Abstract
Staphylococcus aureus reacts to changing environmental conditions such as heat, pH, and chemicals through global regulators such as the sae (S. aureus exoprotein expression) two-component signaling system. Subinhibitory concentrations of some antibiotics were shown to increase virulence factor expression. Here, we investigated the S. aureus stress response to sublethal concentrations of a commonly used biocide (Perform), by real-time quantitative PCR (qRT-PCR), promoter activity assay, sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and a flow cytometric invasion assay. Perform, acting through the production of reactive oxygen species, generally downregulated expression of extracellular proteins in strains 6850, COL, ISP479C but upregulated these proteins in strain Newman. Upregulated proteins were sae dependent. The Perform component SDS, but not paraquat (another oxygen donor), mimicked the biocide effect. Eap (extracellular adherence protein) was most prominently augmented. Upregulation of eap and sae was confirmed by qRT-PCR. Promoter activity of sae P1 was increased by Perform and SDS. Both substances enhanced cellular invasiveness, by 2.5-fold and 3.2-fold, respectively. Increased invasiveness was dependent on Eap and the sae system, whereas agr, sarA, sigB, and fibronectin-binding proteins had no major effect in strain Newman. This unique response pattern was due to a point mutation in SaeS (the sensor histidine kinase), as demonstrated by allele swapping. Newman saePQRSISP479C behaved like ISP479C, whereas saePQRSNewman rendered ISP479C equally responsive as Newman. Taken together, the findings indicate that a point mutation in SaeS of strain Newman was responsible for increased expression of Eap upon exposure to sublethal Perform and SDS concentrations, leading to increased Eap-dependent cellular invasiveness. This may be important for understanding the regulation of virulence in S. aureus.
Staphylococcus aureus is a facultative pathogen which asymptomatically colonizes the anterior nares of 20 to 30% of the human population, while 60% are intermittently colonized and 20% are never colonized by S. aureus (27). S. aureus is one of the major pathogens of both community-acquired and nosocomial infections. Cell surface-associated proteins as well as extracellular proteins are used for colonization (39), but teichoic acids also play an important role in nasal colonization (51). For carriers it is likely that endogenous strains are the source of infection (50), ranging from superficial wound infections to life-threatening invasive infections such as osteomyelitis, endocarditis, and sepsis (46).
S. aureus can persist outside the host on surfaces and in dust, soil, water, and other environments. Its versatility to adjust to changing conditions (7) is due to its equipment of global regulators such as the alternative sigma factor SigB, the SarA protein family, or 16 two-component regulatory systems (TCS) such as Agr or Sae (29). Multiple overlapping and interacting feedback networks of these regulators coordinate the expression of virulence factors such as adhesins and toxins. Perform is a commonly used biocide for disinfection and cleaning of medical products and surfaces. It has a broad biocidal spectrum that includes S. aureus, and it is therefore suitable for use in high-risk areas, such as hospitals. Its mode of action is mediated by reactive oxygen species (ROS) that emerge after dilution of the powder in water. Among the listed ingredients are pentapotassium bis(peroxymonosulfate) bis(sulfate) (PPMS) (20%), sodium benzoate (15%), anionic surfactants (5 to 15%), tartaric acid (10%), nonionic surfactants (5%), soap (5%), phosphonate, and perfumes.
S. aureus produces two types of adhesins. The MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) are anchored to the staphylococcal cell wall (33). Among the most prominent representatives of the 21 known MSCRAMMs are protein A, the fibronectin-binding proteins (FnBPs), and the clumping factors (41). The second group of adhesins is the SERAMs (secreted expanded-repertoire adhesive molecules), which are secreted but partially rebind to the bacterial surface. Eap (extracellular adhesive protein), Emp (extracellular matrix protein), Efb (extracellular fibrinogen-binding protein), and coagulase belong to the SERAMs. Eap, an sae-regulated protein, has a gene prevalence of 100% of all tested S. aureus isolates (24, 25). It has a broad capacity for binding to several host matrix and plasma proteins such as Fn, fibrinogen, vitronectin, and collagen (34). Due to strain differences, its size varies between 60 and 72 kDa, corresponding to four to six tandem repeat domains (23). Eap also has immunomodulatory functions. With its ability to form oligomers and to rebind to the staphylococcal cell surface, Eap further mediates bacterial aggregation. Since Eap binds to the staphylococcal surface and to eukaryotic compounds, it improves binding to epithelial cells and fibroblasts (36). Strain Newman strongly expresses Eap and in this way compensates for its loss of functional FnBPs. Due to a point mutation resulting in a stop codon, FnBPs lack the LPXTG motif in Newman. Thus, they are not anchored to the cell wall and are completely secreted into the culture medium (19).
Giraudo et al. first described the sae locus (sae system) and postulated a decreased virulence of sae-deficient mutants (15). The regulatory sae operon itself consists of four open reading frames (saePQRS), with SaeR and SaeS exhibiting strong sequence homology to response regulators and sensor histidine kinases, respectively, of a classical TCS (12). The open reading frames saeP and saeQ are located upstream of saeRS, and their respective gene products probably interfere with the saeRS-dependent regulatory mechanism. SaeQ seems to be membrane associated, while SaeP is a putative lipoprotein (35, 48). Sae activation leads to transcription of a wide range of virulence factors, such as hemolysins, coagulase, FnBPs, and Eap (13, 21, 42, 48). Mutations in saeRS do not affect the transcription of agr, sigB, sarA, or rot (13, 17, 32). In contrast, agr activates, whereas sigB represses, sae transcription, at least in some strains (10, 14, 35), placing sae as a central downstream regulator within the S. aureus regulatory network. The sae operon is transcribed from two promoters. The main promoter P1 is strongly autoregulated (1, 10) and can be activated by phagocytosis-related effector molecules such as H2O2, low pH, and subinhibitory concentrations of α-defensins (10). Additionally, subinhibitory concentrations of antibiotics can activate (glycopeptides and β-lactams) (28) as well as inhibit (clindamycin) (35) the sae system. The sae-dependent activation of toxins and immune-modulatory genes probably leads to immune evasion and to the destruction of polymorphonuclear leukocytes after phagocytosis. Interestingly, there are strong strain-dependent differences regarding sae regulation (16, 28, 35, 42). The prototype S. aureus strain Newman shows a high, constitutive expression of the sae operon due to an amino acid substitution within the first membrane-spanning domain of the sensor histidine kinase SaeS (1, 10, 48).
This study investigated the response of a set of S. aureus strains to sublethal concentrations of a commonly used biocide (Perform). Increased expression of fnbA in response to stress by subinhibitory concentrations of ciprofloxacin has been reported (4, 5). Thus, we hypothesized that a similar phenomenon might also occur for biocides.
MATERIALS AND METHODS
Reagents.
Perform was obtained from Schülke & Mayr (Norderstedt, Germany). Tetracycline, rifampin (rifampicin), paraquat, and Triton X-100 were purchased from Sigma-Aldrich. Sodium dodecyl sulfate (SDS) and sodium benzoate were purchased from AppliChem. Tween 20, tartaric acid, and PPMS were purchased from Merck.
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Cultures of staphylococci were routinely grown at 37°C under aerobic conditions (200 rpm) in tryptic soy broth (TSB) (BD) with or without biocide/SDS and on sheep blood (Biomerieux) and Caso-agar (BD). Antibiotics were used for selection when appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Properties | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| Newman | Wild type | 8 |
| AS3 | Newman sae::Tn917 (Emr) | 17 |
| ALC355 | Newman agr::tetM | 52 |
| ALC637 | Newman sar::Tn917 LTV1 | 53 |
| Newman-29 | Newman Δsae::kanA | 10 |
| DU5886 | Newman fnbA::TcrfnbB::Emr | 18 |
| mAH12 | Newman eap::Eryr | 22 |
| MS64 | Newman sigB1(Am) Tcr | 45 |
| UAMS-1 | Osteomyelitis isolate | 11 |
| Cowan I | Wild type | ATCC 12598 |
| COL | Wild type | 9 |
| 6850 | Wild type | 3 |
| ISP479C | Derivative of 8325-4 | 37 |
| ISP479R | 8325-4, rsbU repaired | 49 |
| ISP479C-29 | ISP479C Δsae::kanA | 10 |
| S. carnosus TM 300 | Wild type | 44 |
| Plasmids | ||
| pCWSAE28 | saePQRSISP479C cloned into pCL84 | This work |
| pCWSAE33 | saePQRSNewman cloned into pCL84 | This work |
| pCG7 | sae P1 promoter-lacZ fusion, integration vector | 10 |
Sequencing of saeS and construction of sae complementations.
The PCR product of saeS of strain 6850 was sequenced (using primers SaeS-Seq-Dw [GTA TTA AGG AAT TTG AGT TA] and SaeS-Seq-Up [GCT TGT AAT TAT TGT CGT TA]), according to a standard PCR protocol. Products were run on an ABI Prism 3130 Genetic Analyzer, and chromatograms were analyzed using Bioedit software (version 7.0.9.0).
The saePQRS operon was amplified with oligonucleotides (sae-1U GCG [TGA ATT CTT ATT GTG GCA AAA GGT TT] and sae-3515L [CCC CGA ATT CCT GTA TGC CGC CTA ATA AT]) using DNA from strains Newman and ISP479C, respectively. The resulting PCR amplicons were cloned into the EcoRI site (underlined) of the integration vector pCL84 (31), gaining plasmids pCWSAE33 and pCWSAE28, respectively. Plasmids were verified by sequencing of the whole insert and transformed into the restriction-deficient S. aureus strain RN4220. The plasmids were then transduced into sae-deficient mutants (AS3 and ISP479C-29) using θ11 lysates. Thus, the whole saePQRS operon was integrated into the chromosomes of the sae-deficient mutants.
SDS-PAGE.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously utilizing 12% polyacrylamide gels (30). For sample preparation, 1 ml of culture (optical density at 600 nm [OD600] of 1.5) was pelleted at 13,000 rpm for 5 min and subsequently resuspended in 40 μl Laemmli buffer. After being heated at 100°C for 30 min, the suspensions were centrifuged at 5,000 rpm for 5 min, and the supernatant was loaded on a polyacrylamide gel. After separation, the proteins were silver stained and the GS800 calibrated densitometer (Bio-Rad) was used for final documentation.
In-gel tryptic digestion and liquid chromatography-tandem mass spectrometry (MS/MS) analysis.
The protein bands were excised with a scalpel and destained with a freshly prepared 1:1 mixture of a 1% aqueous solution of potassium ferricyanide [K3Fe(CN)6] and a 1.6% aqueous solution of sodium thiosulfate. After being washed with water, the sample was reduced, alkylated, and digested with trypsin (6) using a Digest pro MS liquid handling system (Intavis AG, Germany). Tryptic peptides were extracted from the gel pieces with 50% acetonitrile-0.1% trifluoroacetic acid (TFA), concentrated nearly to dryness in a SpeedVac vacuum centrifuge, and diluted to a total volume of 30 μl with 0.1% TFA. A 25-μl portion of each sample was analyzed with a nano-high-pressure liquid chromatography system (Ultimate; Dionex, The Netherlands; equipped with a Famos autosampler) coupled to an ESI QTOF hybrid mass spectrometer (Allied Biosystems). Samples were loaded on a C18 trapping column (Inertsil; LC Packings) with a flow rate of 20 μl/min 0.1% TFA. Peptides were eluted and separated on an analytical column (75 μm by 150 mm) packed with Inertsil 3-μm C18 material (LC Packings) with a flow rate of 200 nl/min in a gradient of buffer A (0.1% formic acid, 5% acetonitrile) and buffer B (0.1% formic acid, 80% acetonitrile) (0 to 2 min, 5% B; 2 to 50 min, 5 to 40% B; 50 to 60 min, 40 to 60% B; 60 to 63 min, 60 to 90% B). The column was connected with a nano-ESI emitter (New Objectives). A total of 2,000 V was applied via liquid junction. The QTOF operated in positive-ion mode. One MS survey scan (0.7 s) was followed by one information-dependent product ion scan (3 s).
Identification of MS/MS spectra by database searches.
The uninterpreted MS/MS spectra were searched against the NCBInr database using bacteria as a taxonomy filter (2,145,526 entries; downloaded 4/1/2007) using the Mascot software (Matrix Science). The algorithm was set to use trypsin as the enzyme, allowing for one missed cleavage site and assuming carbamidomethyl as a fixed modification of cysteine and oxidized methionine and deamidation of asparagines and glutamine as variable modifications. Mass tolerances were set to 1.1 Da and 0.1 Da for MS and MS/MS, respectively. Proteins identified by a single peptide are listed in the tables, if the following criteria were met: (i) the scoring value exceeded the Mascot homology threshold, (ii) manual interpretation of the fragment spectrum resulted in a continuous stretch of at least four amino acids, and (iii) a database search with this stretch of amino acids using the Mascot query returned the same protein.
Cell culture.
Cells were cultured as described previously (47). All cells were split twice weekly 1:5 by trypsinization. Cell culture media and supplements were obtained from Gibco. At intervals of 6 to 8 weeks the cell lines were tested for mycoplasma infection using the PCR mycoplasma test kit (AppliChem).
Real-time quantitative PCR (qRT-PCR).
For RNA isolation, bacteria were grown in TSB with or without biocide/SDS to OD600 of 0.8 to 1. After pelleting, the staphylococci were mechanically disrupted with FastPrep instrument FP120 (Thermo Savant), and RNA was isolated via the RNeasy mini kit (Qiagen). After two treatments with recombinant DNase I, RNase-free RNA (Roche) was transcribed into cDNA with the Omniscript RT kit (Qiagen). Subsequently 5, 25, or 50 ng cDNA was used for real-time amplification with specific primers (summarized in Table 2). The transcriptional levels of target genes were normalized against the expression of 16S rRNA as internal control. The quantification of mRNA expression was performed according to Pfaffl (38) with transcript amounts expressed as n-fold difference relative to the control (2ΔCT; ΔCT represents the difference in threshold cycle between the target and control genes).
TABLE 2.
Primers for qRT-PCR
| Primer | Sequence (5′ → 3′) |
|---|---|
| eapRT For | AAG CGT CTG CCG CAG CTA |
| eapRT Rev | TGC ATA TGG AAC ATG GAC TTT AGA A |
| saeRT For | AAA CTT GCT TGA TAA TGC GCT AAA |
| saeRT Rev | GTT CTG GTA TAA TGC CAA TAC CTT CA |
| gyrB For | TTA GTG TGG GAA ATT GTC GA |
| gyrB Rev | CCG CCG AAT TTA CCA CCA GC |
| 16S rRNA For | CCA TAA AGT TGT TCT CAG TT |
| 16S rRNA Rev | CAT GTC GAT CTA CGA TTA CT |
Promoter activity assay.
Promoter activity assays were performed following the protocol of Geiger et al. (10). Initially bacteria were grown in TSB with or without biocide/SDS at 37°C (200 rpm) for 2 h, and then 1 ml of culture at an OD600 of 1 was centrifuged for 5 min at 5,000 × g. The bacterial pellet was resuspended in 1 ml 0.1 M phosphate buffer (pH 7.4) and mechanically disrupted using 0.350 ml zirconia/silica beads (0.1 mm diameter) in a high-speed homogenizer FastPrep-24 (MP) for 11 s at 6,500 rpm. Subsequently the β-galactosidase activity was determined using the FluoReporter galactosidase quantitation kit (Invitrogen). The emerging fluorescence emission was measured in white 96-well microtiter plate (Nunc) with a LS50B luminescence spectrometer (Perkin Elmer). The corresponding promoter activities are expressed as ng/ml β-galactosidase (Sigma-Aldrich) according to the manufacturer's instructions for the FluoReporter galactosidase quantitation kit.
Preparation of FITC-labeled bacteria and flow cytometric invasion assay.
Preparation of fluorescein isothiocyanate (FITC)-labeled bacteria was performed as described previously with minor modifications (26, 47). Fifty-milliliter cultures grown in TSB with or without biocide/SDS (2 h at 37°C and 200 rpm in 100-ml Erlenmeyer flasks) were harvested by centrifugation at 4,000 rpm for 5 min at 4°C and washed in phosphate-buffered saline (PBS). After an additional centrifugation step, the bacteria were labeled with 3 ml FITC in dimethyl sulfoxide for 30 min at 37°C, followed by a wash step in PBS. The flow cytometric invasion assay was performed as described previously (47). 293 human embryonic kidney cells were plated in 24-well plates at 3 × 105 cells/well the day before the assay. The cells were washed with preheated invasion medium consisting of Dulbecco's modified Eagle's medium containing 1% human serum albumin and 10 mM HEPES. After addition of 0.5 ml invasion medium, cells were cooled on ice for 10 min, and then 50 μl of FITC-labeled bacteria were added. For sedimentation, the culture dishes were preincubated for 1 h at 4°C and then shifted to 37°C for 2 h of invasion. After invasion, cells were washed with 1 ml PBS. The cells were then harvested and treated with monensin to neutralize fluorescence quenching. After addition of propidium iodide to differentiate between live and dead cells, the cells were analyzed by using a FACSCalibur (BD) flow cytometer. Results for cellular invasiveness are given as percent invasiveness relative to a reference strain (Cowan I).
Statistics.
Results are presented as means ± standard deviations (SDs) or standard errors of the means (SEMs), as detailed in figure legends. Statistical analysis was performed with Student's two-tailed t test type three. Statistical significance was assumed at P values of <0.05.
RESULTS
Exposure to sublethal biocide concentrations causes a differential response in the protein expression pattern.
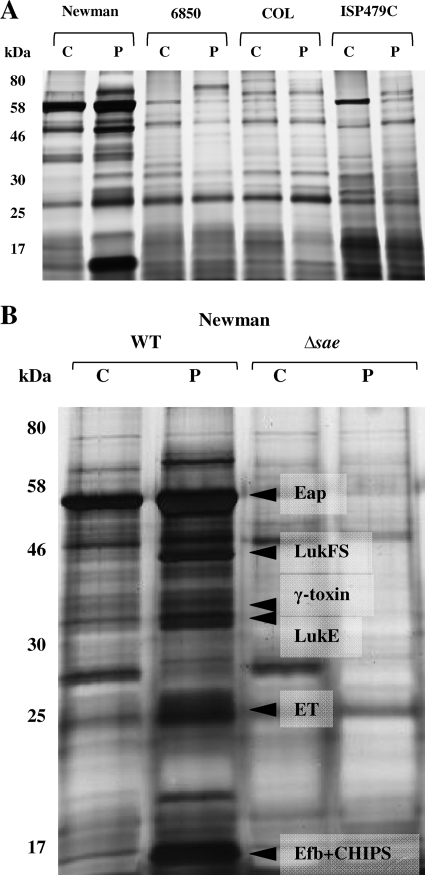
A set of well-studied S. aureus strains (Newman, 6850, COL, and ISP479C) was investigated for their response to sublethal concentrations (i.e., 30% of the MIC) of the commonly used biocide Perform by SDS-PAGE. In general, protein expression appeared to be considerably altered and mostly reduced upon biocide exposure (Fig. 1A). However, biocide treatment of S. aureus strain Newman resulted in increased production of a number of proteins (Fig. 1B). The analysis of the most prominent protein bands in strain Newman by mass spectrometry showed an increased expression of Eap, LukFS, γ-hemolysin component C, LukE, putative staphylococcal enterotoxin, Efb, and chemotaxis inhibitory protein of S. aureus (CHIPS) in the biocide-treated staphylococci (see the legend to Fig. 1B). All of these identified proteins are described to be under the control of the global regulator Sae (21, 42, 43). Sae dependence of expression could be confirmed since they were not expressed in an sae-deficient mutant and were not upregulated in the presence of Perform. These results suggest that the Perform-mediated influence on extracellular protein expression is mediated by the global regulator sae.
FIG. 1.
SDS-PAGE analysis (with total protein extracts) of S. aureus strains Newman, 6850, COL, and ISP479C (A) and Newman versus Newman Δsae (B). (A) Changes in the total protein patterns of S. aureus strains Newman, 6850, COL, and ISP479C after treatment with Perform. Generally, a large number of proteins were downregulated, except in strain Newman. (B) Changes in the protein pattern of S. aureus strain Newman after treatment with Perform are dependent on the sae system. The upregulated proteins in SDS-PAGE were analyzed by mass spectrometry (Eap, extracellular adherence protein; LukFS, leukocidin FS; γ-toxin, γ-hemolysin component C; LukE, leukocidin E; ET, putative staphylococcal enterotoxin; Efb, extracellular fibrinogen-binding protein). Total staphylococcal protein extracts were separated by SDS-PAGE (12%). WT, wild type; C, control; P, 0.04% Perform.
Changes in the protein expression pattern can be mimicked by SDS and are due to an active stress response.
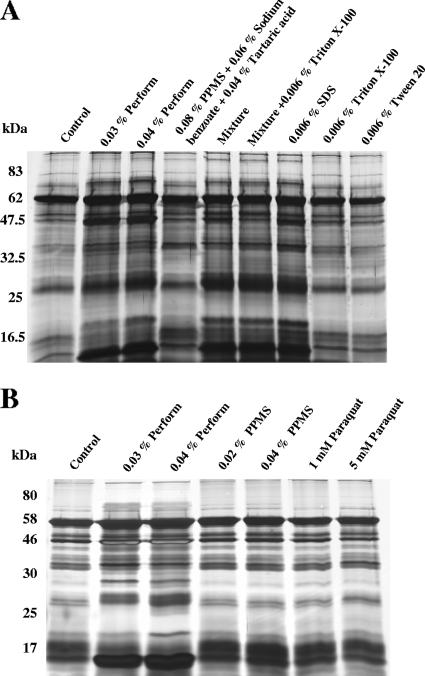
Since Perform is a mixture of several substances, the aim was to identify the substance responsible for the alteration of the protein profile. Therefore, strain Newman was separately treated with components of Perform, among others a mixture of PPMS (an ROS donor), tartaric acid, and sodium benzoate and the detergent SDS. Surprisingly, only SDS, and not the ROS-generating mixture, mimicked the effect of Perform at a concentration corresponding to that in the complete product (Fig. 2A). In contrast, treatment with other detergents, such as Triton X-100 and Tween 20, did not alter the protein expression patterns (Fig. 2A). Furthermore, treatment of strain Newman with another ROS donor, paraquat, and PPMS (without enhancing components of the mixture) had no effect (Fig. 2B). Taken together, these results suggest an oxygen-independent stress by Perform treatment.
FIG. 2.
SDS-PAGE analysis of S. aureus strain Newman. (A) The effect of Perform on the protein pattern (total protein extracts) resembles the effect of SDS in SDS-PAGE. Cell extracts of S. aureus strain Newman were treated with Perform and its ingredients. Mixture, PPMS plus sodium benzoate plus tartaric acid plus SDS, corresponding to 0.04% Perform. (B) The altered protein pattern was not due to oxidative stress but to detergent activity. Another ROS donor, paraquat, had no effect. For experimental controls, see Fig. S1 in the supplemental material.
To ensure that the biocide's effect on the protein pattern was not the result of detergent solubilization, we inhibited transcription with rifampin (final concentration, 1 mg/ml) and translation with tetracycline (final concentration, 200 μg/ml). Autoclaved and mechanically lysed staphylococci were subsequently treated with Perform and SDS, and gel electrophoresis was performed. The resulting protein expression patterns were similar to those from control extracts which were not treated with Perform and SDS (see Fig. S1 in the supplemental material). Consequently, the differential protein pattern (total protein extract) in the presence of Perform or SDS was not due to solubilization or passive accumulation but was the result of an active, target-oriented mechanism depending on live bacteria.
Transcription of saeS and eap and promoter activity of sae P1 are increased by Perform and SDS treatment.
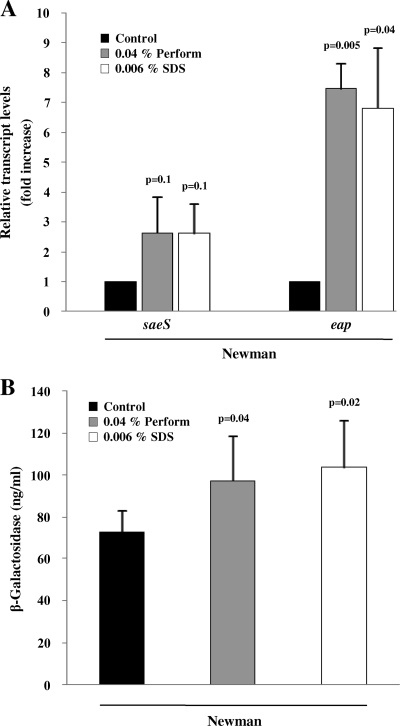
Protein expression for Eap data were validated by qRT-PCR, normalized against 16S rRNA. We detected a 2.5-fold-enhanced transcription of saeS and an up to 7-fold-increased transcription of eap after incubation with Perform and SDS for strain Newman (Fig. 3A). Similar results were obtained with gyrB as a reference gene (data not shown). These data were consistent with the protein expression analyses.
FIG. 3.
Transcriptional activity of strain Newman upon Perform and SDS treatment. (A) The transcript levels of saeS and eap are raised by treatment with Perform and SDS. As an internal control, 16S rRNA was used. The relative transcript levels are expressed as fold increases with respect to the control. The results are means plus SDs from three independent experiments. The statistical significance of individual conditions was in comparison with the respective control condition. (B) The activity of the sae promoter P1 is increased by incubation with Perform and SDS. Equal numbers of bacteria were lysed, and promoter activity is expressed as ng/ml β-galactosidase. Of note, strain Newman has a strongly activated basal sae P1 activity (e.g., 3.2-fold versus strain UAMS-1 and 33-fold versus strain ISP479R). The results are means plus SDs from three independent experiments, each consisting of two independent cultures, analyzed in triplicates.
To confirm the role of the sae system in response to biocides, we analyzed the activity of the sae promoter P1 in a β-galactosidase activity assay. For strain Newman, P1 activity was increased up to 140% of control conditions (from 73.11 ± 9.93 ng/ml to 97.42 ± 21.05 and 103.68 ± 22.08 ng/ml β-galactosidase ng/ml) by treatment with Perform and SDS, respectively (Fig. 3B). Of note, strain Newman has a strongly activated basal sae P1 activity, e.g., 3.2-fold versus strain UAMS-1 and 33-fold versus strain ISP479R. Interestingly, in these two strains the P1 promoter activity was decreased by growth in Perform or SDS (data not shown). This is consistent with the strain-dependent differences observed in the phenotypic analysis, such as protein profiling and cell invasion assay.
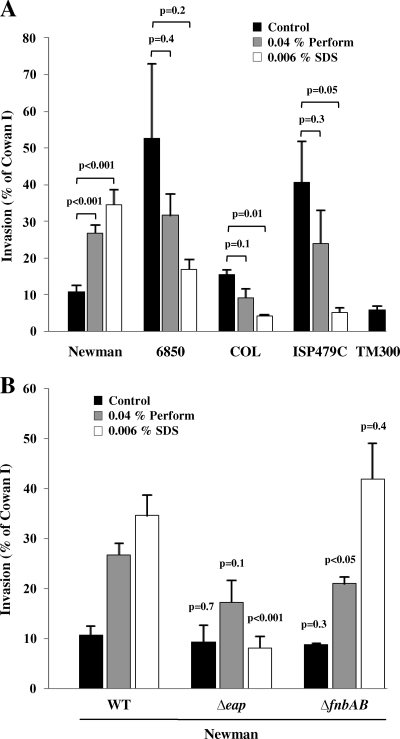
Biocide/SDS increases invasiveness of S. aureus strain Newman but not of other strains tested.
Since the most prominent protein band in strain Newman corresponded to Eap, which is a multifunctional protein with immunomodulatory, adhesive, and invasive properties, we investigated whether increased Eap expression also affects an Eap-dependent function, invasion of host cells. The cellular invasiveness of biocide- and SDS-treated strain Newman was significantly enhanced, as tested for 293 cells. Treatment of strain Newman with Perform and SDS increased invasiveness from 10.7% of the reference for untreated controls to 26.8% and 34.6%, respectively (Fig. 4A). SDS showed a bell-shaped dose response curve for invasiveness, with a maximal increase at between 0.003% and 0.006% (wt/vol) SDS (data not shown).
FIG. 4.
Invasiveness of S. aureus strains Newman, 6850, COL, and ISP479C for 293 cells. (A) Unlike that of other staphylococcal wild-type strains, the invasiveness of S. aureus strain Newman grown in Perform and SDS containing medium increased. The results are means plus SEMs from at least three independent experiments run in duplicate and are expressed as relative invasiveness compared to strain Cowan I. The statistical significance of individual conditions in each strain background was determined. (B) Increased invasiveness of S. aureus strain Newman after treatment with Perform and SDS is dependent on Eap but independent of FnBPs. The results are means plus SEMs from 13 (wild type [WT]), 5 (Δeap), and 3 (ΔfnbAB) independent experiments run in duplicate and are expressed as relative invasiveness compared to strain Cowan I. The statistical significance of individual conditions in each strain background was in comparison with the respective conditions in the WT background.
Consistent with the observed protein patterns (Fig. 1A), further tested wild-type strains showed a decrease of host cell invasion. Invasiveness of COL was reduced from 15.3% to 9.1% and 4.2% in the presence of Perform and SDS, respectively. Strains 6850 and ISP479C displayed a similar invasion pattern, with a decrease from 52.5% to 31.6% and 16.9% invasiveness and a decrease from 40.5% to 23.9% and 5.2% invasiveness by Perform and SDS treatment, respectively (Fig. 4A).
In order to investigate the dependence of host cell invasion on Eap under the conditions investigated, we tested an eap-deficient mutant. Treatment with Perform resulted in a slight increase (from 9.3% to 17.2% invasiveness), whereas SDS treatment further decreased invasiveness to 8.1% (Fig. 4B). These findings implicate that the observed effect of increased invasiveness after Perform and SDS treatment is mediated by Eap. In contrast, a mutant (ΔfnbAB) without FnBPs showed a pattern comparable to that of the wild type (Fig. 4B). These results are in accordance with the known properties of strain Newman, where FnBPs are secreted (19) and therefore should not play a role in host cell invasion. However, Eap has been described to partially compensate for the lack of functionally intact FnBPs in strain Newman (20).
A point mutation in saeS substantially alters the stress response to Perform or SDS exposure.
The sequence for the saeS gene is highly conserved and is nearly identical for all strains sequenced so far. Strain Newman is known to carry a point mutation in saeS (coding for a sensor histidine kinase) (Fig. 5A), which results in a constitutively activated Sae system (1, 10).
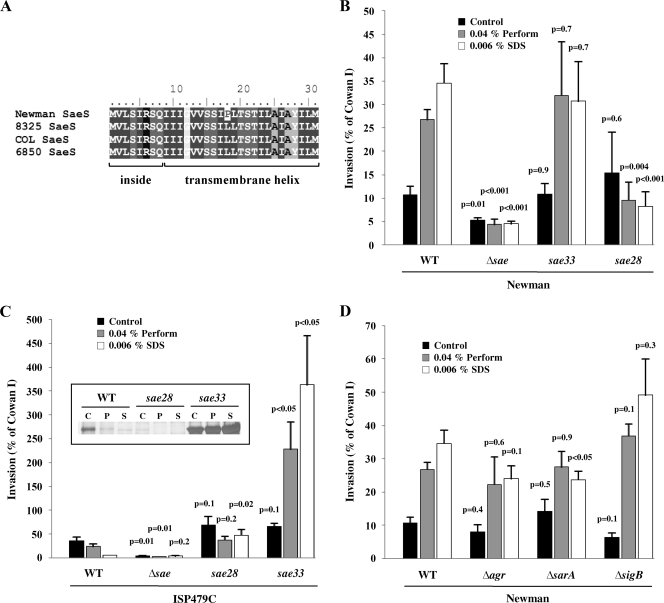
FIG. 5.
SaeS alignment and invasiveness of S. aureus strains Newman and ISP479C for 293 cells. (A) Only strain Newman harbors a mutation in SaeS. (B) Increased invasiveness of S. aureus strain Newman after treatment with Perform and SDS is dependent on the sae system. The results are means plus SEMs from 13 (wild type [WT]), 4 (Δsae), and 3 (sae33) independent experiments run in duplicate and are expressed as relative invasiveness compared to strain Cowan I. The statistical significance of individual conditions in each strain background was in comparison with the respective conditions in the WT background. (C) Increased invasiveness of S. aureus strain ISP479C after treatment with Perform and SDS is dependent on the Newman sae system. The results are means plus SEMs from four independent experiments run in duplicate and are expressed as relative invasiveness compared to strain Cowan I. The statistical significance of individual conditions in each strain background was in comparison with the respective conditions in the WT background. Inset, SDS-PAGE for allele swapping of sae28 (saePQRSISP479C) and sae33 (saePQRSNewman) in strain ISP479C. Eap expression is most prominently increased, paralleling enhanced cellular invasiveness. (D) Increased invasiveness of S. aureus strain Newman after treatment with Perform and SDS is independent of agr and sarA. A sigB knockout seems to further enhance the invasion. The results are means plus SEMs from 13 (WT), 3 (Δagr), 3 (ΔsarA), and 3 (ΔsigB) independent experiments run in duplicate and are expressed as relative invasiveness compared to strain Cowan I. The statistical significance of individual conditions in each strain background was in comparison with the respective conditions in the WT background.
In order to determine whether the observed functional modifications are indeed due to the allelic variant in strain Newman, we tested mutants of strain Newman complemented either with the Newman-specific sequence (pCWSAE33) or with the commonly found sequence (e.g., in the 8325 family), pCWSAE28. For complementation, the whole saePQRS operon was integrated into the chromosomes of the sae-deficient mutants. An sae-deficient mutant of Newman was nearly not invasive (i.e., comparable to the negative control TM300), consistent with the observed protein expression pattern (Fig. 1B). Treatment with Perform or SDS did not change the invasion efficiency, supporting the crucial role of the Sae system (Fig. 5B). A lysostaphin protection assay corroborated the results of the flow cytometric assay, confirming the increased cellular invasiveness after treatment with Perform and SDS (data not shown). Complementation of Newman Δsae with saePQRSNewman (sae33) resulted in complete restoration of the Perform and SDS response. Untreated controls showed 10.9% invasiveness, and treatment with Perform and SDS increased invasiveness to 32% and 30.7%, respectively (Fig. 5B). In contrast, cloning of saePQRSISP479C (sae28) in the Newman sae mutant background resulted in an increased invasiveness compared to that for the wild-type strain, and invasion was significantly inhibited by Perform and SDS.
Strain ISP479C showed 40.5% invasiveness under control conditions. In line with results for the other tested wild-type strains, treatment with Perform and SDS decreased invasiveness of ISP479C to 23.9% and 5.2%, respectively. Like in the Newman background, an sae-deficient mutant of strain ISP479C was nearly not invasive, confirming the importance of the sae system for the invasive phenotype. However, complementation of strain ISP479C with the Newman sae system markedly affected the invasiveness of the mutant. The ISP479C pCWSAE33 derivate was already more invasive (66.2% of the reference value) than the ISP479C wild-type strain. Treatment with Perform and SDS dramatically increased the invasiveness of this ISP479C Δsae mutant complemented with saePQRSNewman (sae33), to 228.2% and 363.2%, respectively (Fig. 5C). The observed protein expression pattern was consistent with the observed functional differences (Fig. 5C, inset).
Other tested global regulators play a minor role in response to biocides and SDS.
Mutations in other global regulators, such as agr, sarA, and sigB, in strain Newman showed no major difference from the parental strain. The Newman sarA mutant tended to be less invasive in response to SDS, but invasiveness was not different for the agr mutant. The sigB-deficient mutant was found to be slightly more invasive in response to Perform and SDS treatment, but this was not statistically significant (Fig. 5D).
DISCUSSION
S. aureus is known to rapidly adapt to changing environmental conditions (7). An increased expression of fnbA in response to subinhibitory concentrations of ciprofloxacin has been reported (4, 5). Therefore, in this study, we investigated the response of a set of S. aureus strains to sublethal concentrations of a commonly used biocide (Perform), which acts through ROS. Sublethal concentrations of Perform considerably altered the protein expression pattern (total protein extract) in a set of well-defined laboratory strains. Strain Newman showed a unique response pattern, with a number of proteins upregulated, rather than downregulated as observed in other strains. Surprisingly, the biocide effect could be mimicked only by SDS, but not by the ROS donor paraquat. The altered protein expression, as tested for Eap, was functionally relevant, as shown by invasion of host cells, which is Eap dependent in strain Newman. Using mutants and allele swapping, we could show that the specific stress response in strain Newman was due to a single point mutation in saeS, the sensor histidine kinase of the saeRS TCS.
Strain Newman showed a response pattern to Perform and SDS exposure that was different from that of other strains. The most prominently upregulated protein was Eap (Fig. 1B), a multifunctional protein which is known to be expressed in an sae-dependent manner (21). There are two known alterations: (i) strain Newman secretes the major staphylococcal invasion proteins, FnBPs, due to a stop codon (19), but high expression of Eap can compensate for the loss of FnBPs (20); and (ii) a point mutation in SaeS, the sensor histidine kinase (Fig. 5A), has been shown to render it constitutively active (1, 10). The point mutation in saeS appears to be generally present in strain Newman. It can be found in the published genome sequence for strain Newman (GenBank accession numbers NC_009641 and AP009351 [2]). It has been described in further reports (1), including two deposited sequences, as GenBank/EMBL accession numbers AJ556794 (48) and AF129010 (12). In addition, we verified the saeS point mutation in the isolate used for this study (Fig. 5A). A complete list of the publicly available sequences can be found in Uniprot under accession number Q840P7 (http://www.uniprot.org/uniprot/).
Consistent with the known Eap-dependent invasion mechanism, our data on invasion of strain Newman after treatment with Perform and SDS argue for an FnBP-independent effect, since the results for a ΔfnbAB mutant of Newman were not different from those for the wild type (Fig. 4B). Eap-dependent host cell invasion was even more pronounced after overnight treatment with Perform or SDS (data not shown). Increased Eap production has been shown to increase aggregation (20, 36), which could explain the enlarged clusters observed in the presence of Perform and SDS (data not shown). However, these larger aggregates did not lead to Perform- and SDS-dependent increased invasiveness. Aggregates could be minimized by sonication, and sonicated and untreated staphylococci did not differ significantly in host cell invasion (data not shown).
In S. aureus strain Newman, Perform as well as SDS treatment significantly enhanced the transcription and promoter activity of the global regulator sae (Fig. 3); the expression of proteins such as Eap, LukFS, γ-hemolysin component C, LukE, putative staphylococcal enterotoxin, Efb, and CHIPS (Fig. 1B); and cellular invasiveness (Fig. 4 and 5). This is in line with previous findings, demonstrating these proteins to be under the control of the sae system (21, 42, 43). Other wild-type strains, however, showed mostly a decreased expression of the above-mentioned proteins and were less invasive in response to Perform and SDS (Fig. 4A). As a consequence, stress induced by Perform and SDS is most likely sensed by the sae TCS, inducing an sae-specific stress response. This assumption is supported both by qRT-PCR data and by promoter activity assays. While strain Newman showed an enhanced transcription of sae and an increased activity of the sae promoter P1 upon Perform and SDS treatment, other wild-type strains tested (i.e., strains UAMS-1 and ISP479R) showed decreased sae P1 activities after treatment with these substances (data not shown). The statistically significant increase of sae P1 promoter activity in strain Newman (i.e., from 73.11 ± 9.93 to 97.42 ± 21.05 and 103.68 ± 22.08 ng/ml β-galactosidase by Perform and SDS, respectively [means ± SDs]) appears to be also biologically significant due to the following reasons: (i) strain Newman showed a very high basal sae P1 activity (in control experiments, 3.2-fold versus strain UAMS-1 and 33-fold versus strain ISP479R), (ii) the qRT-PCR data demonstrate a more than 2.5-fold increase in saeS transcript levels (Fig. 3A), (iii) the activity is paralleled by function (i.e., increased Eap production and cellular invasiveness), and (iv) the sae system in strain Newman has been reported to be constitutively nearly maximally active (1, 10).
We hypothesized that the specific response pattern in strain Newman is dependent on the sequence of SaeS. Strain Newman harbors an amino acid substitution within the first membrane-spanning domain (Fig. 5A) of the sensor histidine kinase SaeS (1, 10, 48). Transforming the sae operon from strain Newman (sae33) into strain ISP479C led to elevated invasiveness after Perform and SDS treatment, as found in wild-type strain Newman (Fig. 5B). This suggests that the point mutation alters the response to biocides. The even larger effect on invasiveness in strain ISP479C (sae33) is likely to be due to a combined upregulation of both Eap and FnBPs by sae (28, 48), since strain ISP479C possesses functional FnBPs. The exact mode of sensing by SaeS is currently unclear. However, it appears not to depend on ROS under these experimental conditions. Since the effect appears not to be elicited by detergents other than SDS, such as Triton X-100 and Tween 20, it may be different from a global membrane stress. SDS is known to affect the tertiary structure of proteins. It has been reported that the L18P substitution leads to a constitutive activation due to a predicted disruption of the surrounding α-helical configuration, changing it to a β-sheet (1). Thus, conformational changes induced by SDS appear to be potentially responsible, altering the signaling properties of SaeS.
Other global regulators seem to play a minor or indirect role in the response to biocide or SDS. Interestingly, activity and sae-dependent functions were markedly increased by biocide and SDS treatment, even over strongly activated baseline levels in strain Newman (Fig. 1, 3, 4, and 5). This is in contrast to results other studies of the sae system (1, 10), indicating that the constitutive activation of the Newman sae system is already maximal (i.e., that it cannot be further influenced by external stimuli). The observed regulatory role of the sae point mutation appears not to generally affect all aspects of the stress response, since biofilm formation did not follow the observed response pattern (data not shown). This supports the specificity of the observed effect on protein expression and invasiveness.
In summary, these data may help in better understanding and dissecting the virulence mechanism in strain Newman. Several defects in strain Newman are now known, such as the loss of cell wall-anchored FnBPs, which have been shown to be crucial for pathogenesis of endocarditis (40). Nevertheless, this strain still has a very high virulence potential, as observed by systemic challenge (Knut Ohlsen et al., personal communication). This may, at least partially, be dependent on the saeS point mutation in strain Newman and thus could help to further clarify virulence mechanisms and their regulation in S. aureus.
Supplementary Material
Acknowledgments
We thank Nadine Leitschuh and Karina Lamprecht for expert technical assistance, Ferenc Kiss for help with protein data bank queries, and Christoph Schoen for statistical advice.
This work has been funded by the Deutsche Forschungsgemeinschaft (grant SFB-TR34 C6) and in part by grants SFB-TR34 B1 and Wo578/5-2.
Footnotes
Published ahead of print on 25 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adhikari, R. P., and R. P. Novick. 2008. Regulatory organization of the staphylococcal sae locus. Microbiology 154:949-959. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balwit, J. M., P. van Langevelde, J. M. Vann, and R. A. Proctor. 1994. Gentamicin-resistant menadione and hemin auxotrophic Staphylococcus aureus persist within cultured endothelial cells. J. Infect. Dis. 170:1033-1037. [DOI] [PubMed] [Google Scholar]
- 4.Bisognano, C., W. L. Kelley, T. Estoppey, P. Francois, J. Schrenzel, D. Li, D. P. Lew, D. C. Hooper, A. L. Cheung, and P. Vaudaux. 2004. A RecA-LexA-dependent pathway mediates ciprofloxacin-induced fibronectin binding in Staphylococcus aureus. J. Biol. Chem. 279:9064-9071. [DOI] [PubMed] [Google Scholar]
- 5.Bisognano, C., P. E. Vaudaux, D. P. Lew, E. Y. Ng, and D. C. Hooper. 1997. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Catrein, I., R. Herrmann, A. Bosserhoff, and T. Ruppert. 2005. Experimental proof for a signal peptidase I like activity in Mycoplasma pneumoniae, but absence of a gene encoding a conserved bacterial type I SPase. FEBS J. 272:2892-2900. [DOI] [PubMed] [Google Scholar]
- 7.Clements, M. O., and S. J. Foster. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458-462. [DOI] [PubMed] [Google Scholar]
- 8.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 9.Dyke, K. G., M. P. Jevons, and M. T. Parker. 1966. Penicillinase production and intrinsic resistance to penicillins in Staphylococcus aureus. Lancet i:835-838. [DOI] [PubMed] [Google Scholar]
- 10.Geiger, T., C. Goerke, M. Mainiero, D. Kraus, and C. Wolz. 2008. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J. Bacteriol. 190:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giraudo, A. T., A. Calzolari, A. A. Cataldi, C. Bogni, and R. Nagel. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15-22. [DOI] [PubMed] [Google Scholar]
- 13.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 14.Giraudo, A. T., C. Mansilla, A. Chan, C. Raspanti, and R. Nagel. 2003. Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol. 46:246-250. [DOI] [PubMed] [Google Scholar]
- 15.Giraudo, A. T., G. L. Martinez, A. Calzolari, and R. Nagel. 1994. Characterization of a transpositional mutant of Staphylococcus aureus underproducing exoproteins. Rev. Latinoam. Microbiol. 36:171-176. [PubMed] [Google Scholar]
- 16.Goerke, C., U. Fluckiger, A. Steinhuber, V. Bisanzio, M. Ulrich, M. Bischoff, J. M. Patti, and C. Wolz. 2005. Role of Staphylococcus aureus global regulators sae and σB in virulence gene expression during device-related infection. Infect. Immun. 73:3415-3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goerke, C., U. Fluckiger, A. Steinhuber, W. Zimmerli, and C. Wolz. 2001. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40:1439-1447. [DOI] [PubMed] [Google Scholar]
- 18.Greene, C., D. McDevitt, P. Francois, P. E. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin-binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 19.Grundmeier, M., M. Hussain, P. Becker, C. Heilmann, G. Peters, and B. Sinha. 2004. Truncation of fibronectin-binding proteins in Staphylococcus aureus strain Newman leads to deficient adherence and host cell invasion due to loss of the cell wall anchor function. Infect. Immun. 72:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haggar, A., M. Hussain, H. Lonnies, M. Herrmann, A. Norrby-Teglund, and J. I. Flock. 2003. Extracellular adherence protein from Staphylococcus aureus enhances internalization into eukaryotic cells. Infect. Immun. 71:2310-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harraghy, N., J. Kormanec, C. Wolz, D. Homerova, C. Goerke, K. Ohlsen, S. Qazi, P. Hill, and M. Herrmann. 2005. sae is essential for expression of the staphylococcal adhesins Eap and Emp. Microbiology 151:1789-1800. [DOI] [PubMed] [Google Scholar]
- 22.Hussain, M., A. Haggar, C. Heilmann, G. Peters, J. I. Flock, and M. Herrmann. 2002. Insertional inactivation of Eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect. Immun. 70:2933-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain, M., A. Haggar, G. Peters, G. S. Chhatwal, M. Herrmann, J. I. Flock, and B. Sinha. 2008. More than one tandem repeat domain of the extracellular adherence protein of Staphylococcus aureus is required for aggregation, adherence, and host cell invasion but not for leukocyte activation. Infect. Immun. 76:5615-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussain, M., C. von Eiff, B. Sinha, I. Joost, M. Herrmann, G. Peters, and K. Becker. 2008. eap gene as novel target for specific identification of Staphylococcus aureus. J. Clin. Microbiol. 46:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joost, I., D. Blass, M. Burian, C. Goerke, C. Wolz, von Müller, L., K. Becker, K. Preissner, M. Herrmann, and M. Bischoff. 2009. Transcription analysis of the extracellular adherence protein from Staphylococcus aureus in authentic human infection and in vitro. J. Infect. Dis. 199:1471-1478. [DOI] [PubMed] [Google Scholar]
- 26.Juuti, K. M., B. Sinha, C. Werbick, G. Peters, and P. I. Kuusela. 2004. Reduced adherence and host cell invasion by methicillin-resistant Staphylococcus aureus expressing the surface protein Pls. J. Infect. Dis. 189:1574-1584. [DOI] [PubMed] [Google Scholar]
- 27.Kluytmans, J., A. van Belkum, and H. Verbrugh. 1997. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin. Microbiol. Rev. 10:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda, H., M. Kuroda, L. Cui, and K. Hiramatsu. 2007. Subinhibitory concentrations of β-lactam induce haemolytic activity in Staphylococcus aureus through the SaeRS two-component system. FEMS Microbiol. Lett. 268:98-105. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 32.Li, D., and A. Cheung. 2008. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect. Immun. 76:1068-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 34.McGavin, M. H., D. Krajewska-Pietrasik, C. Ryden, and M. Höök. 1993. Identification of a Staphylococcus aureus extracellular matrix-binding protein with broad specificity. Infect. Immun. 61:2479-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novick, R. P., and D. Jiang. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology 149:2709-2717. [DOI] [PubMed] [Google Scholar]
- 36.Palma, M., A. Haggar, and J. I. Flock. 1999. Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 181:2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pattee, P. A. 1981. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J. Bacteriol. 145:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-82. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, NY.
- 40.Que, Y. A., J. A. Haefliger, L. Piroth, P. Francois, E. Widmer, J. M. Entenza, B. Sinha, M. Herrmann, P. Francioli, P. Vaudaux, and P. Moreillon. 2005. Fibrinogen and fibronectin binding cooperate for valve infection and invasion in Staphylococcus aureus experimental endocarditis. J. Exp. Med. 201:1627-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roche, F. M., R. Massey, S. J. Peacock, N. P. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 42.Rogasch, K., V. Rühmling, J. Pané-Farré, D. Höper, C. Weinberg, S. Fuchs, M. Schmudde, B. M. Bröker, C. Wolz, M. Hecker, and S. Engelmann. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooijakkers, S. H., M. Ruyken, J. van Roon, K. P. van Kessel, J. A. van Strijp, and W. J. van Wamel. 2006. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell. Microbiol. 8:1282-1293. [DOI] [PubMed] [Google Scholar]
- 44.Schleifer, K. H., and U. Fischer. 1982. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int. J. Syst. Bacteriol. 32:153-156. [Google Scholar]
- 45.Senn, M. M., P. Giachino, D. Homerova, A. Steinhuber, J. Strassner, J. Kormanec, U. Flückiger, B. Berger-Bächi, and M. Bischoff. 2005. Molecular analysis and organization of the σB operon in Staphylococcus aureus. J. Bacteriol. 187:8006-8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheagren, J. N. 1984. Staphylococcus aureus. The persistent pathogen. N. Engl. J. Med. 310:1368-1373. [DOI] [PubMed] [Google Scholar]
- 47.Sinha, B., P. P. Francois, O. Nüsse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5b1. Cell Microbiol. 1:101-117. [DOI] [PubMed] [Google Scholar]
- 48.Steinhuber, A., C. Goerke, M. G. Bayer, G. Döring, and C. Wolz. 2003. Molecular architecture of the regulatory locus sae of Staphylococcus aureus and its impact on expression of virulence factors. J. Bacteriol. 185:6278-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Debarbouille, J. R. Penades, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187:5318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Eiff, C., K. Becker, K. Machka, H. Stammer, and G. Peters. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11-16. [DOI] [PubMed] [Google Scholar]
- 51.Weidenmaier, C., J. F. Kokai-Kun, S. A. Kristian, T. Chanturiya, H. Kalbacher, M. Gross, G. Nicholson, B. Neumeister, J. J. Mond, and A. Peschel. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243-245. [DOI] [PubMed] [Google Scholar]
- 52.Wolz, C., D. McDevitt, T. J. Foster, and A. L. Cheung. 1996. Influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect. Immun. 64:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.