes is in agreement with the ribosomal tree representing monophyletic branches well separated from the Alphaproteobacteria and Gammaproteobacteria in the lower part of the LOV tree. However, the upper part of the tree is interspersed with Gamma-, Beta-, and a few Alphaproteobacteria, which, in the light of ribosomal phylogeny (Fig. 2), should belong to the lower part of the LOV tree. The same is evident for the Cyanobacteria, which should form a monophyletic group diverging before the separation of the other bacterial phyla. The LOV sequence of the cyanobacterium Acaryochloris marina MBIC11017 (Cyano14) groups with plant phototropin LOV sequences in the LOV1 part of the lower subtree. Such incongruent placements between the LOV and ribosomal trees might be explained by invoking an early LOV duplication event and subsequent gene loss in many bacterial lineages. However, given the scattered distribution of the LOV sensor module, an alternative and probably more parsimonious explanation involves frequent horizontal gene transfer of the LOV module throughout the Bacteria, many times originating from proteobacterial genera. To qualitatively delineate between the two scenarios, a duplication and deletion analysis to reconcile the LOV tree with the SSU rRNA tree was performed (see the supplemental material for details). This analysis revealed rate heterogeneities between the plant and bacterial subtrees as well as an unexpectedly high deletion rate determined for the bacterial subtree, which suggests that, in addition to duplication and deletion events, frequent horizontal gene transfer must have contributed to the distribution of LOV domains among the prokaryotes. Unfortunately, it is currently not possible to computationally infer the occurrence of horizontal gene transfer in the presence of gene duplication and loss events. Hence, we cannot estimate the magnitude and direction of such events, which have undoubtedly contributed to the evolution and distribution of the LOV domain.
Taken together, the topology of the LOV tree as well as congruencies and incongruencies observed between the LOV and SSU rRNA trees argue for a bacterial origin of the eukaryotic LOV signaling modules, with two distinct endosymbiotic gene transfer events accounting for the presence of the LOV signaling module in the eukaryotes. This, in conclusion, argues for an independent evolution of the plant circadian photoreceptor LOV domains (ZTL/ADO/FKF1) on the one hand and the remaining eukaryotic LOV domains on the other. Hence, the LOV photoreceptor families in plants (ZTL/ADO/FKF1-LOVs, phototropins, neochromes, and aureochromes) probably underwent independent divergent evolution toward distinct functions, namely, light-dependent regulation of (i) circadian rhythmicity (ZTLs) and (ii) phototropic responses (including photomorphogenesis) (phototropins, WC-1, neochromes, and aureochromes). In contrast, the plant and fungal circadian LOV photoreceptor systems, i.e., ZTL/ADO/FKF1 LOVs and WC-1 LOVs, which we speculate to have originated from two different endosymbiotic transfer events, convergently evolved to use the same light-sensitive domain for the control of similar cellular processes, namely, the light entrainment of the circadian clock.
Conclusion.
Our phylogenetic analysis suggests that the LOV domains of all so-far-known eukaryotic LOV photoreceptor families have originated from two distinct endosymbiotic gene transfer events from cyanobacterial or alphaproteobacterial lineages, respectively. Consequently, the respective endosymbiotic transfer event should mark the time of the appearance of LOV domains among the eukaryotes. Moreover, incongruencies between the LOV and SSU rRNA trees as well as a duplication/deletion analysis performed on the bacterial part of the LOV tree are indicative of the presence of frequent horizontal gene transfer, massive gene duplication, and gene loss in the Bacteria. In conclusion, throughout evolution, the LOV signaling module has been readily distributed within and between the pro- and eukaryotic worlds, most probably many times, involving direct gene transfer processes. This together, with the diversity of functional light responses (and fused effector domains) that are triggered by LOV sensor domains in all kingdoms of life, implicates the LOV domain as a versatile light sensor module that can easily be integrated and adapted into the existing cellular signaling machinery. The wide distribution throughout the prokaryotes, moreover, highlights the possibility that blue light has in the past represented, and still is representing now, an important environmental stimulus even for chemotrophic (nonphotosynthetic) microorganisms.
Supplementary Material
Acknowledgments
This work was supported in part by the Deutsche Forschungsgemeinschaft (DFG) (Forschergruppe “Blue-Light Photoreceptors,” FOR526). Financial support from the Vienna Science and Technology Fond (WWTF) to A.v.H. and B.Q.M. is also greatly appreciated.
Footnotes
Published ahead of print on 25 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballario, P., P. Vittorioso, A. Magrelli, C. Talora, A. Cabibbo, and G. Macino. 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15:1650-1657. [PMC free article] [PubMed] [Google Scholar]
- 4.Battistuzzi, F. U., A. Feijao, and S. B. Hedges. 2004. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boer, P. H., and M. W. Gray. 1988. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell 55:399-411. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, W. R., and J. M. Christie. 2002. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7:204-210. [DOI] [PubMed] [Google Scholar]
- 7.Brudler, R., K. Hitomi, H. Daiyasu, H. Toh, K. Kucho, M. Ishiura, M. Kanehisa, V. A. Roberts, T. Todo, J. A. Tainer, and E. D. Getzoff. 2003. Identification of a new cryptochrome class. Structure, function, and evolution. Mol. Cell 11:59-67. [DOI] [PubMed] [Google Scholar]
- 8.Cao, Z., V. Buttani, A. Losi, and W. Gärtner. 2008. A blue light inducible two component signal transduction system in the plant pathogen Pseudomonas syringae pv. tomato. Biophys. J. 94:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 10.Christie, J. M., P. Reymond, G. K. Powell, P. Bernasconi, A. A. Raibekas, E. Liscum, and W. R. Briggs. 1998. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282:1698-1701. [DOI] [PubMed] [Google Scholar]
- 11.Christie, J. M., M. Salomon, K. Nozue, M. Wada, and W. R. Briggs. 1999. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. USA 96:8779-8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosson, S., S. Rajagopal, and K. Moffat. 2003. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42:2-10. [DOI] [PubMed] [Google Scholar]
- 13.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do, C. B., M. S. Mahabhashyam, M. Brudno, and S. Batzoglou. 2005. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 15:330-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esser, C., W. Martin, and T. Dagan. 2007. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol. Lett. 3:180-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 18.Fredricks, D. 2006. Introduction to the Rickettsiales and other intracellular prokaryotes, p. 457-466. In M. Dworkin et al. (ed.), The prokaryotes. Springer, New York, NY.
- 19.Froehlich, A. C., Y. Liu, J. J. Loros, and J. C. Dunlap. 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297:815-819. [DOI] [PubMed] [Google Scholar]
- 20.Goldman, N., J. P. Anderson, and A. G. Rodrigo. 2000. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 49:652-670. [DOI] [PubMed] [Google Scholar]
- 21.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497-500. [DOI] [PubMed] [Google Scholar]
- 22.Harper, S. M., L. C. Neil, and K. H. Gardner. 2003. Structural basis of a phototropin light switch. Science 301:1541-1544. [DOI] [PubMed] [Google Scholar]
- 23.Huala, E., P. W. Oeller, E. Liscum, I. S. Han, E. Larsen, and W. R. Briggs. 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278:2120-2123. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 25.Kagawa, T., M. Kasahara, T. Abe, S. Yoshida, and M. Wada. 2004. Function analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol. 45:416-426. [DOI] [PubMed] [Google Scholar]
- 26.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 28.Krauss, U., A. Losi, W. Gärtner, K. E. Jaeger, and T. Eggert. 2005. Initial characterization of a blue-light sensing, phototropin-related protein from Pseudomonas putida: a paradigm for an extended LOV construct. Phys. Chem. Chem. Phys. 7:2804-2811. [DOI] [PubMed] [Google Scholar]
- 29.Kumauchi, M., M. T. Hara, P. Stalcup, A. Xie, and W. D. Hoff. 2008. Identification of six new photoactive yellow proteins—diversity and structure-function relationships in a bacterial blue light photoreceptor. Photochem. Photobiol. 84:956-969. [DOI] [PubMed] [Google Scholar]
- 30.Losi, A., E. Polverini, B. Quest, and W. Gärtner. 2002. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys. J. 82:2627-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Losi, A. 2004. The bacterial counterparts of plants phototropins. Photochem. Photobiol. Sci. 3:566-574. [DOI] [PubMed] [Google Scholar]
- 32.Losi, A. 2006. Flavin-based photoreceptors in bacteria, p. 223-276. In E. Silva and A. M. Edwards (ed.), Flavin photochemistry and photobiology, vol. 6. The Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 33.Losi, A., and W. Gärtner. 2008. Bacterial bilin- and flavin-binding photoreceptors. Photochem. Photobiol. Sci. 7:1168-1178. [DOI] [PubMed] [Google Scholar]
- 34.Madigan, M. T. 2009. Brock biology of microorganisms, 12th ed. Pearson/Benjamin Cummings, San Francisco, CA.
- 35.McFadden, G. I. 2001. Chloroplast origin and integration. Plant Physiol. 125:50-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mereschkowsky, C. 1905. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Centralbl. 25:593-604, 689-691. [Google Scholar]
- 37.Nelson, D. C., J. Lasswell, L. E. Rogg, M. A. Cohen, and B. Bartel. 2000. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101:331-340. [DOI] [PubMed] [Google Scholar]
- 38.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 39.Ohgishi, M., K. Saji, K. Okada, and T. Sakai. 2004. Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 101:2223-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:R674-R677. [DOI] [PubMed] [Google Scholar]
- 41.Ponting, C. P., J. Schultz, F. Milpetz, and P. Bork. 1999. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 27:229-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 43.Pruesse, E., C. Quast, K. Knittel, B. M. Fuchs, W. Ludwig, J. Peplies, and F. O. Glockner. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purcell, E. B., D. Siegal-Gaskins, D. C. Rawling, A. Fiebig, and S. Crosson. 2007. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl. Acad. Sci. USA 104:18241-18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salomon, M., J. M. Christie, E. Knieb, U. Lempert, and W. R. Briggs. 2000. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39:9401-9410. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, H. A. 2009. Testing tree topologies, p. 381-404. In M. S. Philippe Lemey, and Anne-Mieke Vandamme (ed.), The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 48.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selby, C. P., and A. Sancar. 2006. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 103:17696-17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimodaira, H., and M. Hasegawa. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114-1116. [Google Scholar]
- 51.Shimodaira, H., and M. Hasegawa. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics 17:1246-1247. [DOI] [PubMed] [Google Scholar]
- 52.Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51:492-508. [DOI] [PubMed] [Google Scholar]
- 53.Somers, D. E., T. F. Schultz, M. Milnamow, and S. A. Kay. 2000. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101:319-329. [DOI] [PubMed] [Google Scholar]
- 54.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688-2690. [DOI] [PubMed] [Google Scholar]
- 55.Stamatakis, A., P. Hoover, and J. Rougemont. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758-771. [DOI] [PubMed] [Google Scholar]
- 56.Suetsugu, N., F. Mittmann, G. Wagner, J. Hughes, and M. Wada. 2005. A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc. Natl. Acad. Sci. USA 102:13705-13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swartz, T. E., T. S. Tseng, M. A. Frederickson, G. Paris, D. J. Comerci, G. Rajashekara, J. G. Kim, M. B. Mudgett, G. A. Splitter, R. A. Ugalde, F. A. Goldbaum, W. R. Briggs, and R. A. Bogomolni. 2007. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science 317:1090-1093. [DOI] [PubMed] [Google Scholar]
- 58.Takahashi, F., D. Yamagata, M. Ishikawa, Y. Fukamatsu, Y. Ogura, M. Kasahara, T. Kiyosue, M. Kikuyama, M. Wada, and H. Kataoka. 2007. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl. Acad. Sci. USA 104:19625-19630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, B. L., and I. B. Zhulin. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Timmis, J. N., M. A. Ayliffe, C. Y. Huang, and W. Martin. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5:123-135. [DOI] [PubMed] [Google Scholar]
- 62.van der Horst, M. A., J. Key, and K. J. Hellingwerf. 2007. Photosensing in chemotrophic, non-phototrophic bacteria: let there be light sensing too. Trends Microbiol. 15:554-562. [DOI] [PubMed] [Google Scholar]
- 62a.Vinh L. S., and A. von Haeseler. 2004. IQPNNI: moving fast through tree space and stopping in time. Mol. Biol. Evol. 21:1565-1571. [DOI] [PubMed] [Google Scholar]
- 63.Wallace, I. M., O. O'Sullivan, D. G. Higgins, and C. Notredame. 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 34:1692-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wuyts, J., G. Perriere, and Y. Van De Peer. 2004. The European ribosomal RNA database. Nucleic Acids Res. 32:D101-D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 66.Yang, Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39:306-314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.