Abstract
The PhoP/PhoQ two-component system controls several physiological and virulence functions in Salmonella enterica. This system is activated by low Mg2+, acidic pH, and antimicrobial peptides, but the biological consequences resulting from sensing multiple signals are presently unclear. Here, we report that the PhoP/PhoQ system regulates different Salmonella genes depending on whether the inducing signal is acidic pH or low Mg2+. When Salmonella experiences acidic pH, the PhoP/PhoQ system promotes Fe2+ uptake in a process that requires the response regulator RstA, activating transcription of the Fe2+ transporter gene feoB. In contrast, the PhoP-induced RstA protein did not promote feoB expression at neutral pH with low Mg2+. The PhoP/PhoQ system promotes the expression of the Mg2+ transporter mgtA gene only when activated in bacteria starved for Mg2+. This is because mgtA transcription promoted at high Mg2+ concentrations by the acidic-pH-activated PhoP protein failed to reach the mgtA coding region due to the mgtA leader region functioning as a Mg2+ sensor. Our results show that a single two-component regulatory system can regulate distinct sets of genes in response to different input signals.
Signal transduction mediated by two-component regulatory systems enables bacterial cells to rapidly adapt to and survive various stressful conditions. The PhoP/PhoQ two-component system is comprised of the response regulator PhoP and the sensor PhoQ. The PhoQ protein has been demonstrated to sense Mg2+ concentrations as a specific signal (14). When the environmental Mg2+ concentration is lowered to micromolar levels, PhoQ promotes the phosphorylated state of the PhoP protein (14, 32). In Salmonella enterica, the low-Mg2+-activated PhoP protein directly regulates the transcription of many genes that are necessary for virulence in mammalian hosts, as well as other physiological processes (15). In particular, consistent with the environment where the system is activated, PhoP/PhoQ allows Salmonella to grow at limited concentrations of Mg2+ (14), which results from PhoP-activated expression of the mgtA and mgtB genes, encoding Mg2+ transporters (34). The PhoQ-mediated PhoP phosphorylation also occurs at acidic pH (31), and transcription levels of the PhoP-activated genes pagA, phoN, and pmrD increase in Salmonella experiencing acidic conditions (3, 28, 31). Taking into account that certain antimicrobial peptides promote the expression of a subset of the PhoP-regulated genes through the PhoP and PhoQ proteins (2), the activity of the PhoP/PhoQ system appears to respond to at least three different signals.
The PhoP/PhoQ system also regulates gene expression by controlling the levels and/or activity of other regulators (22). The RstA/RstB two-component system, which consists of a response regulator RstA and its partner sensor RstB, is the one whose expression is regulated by the PhoP/PhoQ system. In Escherichia coli, the PhoP protein binds to the rstA promoter, and transcription of the rstA gene is repressed by the PhoP/PhoQ system in cells grown at a high concentration of Mg2+ (26). The RstA protein promotes transcription of the asr gene, coding for a product necessary for adaptation to acidic stress (27). Consequently, at acidic pH, transcription of the asr gene is not fully activated in a strain lacking the phoP gene (27), due to the reduced levels of the RstA protein. In addition, the rstA gene has been identified as a multicopy suppressor of the essential genes yjeE, yeaZ, and ygjD (5, 16).
The Salmonella PhoP protein also directly binds to and activates the rstA promoter at low Mg2+ (Fig. 1) (I. Zwir et al., unpublished data). We have recently demonstrated that when overexpressed from a plasmid, the RstA protein specifically binds to the feoA promoter and promotes transcription of the feoAB operon encoding the ferrous iron (Fe2+) transporter FeoB (Fig. 1) (20).
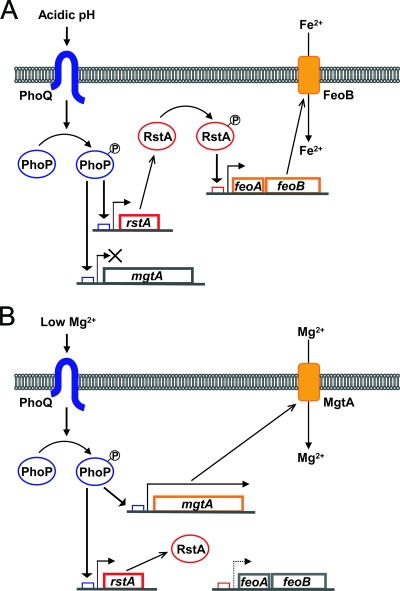
FIG. 1.
Model illustrating Fe2+ and Mg2+ uptake regulated by the PhoP/PhoQ system and RstA protein in Salmonella. (A) When activated at millimolar Mg2+ concentrations by the acidic-pH signal, the phosphorylated PhoP protein promotes transcription of the rstA and mgtA genes. The induced RstA protein is activated, possibly by phosphorylation. The increase in feoB expression resulting from RstA binding to the feoA promoter enhances the FeoB-mediated Fe2+ uptake. In contrast, mgtA transcription fails to proceed due to Mg2+ binding to the mgtA leader. (B) The low-Mg2+ signal activates the PhoP/PhoQ system at neutral pH. Activation of mgtA transcription by phosphorylated PhoP increases production of the full length of the mgtA mRNA, leading to expression of the MgtA protein, which promotes Mg2+ uptake. In contrast, the RstA protein induced at neutral pH lacks the activity to promote transcription of the feoB gene.
The PhoP/PhoQ system promotes the expression of the RstA protein when activated at either low Mg2+ or acidic pH (Fig. 1). We now report that the RstA protein promotes transcription of the feoB gene exclusively when activated at acidic pH, thereby enhancing the growth of Salmonella in environments with limited iron. We show that, in contrast to the PhoP-controlled Fe2+ uptake, the expression of the Mg2+ transporter MgtA occurs only when the PhoP/PhoQ system is activated at low Mg2+. Thus, depending on the input signals, a single signal transduction system can differentially regulate its target genes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Salmonella enterica serovar Typhimurium strains are derived from strain 14028s. Phage P22-mediated transductions were performed as described previously (11). Bacteria were grown at 37°C in N-minimal medium (33), pH 7.7 or pH 5.7, supplemented with 0.1% Casamino Acids, 38 mM glycerol, and different concentrations of MgCl2. Ampicillin, chloramphenicol, and kanamycin were used at 50 μg/ml, 25 μg/ml, and 50 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| S. enterica serovar Typhimurium strains | ||
| 14028s | Wild type | 13 |
| DS267 | ΔphoPQ::Cmr, same as EG15599 | 32 |
| JH101 | ΔrstA::Kmr | 20 |
| DS603 | ΔrstB::Cmr | This study |
| DS604 | rstA-FLAG | This study |
| EN267 | ΔphoPQ::Cmr rstA-FLAG | This study |
| EN293 | ΔphoPQ::Cmr ΔrstA::Kmr | This study |
| EN252 | ΔackA-pta::Kmr | This study |
| EN323 | Δ5′UTR-PmgtA | This study |
| EN331 | Δ5′UTR-PmgtA ΔphoPQ::Cmr | This study |
| JH380 | ΔsitABCD ΔmntH | This study |
| EN256 | ΔsitABCD ΔmntH ΔrstA::Kmr | This study |
| EN258 | ΔsitABCD ΔmntH ΔfeoB::Cmr | This study |
| EN290 | ΔsitABCD ΔmntH ΔphoPQ::Cmr | This study |
| JH352 | Δfur | 20 |
| Plasmids | ||
| pUHE21-2lacIq | reppMBI AprlacIq | 36 |
| pKD3 | repR6Kγ Apr FRT Cmr FRT | 10 |
| pKD4 | repR6Kγ Apr FRT Kmr FRT | 10 |
| pKD46 | reppSC101(Ts) Apr ParaBAD γ β exo | 10 |
| pCP20 | reppSC101(Ts) Apr CmrcI857 λPRflp | 10 |
| pJH4 | reppMBI AprlacIqrstA | 20 |
| pEN105 | reppMBI AprlacIqrstA-FLAG | This study |
| pEN106 | reppMBI AprlacIqrstA(D52A)-FLAG | This study |
| pDS303 | reppMBI AprlacIqphoP* | This study |
UTR, untranslated region; FRT, FLP recombination target.
Construction of bacterial strains.
The one-step gene inactivation method (10) was used for chromosomal gene deletion and epitope tagging. The sequences of primers used are indicated in Table S1 in the supplemental material. For construction of the rstB deletion strain, DS603, the Cmr cassette from plasmid pKD3 (10) was amplified using primers DE-rstB-F and DE-rstB-R and integrated into the rstB region of strain 14028s. The EN323 strain, where 100 base pairs (bp) of the mgtA leader was replaced with 84 bp of the “scar” sequence, was constructed as described previously (9). The Kmr cassette from plasmid pKD4 (10) was amplified using primers DE-mgtA(5′UTR)-F and DE-mgtA(5′UTR)-R and was integrated into the chromosome of strain 14028s. The Kmr cassette was removed using plasmid pCP20 (10). To construct the EN252 strain, where the ackA and pta genes are deleted, the Kmr cassette was amplified using primers DE-ackApta-F and DE-ackApta-R and pKD4 as DNA template, and the purified PCR products were integrated into the chromosome of strain 14028s. The JH380 strain, in which both sitABCD and mntH are deleted, was constructed as follows. First, the Kmr cassette was amplified using primers DE-sitABCD-F and DE-sitABCD-R and pKD4 as DNA template; the Cmr cassette was obtained by PCR amplification of pKD3 with primers DE-mntH-F and DE-mntH-R. The purified PCR products were introduced into strain 14028s, resulting in the ΔsitABCD::Kmr or ΔmntH::Cmr strain, respectively. Second, the ΔmntH::Cmr region was transferred into the ΔsitABCD::Kmr strain by phage P22-mediated transduction. Finally, both the Kmr and Cmr cassettes were removed from the ΔsitABCD::Kmr ΔmntH::Cmr strain using pCP20. Deletion of the corresponding genes was verified by colony PCR. The Salmonella Typhimurium strain DS604 encoding the RstA protein with a FLAG tag at the C terminus in the normal rstA chromosomal location was constructed as follows: the Kmr cassette was amplified by PCR using primers rstA-FLAG-F and rstA-FLAG-R and pKD4 as template and integrated at the 3′ end of the rstA gene. The Kmr cassette was removed from the resulting strain by using pCP20, and the presence of a FLAG tag at the C terminus of RstA was confirmed by nucleotide sequencing.
Plasmid construction.
Plasmid pEN105 expressing the RstA-FLAG protein from the lac promoter was constructed as follows. The rstA-FLAG gene was amplified by PCR using primers CD-rstA-FLAG-F and CD-rstA-FLAG-R and chromosomal DNA from the DS604 strain. The PCR products were purified and introduced between the BamHI and PstI restriction sites of pUHE21-2lacIq (36). Plasmid pEN106 is a derivative of pEN105 and expresses a variant of RstA-FLAG with a D52A substitution. This plasmid was constructed using a QuikChange II site-directed mutagenesis kit (Stratagene) with primers SM-rstA(D52A)-F and SM-rstA(D52A)-R and pEN105 as DNA template. For construction of the pDS303 plasmid in which PhoP*, a variant of the PhoP protein that can promote gene transcription independently of the PhoQ protein, is expressed from the lac promoter, the gene encoding PhoP* was amplified by PCR using primers CD-phoP-F and CD-phoP-R and chromosomal DNA from the EG10232 strain (7). The PCR products were purified and introduced between the BamHI and PstI restriction sites of pUHE21-2lacIq (36). The sequences of the rstA-FLAG gene and PhoP*-encoding regions on the recombinant plasmids were confirmed by nucleotide sequencing. The sequences of the primers used are indicated in Table S1 in the supplemental material.
RNA isolation and quantitative real-time reverse transcription-PCR (qRT-PCR) analysis.
RNA was isolated from mid-exponential-phase culture (optical density at 600 nm [OD600] of 0.5 to 0.6) grown in 20 ml of N-minimal medium. One-half milliliter of the culture was removed and mixed with 1 ml of RNAprotect bacterial reagent (Qiagen), and RNA was isolated using an RNeasy mini kit (Qiagen). The RNA sample was treated further with RNase-free DNase (Ambion). By using Omnitranscript reverse transcription reagents (Qiagen) and random primers (Invitrogen), cDNA was synthesized from 0.5 μg of RNA. Transcripts were quantified by real-time PCR using SYBR green PCR master mix (Applied Biosystems) on an ABI7300 sequence detection system (Applied Biosystems). The primers used for detection of transcripts of each gene are listed in Table S2 in the supplemental material. The transcription levels of each gene were calculated from a standard curve obtained by PCR with the same primers and serially diluted genomic DNA. The mRNA levels of target genes were normalized to 16S rRNA levels.
Western blot analysis.
Salmonella strains harboring the rstA-FLAG gene were grown in 20 ml of N-minimal medium. When the cells’ OD600 reached ∼0.5, bacterial cells were washed once with phosphate-buffered saline, suspended in 0.5 ml of phosphate-buffered saline, and opened by sonication. Total protein concentrations were determined by the bicinchoninic acid method. Whole-cell lysates containing 25 μg of total proteins were resolved on 12% sodium dodecyl sulfate-polyacrylamide gels, transferred to nitrocellulose membranes, and analyzed by Western blotting using monoclonal anti-FLAG (Sigma) or anti-DnaK antibody (Stressgen). Blots were developed by using horseradish peroxidase-linked anti-mouse immunoglobulin G antibody (GE Healthcare) and an ECL detection system (GE Healthcare). We verified that signals corresponding to the RstA-FLAG proteins were within the linear range of detection by conducting the experiments with cell lysates containing 12.5, 25, and 50 μg of total protein.
RESULTS
The PhoP/PhoQ system promotes RstA expression at similar levels when activated at either low Mg2+ or acidic pH.
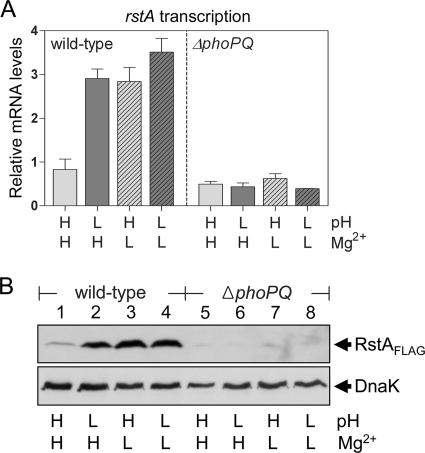
We explored whether rstA expression could be controlled by acidic pH because low pH also plays a role as a signal activating the PhoP/PhoQ system (31). We isolated RNA from Salmonella strains grown in minimal medium containing activating (i.e., micromolar Mg2+ or acidic pH) or repressing (i.e., neutral pH with millimolar Mg2+) signals for the PhoP/PhoQ system. The results of qRT-PCR revealed that the rstA mRNA levels were threefold higher in the wild-type strain grown at pH 7.7 with 50 μM Mg2+ or at pH 5.7 with 2 mM Mg2+ than at pH 7.7 with 2 mM Mg2+ (Fig. 2A). This activation was dependent on the PhoP/PhoQ system, because there was no rstA transcription in a strain with the phoPQ operon deleted (Fig. 2A). In agreement with the transcription data, Western blot analysis using the cell extracts prepared from strains expressing the RstA-FLAG protein from the normal chromosomal location showed the RstA protein in the wild-type strain only following growth in low Mg2+ or acidic pH (Fig. 2B, lanes 2 and 3). The RstA protein was hardly detected in the phoPQ deletion mutant, regardless of the Mg2+ concentration and pH value (Fig. 2B, lanes 5 to ∼7), or in the wild-type strain that was grown at pH 7.7 with 2 mM Mg2+ (Fig. 2B, lane 1). Therefore, the data showed that both the low-Mg2+ and acidic-pH signal can promote RstA expression to similar levels in a process that is dependent on the PhoP/PhoQ two-component system.
FIG. 2.
The PhoP/PhoQ system activated by acidic pH or low Mg2+ promotes expression of the rstA gene. Salmonella strains were grown to an OD600 of 0.5 to 0.6 in N-minimal medium containing 2 mM (H, high) or 50 μM (L, low) Mg2+ and adjusted to pH 7.7 (H, high) or 5.7 (L, low). (A) The rstA mRNA levels in wild-type (14028s) and phoPQ deletion (DS267) strains were determined using qRT-PCR. Shown are the mean values and standard deviations from three independent experiments. (B) Western blot analysis of crude extracts prepared from wild-type (DS604) and phoPQ mutant (EN267) strains and probed with anti-FLAG and anti-DnaK antibodies.
The RstA protein, induced at acidic pH by the PhoP/PhoQ system, promotes transcription of the feoB gene.
We have recently reported that overexpression of the RstA protein from a plasmid activates transcription of the feoAB operon encoding the ferrous iron transporter FeoB by direct RstA binding to the feoA promoter (20). Thus, we explored whether the RstA protein induced at low Mg2+ and acidic pH by the PhoP/PhoQ system could promote transcription of the feoB gene.
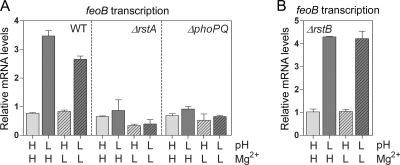
We determined that, when grown in the presence of 2 mM Mg2+, the wild-type strain expressed fourfold higher levels of feoB mRNA at pH 5.7 than at pH 7.7 (Fig. 3A). The feoB activation required the PhoP/PhoQ-dependent RstA protein, because there was no feoB transcription in response to acidic pH in strains with rstA or phoPQ deleted (Fig. 3A). Transcription of the feoB gene was not activated in the wild-type strain grown at pH 7.7 with 50 μM Mg2+ (Fig. 3A). This is in spite of the fact that Salmonella bacteria experiencing low Mg2+ produced the RstA protein at levels similar to the levels in bacteria grown at acidic pH (Fig. 2B, lanes 2 and 3). When both signals were present (i.e., low Mg2+ and acidic pH), feoB transcription took place in an RstA- and PhoPQ-dependent manner (Fig. 3A). This result was not due to differences in the RstA protein levels between Salmonella experiencing pH 5.7 with 50 μM Mg2+ versus pH 7.7 with 50 μM Mg2+ (Fig. 2B, compare lanes 3 and 4). In sum, our experiments demonstrated that acidic pH can promote feoB transcription in an RstA-dependent fashion, whereas the low-Mg2+ signal is unable to elicit feoB transcription.
FIG. 3.
Activation of feoB transcription at acidic pH is dependent on the PhoP/PhoQ and RstA proteins but independent of the RstB protein. Wild-type (WT; 14028s), rstA deletion (JH101), phoPQ deletion (DS267), and rstB deletion (DS603) strains were grown in N-minimal medium at pH 7.7 (H, high) or 5.7 (L, low) with 2 mM (H, high) or 50 μM (L, low) Mg2+. The feoB mRNA levels were determined using qRT-PCR. Shown are the mean values and standard deviations from three independent experiments.
The RstB sensor is dispensable for activation of feoB transcription at acidic pH.
Because the phosphorylation of a response regulator is primarily mediated by its cognate sensor kinase, we next asked whether the RstB sensor is responsible for the acidic-pH-promoted RstA activity. However, the RstB protein does not appear to affect feoB transcription because an rstB deletion mutant grown in medium with high (i.e., 2 mM) or low (i.e., 50 μM) Mg2+ expressed fourfold higher levels of the feoB mRNA at pH 5.7 than at pH 7.7 (Fig. 3B), just like the wild-type strain. Thus, our data suggest that phosphorylation of the RstA protein could be mediated by a phosphodonor other than RstB during the growth of Salmonella at acidic pH.
Acidic pH promotes RstA activity, possibly via phosphorylation.
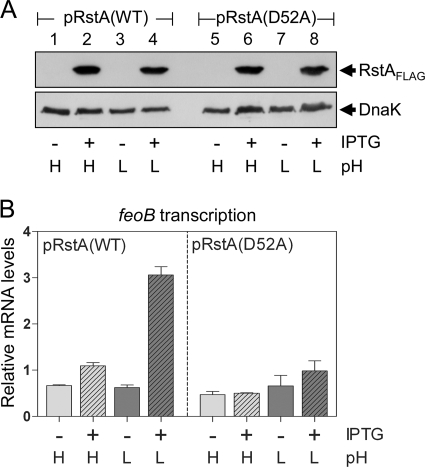
The results presented above suggest that the activity of the RstA protein is promoted at acidic pH. To further explore the activation of the RstA protein, we constructed plasmid pEN105, where expression of the RstA-FLAG protein is under the control of the lac promoter. Western blot analysis revealed that the rstA deletion strain harboring pEN105 expressed the RstA-FLAG protein only in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG), regardless of the pH of the medium (Fig. 4A, lanes 2 and 4). (These RstA-FLAG protein levels were similar to those expressed from a strain with a chromosomal rstA-FLAG gene experiencing acidic pH [data not shown]). The feoB mRNA levels were threefold higher in organisms grown at pH 5.7 than in those grown at pH 7.7 (Fig. 4B). Because the rstA gene is expressed from the lac promoter, these data indicate that RstA activity (as opposed to RstA level) is increased in acidic pH.
FIG. 4.
RstA-promoted feoB transcription requires the acidic-pH signal and its putative phosphorylation site even when the rstA gene is transcribed from a heterologous promoter. Plasmids pEN105 [pRstA(WT)] and pEN106 [pRstA(D52A)] express the FLAG-tagged wild-type RstA and D52A RstA protein, respectively, from the lac promoter. Strains were grown in N-minimal medium containing 2 mM Mg2+ and buffered at pH 7.7 (H, high) or 5.7 (L, low) in the presence (+) or absence (−) of 0.1 mM IPTG. (A) Western blot analysis of crude extract prepared from the ΔrstA (JH101) strain harboring pEN105 or pEN106 and probed with anti-FLAG and anti-DnaK antibodies. (B) feoB mRNA levels produced by the ΔrstA (JH101) strain harboring pEN105 or pEN106. Shown are the mean values and standard deviations from three independent experiments.
The activation of response regulators usually results in phosphorylation at a conserved aspartic acid residue (38). Thus, we reasoned that RstA phosphorylation might be responsible for the acidic-pH-promoted RstA activity. To test this idea, we constructed plasmid pEN106, expressing a mutant form of RstA-FLAG with a single amino acid substitution in the predicted phosphorylation site (i.e., a D52A substitution). We determined that when the rstA deletion strain carrying pEN106 was grown in the presence of IPTG, the feoB mRNA levels were slightly higher at pH 5.7 than at pH 7.7 but failed to reach the threefold difference exhibited by the isogenic strain with the original RstA-FLAG protein (Fig. 4B). The levels of the mutant RstA-FLAG protein were similar to those of the wild-type RstA-FLAG protein (Fig. 4A, compare lanes 4 and 8), indicating that the differences in feoB mRNA levels were not due to altered RstA-FLAG amounts. Cumulatively, these results suggest that the activity of the RstA protein is promoted under acidic pH conditions, possibly via phosphorylation of the conserved aspartic acid which, in turn, activates transcription of the feoB gene.
The PhoQ protein does not affect the activity of the RstA protein at acidic pH.
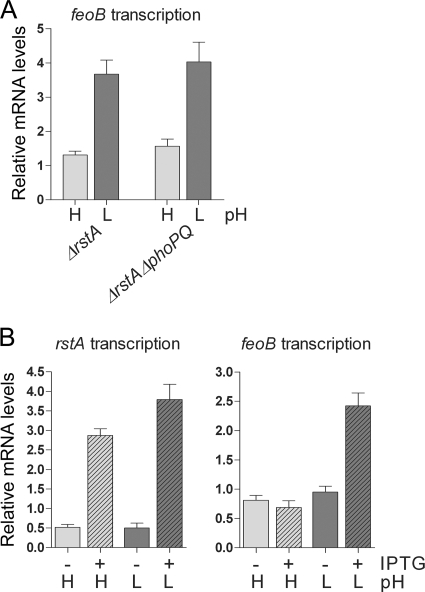
Though it is rare, some response regulators can be phosphorylated by noncognate sensors (24). As RstB was not required for the RstA-promoted transcription of feoB, we wondered whether the PhoQ protein, whose activity is promoted at acidic pH (31), might be responsible for activation of the RstA protein at acidic pH. To test this idea, we compared feoB transcription between isogenic phoPQ+ and phoPQ− strains that had the chromosomal copy of the rstA gene deleted and expressed the rstA gene from the lac promoter. This allowed us to explore the contribution of the PhoP/PhoQ system to RstA activity independently from its role in RstA expression. We determined that, when RstA protein was induced by IPTG, both the ΔrstA and ΔrstA ΔphoPQ strains displayed ∼2.8-fold-higher levels of feoB transcription at pH 5.7 than at pH 7.7 (Fig. 5A), indicating that the PhoP and PhoQ proteins are not required for the acidic-pH-promoted RstA activity.
FIG. 5.
The PhoP/PhoQ system is not required for RstA-promoted feoB transcription at acidic pH. (A) feoB mRNA levels produced by the ΔrstA (JH101) and ΔrstA ΔphoPQ (EN293) strains harboring pJH4, which expresses RstA from the lac promoter. The strains were grown in N-minimal medium containing 2 mM Mg2+ and buffered at pH 7.7 (H, high) or 5.7 (L, low) in the presence of 0.1 mM IPTG. (B) rstA and feoB mRNA levels produced by the ΔphoPQ strain (DS267) harboring pDS303, which expresses the PhoP* protein from the lac promoter. The strain was grown in N-minimal medium containing 2 mM Mg2+ and buffered at pH 7.7 (H, high) or 5.7 (L, low) in the presence (+) or absence (−) of 0.5 mM IPTG. Shown are the mean values and standard deviations from three independent experiments.
PhoP* is a variant of the PhoP protein that can promote gene transcription independently of the PhoQ protein (7). Thus, to further assess the participation of the PhoQ protein in RstA activity, we constructed a strain with the phoPQ operon deleted and carrying the pDS303 plasmid expressing the PhoP* protein from the lac promoter. We determined that the IPTG-induced PhoP* protein increased the expression of the rstA gene at both high and low pH (Fig. 5B). However, activation of feoB transcription was observed only at low pH (Fig. 5B). Note that the rstA expression levels promoted by the PhoP* protein were similar to those achieved by the low-Mg2+- or acidic-pH-activated PhoP protein in the wild-type strain (compare Fig. 2A and 5B). This indicates that the RstA protein was active for feoB transcription in the absence of PhoQ, and it reinforces the notion that the PhoP/PhoQ system only controls the RstA protein levels.
Acidic pH promotes feoB transcription in a strain lacking acetyl phosphate production.
We hypothesized that acetyl phosphate might be necessary for RstA activity at acidic pH because this small molecule serves as a phosphodonor for many response regulators (41). To test this, we determined the feoB transcription levels in a strain with both the ackA and pta genes, encoding the enzymes that are required for acetyl phosphate production, deleted (41). The ΔackA-pta strain still produced threefold-higher levels of feoB mRNA at pH 5.7 than at pH 7.7, though the lack of ackA and pta genes slightly reduced the wild-type levels of feoB transcription at acidic pH (see Fig. S1 in the supplemental material). Thus, this result suggests that the RstA protein can be activated at acidic pH in the absence of acetyl phosphate.
Acidic activation of feoB transcription enhances Salmonella growth under iron-depleted conditions.
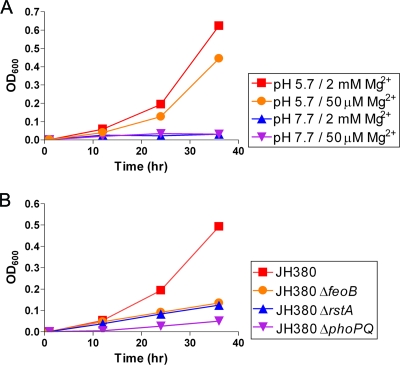
We previously reported that RstA-promoted expression of the feoAB operon increases Fe2+ uptake in Salmonella (20). Thus, we hypothesized that FeoB expression at acidic pH might contribute to Salmonella's ability to grow under Fe2+-depleted conditions. However, wild-type Salmonella grew equally well in minimal medium that contained sodium ascorbate as an iron-reducing agent and the iron chelator diethylenetriaminepentaacetic acid for restriction of Fe2+ and was buffered at pH 7.7 and 5.7 (data not shown). Because Salmonella can also import Fe2+ using other transporters, such as SitABCD and MntH (4, 23), we reasoned that Fe2+ uptake via these routes might mask the effect of FeoB in our experimental conditions. Thus, we reevaluated bacterial growth using strain JH380, which has both the sitABCD and mntH genes deleted. Upon Fe2+ restriction, strain JH380 grew in the medium at pH 5.7 but not at pH 7.7, regardless of the Mg2+ concentration (Fig. 6A). This growth difference is due to FeoB-mediated Fe2+ transport, because deletion of the feoB gene greatly impaired the growth of the JH380 strain in the Fe2+-depleted medium adjusted to pH 5.7 (Fig. 6B). Moreover, consistent with the regulatory roles of the PhoP/PhoQ and RstA proteins in promoting feoB transcription (Fig. 3A), deletion of the rstA or phoPQ genes prevented the growth of JH380 experiencing acidic pH and Fe2+ limitation (Fig. 6B). Cumulatively, these results suggest that activation of the PhoP/PhoQ system and the RstA protein at acidic pH increases FeoB-mediated Fe2+ uptake under iron-restricted conditions.
FIG. 6.
FeoB induction at acidic pH by the PhoP/PhoQ and RstA proteins enhances Salmonella growth upon iron starvation. (A) Growth of Salmonella strain (JH380) lacking both the sitABCD and mntH genes in N-minimal medium at pH 7.7 or 5.7 with 2 mM or 50 μM Mg2+. (B) Growth of the JH380 strain and its isogenic feoB (EN258), rstA (EN256), and phoPQ (EN290) deletion mutants in N-minimal medium at pH 5.7 with 2 mM Mg2+. All media contained 2 mM sodium ascorbate and 10 μM diethylenetriaminepentaacetic acid. The results shown are representative of three experiments.
Iron represses feoB transcription at acidic pH.
When bacteria are grown in the presence of iron, the Fur protein associates with Fe2+ to repress transcription of genes involved in iron acquisition (1). Because the feoAB operon is one of the Fur-regulated targets (20, 21), we explored how transcriptional activation of the feoB gene at acidic pH is affected by iron. On one hand, iron repressed feoB transcription in both the wild-type and the rstA deletion strain, but the effect was much stronger in the wild-type strain, demonstrating the RstA requirement in feoB transcription (see Fig. S2 in the supplemental material). On the other hand, deletion of the fur gene allowed feoB expression at acidic pH in both the presence and the absence of iron (see Fig. S2 in the supplemental material).
The mRNA leader sequence allows expression of the mgtA gene only when the PhoP/PhoQ system is activated at low Mg2+.
Given that feoB expression was promoted exclusively when the PhoP/PhoQ system was activated at acidic pH (Fig. 3A), we wondered whether there might be a PhoP-dependent gene(s) whose expression occurs only at low Mg2+. To explore this possibility, we examined transcription of the Mg2+ transporter mgtA gene, which is directly activated by the PhoP protein (25). It has been previously reported that the β-galactosidase activity produced by a strain harboring a lacZ fusion within the mgtA coding region was 12 times higher in organisms grown in N-minimal medium at pH 7.7 with 10 μM Mg2+ than in those grown at pH 5.8 with 10 mM Mg2+ (35). Why is mgtA not being fully expressed despite the acidic-pH activation of the PhoP protein?
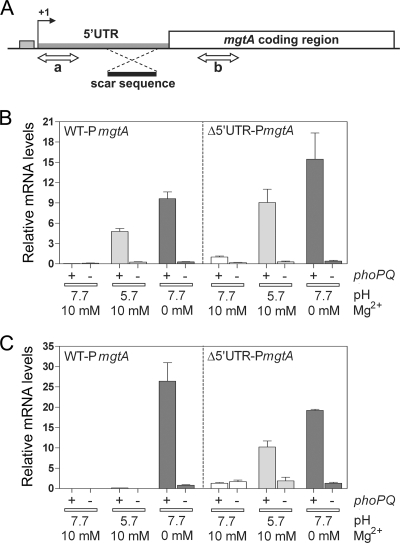
We focused on the recent finding that in response to intracellular Mg2+ levels, the expression of the mgtA gene is controlled by a riboswitch on the leader sequences (9). When Mg2+ concentration attains certain levels in the cytoplasm, Mg2+ binds to the mgtA leader to form a structure that does not favor transcription elongation into the coding region (9). Thus, we hypothesized that mgtA transcription that has been initiated at acidic pH fails to proceed unless cytoplasmic Mg2+ levels are sufficiently low. To test this idea, we examined mgtA transcription by conducting qRT-PCR with two primer sets, one specific to the coding region and the other targeting the first 100 nucleotides of the mgtA leader (Fig. 7A). We found that in the wild-type strain, the mgtA leader mRNA levels were 70- and 140-fold higher at pH 5.7 with 10 mM Mg2+ and at pH 7.7 without Mg2+, respectively, than at pH 7.7 with 10 mM Mg2+ (Fig. 7B). In contrast, the mgtA coding region mRNA level was 1,000-fold higher at pH 7.7 without Mg2+ but only 6-fold higher at pH 5.7 with 10 mM Mg2+ than at pH 7.7 with 10 mM Mg2+ (Fig. 7C). As expected, both low-Mg2+- and acidic-pH-mediated mgtA induction was greatly impaired in the ΔphoPQ mutant (Fig. 7B and C). These results suggest that mgtA transcription initiated in cells experiencing acidic pH fails to reach the coding region.
FIG. 7.
The leader sequence allows the mgtA gene to be expressed only when Salmonella is starved for Mg2+. (A) Schematic representation of the leader and coding regions of the mgtA gene. The double-headed arrows marked as “a” and “b” indicate the regions amplified by qRT-PCR using two pairs of primers. A part of the leader replaced with the “scar” sequence (10) in strain EN323 is also shown. (B) The wild-type Salmonella strain (14028s) and its phoPQ deletion mutant (DS267), mgtA leader mutant (EN323), and EN323 with the phoPQ deletion (EN331) were grown to an OD600 of 0.4 in N-minimal medium at pH 7.7 with 2 mM Mg2+ and transferred to medium at pH 7.7 or 5.7 in the presence or absence of Mg2+. After 30 min, RNA was isolated from the strains, and the levels of mgtA mRNA corresponding to region “a” were determined by qRT-PCR. (C) By conducting qRT-PCR on the same RNA samples used for the experiment described for panel B, the mgtA transcripts corresponding to region “b” were quantified. Shown are the mean values and standard deviations from three independent experiments. UTR, untranslated region; WT, wild type; +, present; −, absent.
To evaluate the role of the mgtA leader region, we constructed a strain where the sequence corresponding to positions 148 to 247 of the mgtA leader was replaced with the 84-bp “scar” sequence (10) (Fig. 7A). This mutation has been previously shown to abolish the Mg2+-sensing ability of the mgtA leader (9). We determined that the mgtA leader mutant strain expressed the mgtA coding region even at pH 5.7 with 10 mM Mg2+ at levels that were 53% of those expressed in organisms grown at pH 7.7 without Mg2+ (Fig. 7C). This was contrary to the behavior of the wild-type strain grown at pH 5.7 with 10 mM Mg2+, where the mgtA coding region mRNA level was only 0.5% of the level produced following growth at pH 7.7 without Mg2+ (Fig. 7C).
The mgtCB operon encodes another Mg2+ transporter, MgtB, whose expression is also determined at the transcription level by the PhoP protein and at the posttranscription level by the leader preceding the first gene in the operon (9, 37). To examine whether the differential pH and Mg2+ regulation displayed by the mgtA gene also applies to mgtB, we determined the mRNA levels for the mgtB coding region in the wild-type and ΔphoPQ strains experiencing different growth conditions. The PhoP-dependent expression of the mgtB gene took place at low Mg2+ but not at acidic pH with high Mg2+ (see Fig. S3 in the supplemental material), indicating that mgtB transcription can reach the coding region only when intracellular Mg2+ levels are sufficiently low. Our results suggest that the leader regions allow the MgtA and MgtB proteins to be produced only when Salmonella faces limiting Mg2+ levels, dramatically decreasing the expression of these Mg2+ transporters when the PhoP/PhoQ system is activated by the acidic-pH signal at high Mg2+ concentrations.
DISCUSSION
Certain two-component regulatory systems are activated by multiple signals. For example, the PhoP/PhoQ system is activated when Salmonella is grown in low Mg2+ (14), in an acidic pH (31), or with certain antimicrobial peptides (2). In the case of the PmrA/PmrB system, both ferric iron (42) and acidic pH (28) promote PmrB activity. These facts raise the question of whether a single regulatory system controls the expression of distinct sets of genes when activated by different signals. We have now demonstrated that the acidic-pH activation of the PhoP/PhoQ system promotes transcription of the Fe2+ transporter-encoding gene feoB, whereas its activation by low Mg2+ promotes the expression of the Mg2+ transporter-encoding mgtA gene (Fig. 1).
Regulation of feoB expression by the PhoP/PhoQ system and the RstA protein.
We demonstrated that the PhoP/PhoQ system promotes the expression of the FeoB Fe2+ transporter when activated by acidic pH but fails to do so when activated at neutral pH by the low-Mg2+ signal (Fig. 3A). This control requires a second response regulator, RstA, which functions as the direct activator of feoB transcription (20). PhoP binds to the rstA promoter region (I. Zwir et al., unpublished) and activates rstA transcription (29, 30). Even though the low-Mg2+ and acidic-pH signals can promote the expression of the RstA protein equally well (Fig. 2), transcription of the RstA-dependent feoB gene took place only when bacteria experienced an acidic pH (Fig. 3A).
We propose that acidic pH controls RstA activity via phosphorylation, because the RstA protein expressed from a heterologous promoter activated feoB transcription at pH 5.7 but not at pH 7.7 and because the activation required RstA's predicted phosphorylation site (i.e., D52) (Fig. 4). Although phosphorylation of a response regulator is generally mediated by its cognate sensor kinase, intriguingly, the acidic-pH activation of feoB transcription still took place in a mutant with a deletion of the rstB gene, which codes for the cognate sensor for RstA (Fig. 3B). That the RstA protein promoted feoB transcription at acidic pH in the absence of PhoQ (Fig. 5) argues against the possibility of cross-phosphorylation of RstA by the noncognate sensor PhoQ. In addition, the finding that acidic pH still activates feoB transcription in a strain that does not synthesize acetyl phosphate rules out the possibility of RstA phosphorylation by this small-molecule phosphodonor (see Fig. S1 in the supplemental material). Therefore, it is likely that a sensor kinase other than RstB and PhoQ might phosphorylate the RstA protein, because some response regulators are phosphorylated by noncognate sensors (24).
PhoP/PhoQ-mediated iron homeostasis.
Under aerobic conditions at neutral pH, iron is present in an oxidized ferric form. Consistent with the notion that iron is reduced to a ferrous form under anaerobic conditions, transcription of the feoAB operon increases when E. coli is grown without oxygen (21). What, then, is the biological significance of the pH-regulated feoB expression? It has been found that Salmonella possesses extracellular enzyme activities to reduce iron (40) and that an acidic pH keeps ferrous iron stable in the presence of oxygen (6). Thus, FeoB induction resulting from activation of the PhoP/PhoQ system and RstA protein could enhance Fe2+ uptake in Salmonella growing in acidic environments with limited iron.
In bacterial cells grown with oxygen, cytoplasmic Fe2+ participates in the Fenton reaction and catalyzes the formation of hydroxyl radicals, which causes DNA damage (39). The Fur protein is a primary regulator that senses intracellular Fe2+ levels (1). When associated with Fe2+, the Fur protein represses the expression of genes for iron acquisition, which minimizes the accumulation of free Fe2+ in the cytoplasm (1, 12). Not surprisingly, the Fur protein repressed feoB transcription even at acidic pH when iron was plentiful (see Fig. S2 in the supplemental material).
The PhoP/PhoQ system is also necessary for the survival of Salmonella under Fe2+-dependent oxidative stress (8). Although the CorA protein is a Mg2+ transporter, it has been reported to import Fe2+ as well (17). When aerobically grown at low Mg2+, a Salmonella phoP mutant displayed increased Fe2+ accumulation in a process dependent on the CorA protein (8). The finding that neither the expression level of the CorA protein nor its membrane location is affected by the PhoP protein implies the presence of a PhoP-regulated gene product(s) regulating CorA activity (8). Indeed, the phoP mutant was hypersensitive to Fe2+-dependent oxidative-stress-mediated killing, which was rescued by inactivation of the corA gene (8). Taking these findings together, the PhoP/PhoQ system is likely to play dual roles in iron homeostasis: PhoP enhances Fe2+ uptake when iron is scarce at acidic pH, whereas downregulation of CorA activity at low Mg2+ protects Salmonella from Fe2+-mediated killing.
PhoP/PhoQ-controlled Mg2+ uptake.
Salmonella imports Mg2+ via three transporters: CorA, MgtA, and MgtB (18, 19, 33). The corA gene is constitutively expressed (8), whereas transcription of the mgtA and mgtB genes is directly activated by the PhoP protein (25, 43). As opposed to the PhoP-controlled Fe2+ uptake at acidic pH, we determined that MgtA expression takes place only at low Mg2+ (Fig. 7). This is because the mgtA leader functions as an RNA sensor (9) such that when the cytoplasmic Mg2+ concentration reaches a certain high level, Mg2+ binds to the mgtA leader to form a structure that prevents RNA polymerase from proceeding to the mgtA coding region (9). The full-length transcript, including the mgtA coding region, is produced when intracellular Mg2+ is low enough to promote the formation of a different structure in the leader region (9).
Supplementary Material
Acknowledgments
This work was supported by a Korea Research Foundation grant funded by the Korean Government (MOEHRD) (KRF-2007-313-C00659). E. Choi was the recipient of a graduate fellowship provided by the Ministry of Education through the Brain Korea 21 Project. E.A.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 2 October 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. [DOI] [PubMed] [Google Scholar]
- 3.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyer, E., I. Bergevin, D. Malo, P. Gros, and M. F. Cellier. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, T. L., C. S. Ederer, A. Allali-Hassani, and E. D. Brown. 2007. Isolation of the rstA gene as a multicopy suppressor of YjeE, an essential ATPase of unknown function in Escherichia coli. J. Bacteriol. 189:3318-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, J., M. R. Woodhall, J. Alvarez, M. L. Cartron, and S. C. Andrews. 2007. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65:857-875. [DOI] [PubMed] [Google Scholar]
- 7.Chamnongpol, S., and E. A. Groisman. 2000. Acetyl phosphate-dependent activation of a mutant PhoP response regulator that functions independently of its cognate sensor kinase. J. Mol. Biol. 300:291-305. [DOI] [PubMed] [Google Scholar]
- 8.Chamnongpol, S., and E. A. Groisman. 2002. Mg2+ homeostasis and avoidance of metal toxicity. Mol. Microbiol. 44:561-571. [DOI] [PubMed] [Google Scholar]
- 9.Cromie, M. J., Y. Shi, T. Latifi, and E. A. Groisman. 2006. An RNA sensor for intracellular Mg2+. Cell 125:71-84. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, R. W., D. Bolstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia Vescovi, E., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 15.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handford, J. I., B. Ize, G. Buchanan, G. P. Butland, J. Greenblatt, A. Emili, and T. Palmer. 2009. Conserved network of proteins essential for bacterial viability. J. Bacteriol. 191:4732-4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hantke, K. 1997. Ferrous iron uptake by a magnesium transport system is toxic for Escherichia coli and Salmonella typhimurium. J. Bacteriol. 179:6201-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hmiel, S. P., M. D. Snavely, J. B. Florer, M. E. Maguire, and C. G. Miller. 1989. Magnesium transport in Salmonella typhimurium: genetic characterization and cloning of three magnesium transport loci. J. Bacteriol. 171:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hmiel, S. P., M. D. Snavely, C. G. Miller, and M. E. Maguire. 1986. Magnesium transport in Salmonella typhimurium: characterization of magnesium influx and cloning of a transport gene. J. Bacteriol. 168:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeon, J., H. Kim, J. Yun, S. Ryu, E. A. Groisman, and D. Shin. 2008. RstA-promoted expression of the ferrous iron transporter FeoB under iron-replete conditions enhances Fur activity in Salmonella enterica. J. Bacteriol. 190:7326-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, A., and E. A. Groisman. 2008. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv. Exp. Med. Biol. 631:7-21. [DOI] [PubMed] [Google Scholar]
- 23.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laub, M. T., and M. Goulian. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121-145. [DOI] [PubMed] [Google Scholar]
- 25.Lejona, S., A. Aguirre, M. L. Cabeza, E. Garcia Vescovi, and F. C. Soncini. 2003. Molecular characterization of the Mg2+-responsive PhoP-PhoQ regulon in Salmonella enterica. J. Bacteriol. 185:6287-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minagawa, S., H. Ogasawara, A. Kato, K. Yamamoto, Y. Eguchi, T. Oshima, H. Mori, A. Ishihama, and R. Utsumi. 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 185:3696-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogasawara, H., A. Hasegawa, E. Kanda, T. Miki, K. Yamamoto, and A. Ishihama. 2007. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J. Bacteriol. 189:4791-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez, J. C., and E. A. Groisman. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 63:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez, J. C., T. Latifi, and E. A. Groisman. 2008. Overcoming H-NS-mediated transcriptional silencing of horizontally acquired genes by the PhoP and SlyA proteins in Salmonella enterica. J. Biol. Chem. 283:10773-10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez, J. C., D. Shin, I. Zwir, T. Latifi, T. J. Hadley, and E. A. Groisman. 2009. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 5:e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prost, L. R., M. E. Daley, V. Le Sage, M. W. Bader, H. Le Moual, R. E. Klevit, and S. I. Miller. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26:165-174. [DOI] [PubMed] [Google Scholar]
- 32.Shin, D., and E. A. Groisman. 2005. Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J. Biol. Chem. 280:4089-4094. [DOI] [PubMed] [Google Scholar]
- 33.Snavely, M. D., S. A. Gravina, T. T. Cheung, C. G. Miller, and M. E. Maguire. 1991. Magnesium transport in Salmonella typhimurium. Regulation of mgtA and mgtB expression. J. Biol. Chem. 266:824-829. [PubMed] [Google Scholar]
- 34.Soncini, F. C., E. Garcia Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soncini, F. C., E. G. Vescovi, and E. A. Groisman. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364-4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spinelli, S. V., L. B. Pontel, E. Garcia Vescovi, and F. C. Soncini. 2008. Regulation of magnesium homeostasis in Salmonella: Mg2+ targets the mgtA transcript for degradation by RNase E. FEMS Microbiol. Lett. 280:226-234. [DOI] [PubMed] [Google Scholar]
- 38.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 39.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 40.Vartivarian, S. E., and R. E. Cowart. 1999. Extracellular iron reductases: identification of a new class of enzymes by siderophore-producing microorganisms. Arch. Biochem. Biophys. 364:75-82. [DOI] [PubMed] [Google Scholar]
- 41.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103:113-125. [DOI] [PubMed] [Google Scholar]
- 43.Zwir, I., D. Shin, A. Kato, K. Nishino, T. Latifi, F. Solomon, J. M. Hare, H. Huang, and E. A. Groisman. 2005. Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. USA 102:2862-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.