Abstract
Ethanolamine can be used as a source of carbon and nitrogen by phylogenetically diverse bacteria. Ethanolamine-ammonia lyase, the enzyme that breaks ethanolamine into acetaldehyde and ammonia, is encoded by the gene tandem eutBC. Despite extensive studies of ethanolamine utilization in Salmonella enterica serovar Typhimurium, much remains to be learned about EutBC structure and catalytic mechanism, about the evolutionary origin of ethanolamine utilization, and about regulatory links between the metabolism of ethanolamine itself and the ethanolamine-ammonia lyase cofactor adenosylcobalamin. We used computational analysis of sequences, structures, genome contexts, and phylogenies of ethanolamine-ammonia lyases to address these questions and to evaluate recent data-mining studies that have suggested an association between bacterial food poisoning and the diol utilization pathways. We found that EutBC evolution included recruitment of a TIM barrel and a Rossmann fold domain and their fusion to N-terminal α-helical domains to give EutB and EutC, respectively. This fusion was followed by recruitment and occasional loss of auxiliary ethanolamine utilization genes in Firmicutes and by several horizontal transfers, most notably from the firmicute stem to the Enterobacteriaceae and from Alphaproteobacteria to Actinobacteria. We identified a conserved DNA motif that likely represents the EutR-binding site and is shared by the ethanolamine and cobalamin operons in several enterobacterial species, suggesting a mechanism for coupling the biosyntheses of apoenzyme and cofactor in these species. Finally, we found that the food poisoning phenotype is associated with the structural components of metabolosome more strongly than with ethanolamine utilization genes or with paralogous propanediol utilization genes per se.
Many bacteria can use diols, such as 1,2-propanediol, or their substituted analogs, such as ethanolamine, as sources of carbon and energy and, in the case of ethanolamine, also as the nitrogen source (27). The ethanolamine degradation is enabled by the enzyme ethanolamine-ammonia lyase (EC 4.3.1.7), which cleaves ethanolamine to ethanol and ammonia (8) and is typically encoded by two genes, which are named eutB and eutC in Salmonella enterica serovar Typhimurium. Ethanolamine-ammonia lyase requires adenosylcobalamin (28), which in different species may be imported from the environment, produced de novo, or synthesized from precursors, such as cyanocobalamin or hydroxycobalamin. The excess of these precursors inhibits ethanolamine-ammonia lyase, and the reactivating factor EutA is used in several species to prevent EutBC inhibition. Ethanolamine lyase produces acetaldehyde, which is converted by the oxidoreductase EutE to acetyl-coenzyme A, which enters the carbon pool of the cell. Alternatively, acetaldehyde can be converted to alcohol by another specialized oxidoreductase, EutG. A phosphotransacetylase, EutD, converts acetyl-coenzyme A to acetylphosphate, which is then converted to acetate, with the production of an ATP molecule (27). Two known types of ethanolamine transporters, i.e., the ethanolamine facilitator EutH (TC 9.A.28.1._) and Eat from the amino acid-polyamine-organocation family (TC 2.A.3.5._), are also part of the ethanolamine utilization systems.
In S. Typhimurium, all these genes are part of the eut operon (15), along with the transcriptional regulator (eutR) and the genes that encode the structural components of metabolosome, a bacterial microcompartment thought to play a role in preventing the escape of gaseous aldehyde but not strictly required for ethanolamine cleavage (7). In addition to these proteins, the eut operons of the Enterobacteriaceae and Firmicutes typically encode several other proteins, including EutP, EutQ, and EutJ. The orthologs of EutBC are sporadically distributed in different lineages of bacteria, in particular in Proteobacteria and Firmicutes, and are not found in archaea or eukaryotes (24). The genes carried by the eut operon and their molecular functions are summarized in Table S1 in the supplemental material.
The evolutionary origin of the ethanolamine utilization system is unclear. The structural proteins of the metabolosome complex are paralogous to the shell proteins of carboxysome (15), an organelle that concentrates CO2 for fixation by ribulose-bisphosphate carboxylase in cyanobacteria and sulfur-oxidizing bacteria (21), but the phylogeny of these shell components remains to be investigated in detail. The amino acid sequences of the main enzyme of the ethanolamine utilization pathway, EutBC, retrieve only closely related, orthologous proteins in database searches, and there is no plausible evolutionary scenario explaining the current phylogenetic distribution of ethanolamine lyase.
The regulation of ethanolamine lyase is most extensively studied for S. Typhimurium, where cobalamin and ethanolamine are both required for the full expression of the eut operon, which is transcriptionally activated by the positive regulator EutR, encoded by the operon itself (15). There is no information on the molecular determinants of the activation of the eut operon by EutR, and the understanding of the coordination of apoenzyme and coenzyme biosynthesis is incomplete. A better-studied paralogous pdu operon of propanediol utilization in S. Typhimurium is controlled by the positive regulator PocR, which, like EutR, belongs to the AraC sequence family. PocR also positively controls the cobalamin synthesis pathway by direct transcriptional activation of the cob operon (6, 25).
The ethanolamine utilization pathway is of practical concern, as it is present in many human and animal pathogens linked to food poisoning. A probabilistic search of the database of phyletic vectors by using the food poisoning phenotype identifies five genes of ethanolamine utilization as near-perfect genotype-to-phenotype matches (18). A more complex machine-learning approach, which examines genome context and cooccurrence of scientific terms in the literature, has connected food poisoning with both ethanolamine and 1,2-propanediol utilization pathways (16). Here, again, however, the molecular basis of the observed biological phenomenon is not known.
In this work, we employed the complete genome sequences of several hundred bacterial species and used computational approaches to answer questions about the regulation of eut genes, the evolution of eut operons, the structure of the crucial EutBC proteins, and the connection between diol utilization pathways and food poisoning.
MATERIALS AND METHODS
Complete bacterial genomes were obtained from GenBank in August 2008, and the information about pathogenicity was compiled from the descriptions of each genome project. Homologous proteins were collected by PSI-BLAST (2), and orthologs were identified by reciprocal best-match criteria (34), verified by examining phylogenetic trees and operon structure, and labeled by the gene names taken from S. Typhimurium. Distant sequence similarities were validated and secondary structures were predicted using the HHPred suite of programs (33).
For phylogenetic inference, the maximum likelihood method with the Jones-Taylor-Thornton model implemented in the Proml program of the PHYLIP package (11) or Bayesian estimation of phylogeny implemented in MRBAYES 3.0 (26) with the fixed-rate Poisson model with unconstrained topology was used. Ancestral states were reconstructed using the parsimony model implemented in the Mesquite suite (19). Regulatory regions were aligned using CLUSTAL_X (35) and MACAW (30), the profiles were built from the most-conserved blocks by using the GenomeExplorer program (20), and genome scans were performed, with the threshold set at the lowest score observed in the training set (13). Sequence logos were produced by the Weblogo program (10), and phylogenetic trees were drawn using the iTOL server (17). The enrichment statistics was derived using the standard hypergeometric distribution formula implemented in the R package (14). The P value was calculated based on that distribution function, using the Phyper function in R.
RESULTS
Diversity of the eutBC genomic contexts in bacteria.
The eutBC pair of genes is found in almost 100 fully sequenced bacterial genomes (not counting closely related strains) (see Table S2 in the supplemental material). The set of genes with experimentally shown or computationally predicted roles in ethanolamine degradation, together with genes localized within the known eut operons, consists of 16 genes in addition to eutBC. The eutBC genes are always found next to each other on the chromosome, indicating that they are coregulated and expressed from the same transcript. The genome context of this eutBC pair is, however, variable.
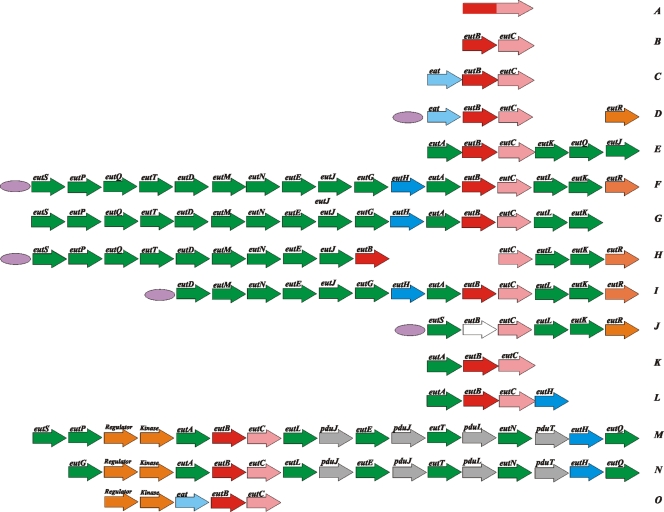
In Actinobacteria and in most Proteobacteria, the eut operon consists only of eutBC and usually the transporter eat. Some Proteobacteria additionally contain an ortholog of the transcription regulator eutR at a different genomic location but no apparent orthologs of other eut genes (Fig. 1). On the other end of the spectrum, there is “the long operon” found in Enterobacteriaceae, Firmicutes, Nocardioides sp., and Fusobacterium nucleatum. This is an arrangement of up to 17 genes, which may also include some of the putative propanediol utilization genes and the duplicates of the metabolosome genes. In Enterobacteriaceae, some genes of such a complete set may be missing: for example, Shigella sonnei Ss046 has no eutS, -P, -Q, and -T genes; Shigella boydii Sb227 lacks eutG, -H, and -A; and Shigella dysenteriae Sd197 has only eutS, -C, -L, -K, and -R and truncated eutB.
FIG. 1.
Diversity of eutBC genome contexts. Short operons: A, Deltaproteobacteria; B, a subset of Proteobacteria, Chlorophlexi, and Bacteroidetes; C, selected Proteobacteria and Acidobacteria; and D, Betaproteobacteria (eutR is in a different genomic location than eutBC and eat). Long operons: E, Nocardioides sp.; F, Enterobacteriaceae; G, M. aquaeolei; H, S. boydii Sb227; I, S. sonnei Ss046; J, S. dysenteriae Sd197; K, Symbiobacterium thermophilum and P. luminescens; L, P. fluorescens Pf-5; M, Clostridiaceae and F. nucleatum; N, Listeriaceae and Enterococcaceae; and O, C. acetobutylicum. A probable eutB pseudogene is shown in white. The predicted EutR-binding sites are indicated by purple ellipses. See Tables S1 and S2 in the supplemental material for complete lists of gene names and species.
Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Marinobacter aquaeolei, and Pseudomonas fluorescens Pf-5 contain two distinct types of the eut operons. In all three species, one of the two operons is of the “short” variety. K. pneumoniae contains eutBC and eat; M. aquaeolei has eutBC only; and in P. fluorescens, the eut operon consists of only eutBC fused into one open reading frame. The sizes of the other eut operons in these species are different: K. pneumonia has 17 genes, M. aquaeolei contains 16 genes, and P. fluorescens has just eutABC. Interestingly, in P. fluorescens the two sets of eutBC genes belong to the same operon, but the phylogenies of these two sets are different (Fig. 1 and 2) (see Discussion).
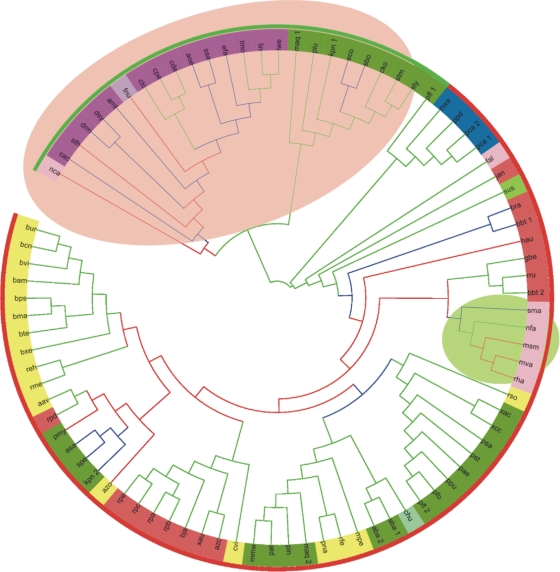
FIG. 2.
Maximum likelihood evolutionary tree of EutB. The bootstrap support of tree partitions is indicated by branch color: green, >70%; blue, 50 to 70%; and red, <50%. The outer color stripe mark genes from the short operon in red and genes from the long operon in green. The inner color circle marks bacterial clades: red, Alphaproteobacteria; orange, Betaproteobacteria; green, Gammaproteobacteria; dark blue, Deltaproteobacteria; dark purple, Firmicutes; pink, Actinobacteria; lime, Acidobacteria; light purple, Fusobacteria; light blue, Chlorophlexi; and light green, Bacteroidetes. The shaded background marks two clades that do not agree with established bacterial phylogeny and suggest horizontal gene transfer events (see text).
Two ethanolamine transporters (Eat and EutH) are typically found to the exclusion of each other, except for those genomes that contain two different types of ethanolamine operons. The eutH transporter tends to cooccur with genes eutT, eutQ, eutA, and eutJ, whereas eat correlates with their absence.
EutB and EutC are paralogous to diol dehydratases/lyases.
The sequences of EutB and EutC are well conserved in evolution, and they are not clearly similar to those of other proteins in typical database searches. Even iterative scans of the protein databases by use of the PSI-BLAST program (2) detect only the orthologs of each protein. The large EutB subunit of ethanolamine-ammonia lyase has been predicted to adopt the eight-stranded TIM barrel-like fold, similar to what was found for other cobalamin-dependent dehydratases with the known structure, such as propanediol dehydratase and glycerol dehydratase (protein data bank [PDB] accession no. 1eex, 1dio, and 1mmf) (32). The interaction with the cofactor must occur predominantly in the “bottom” portion of the barrel, corresponding to the C termini of the predicted beta strands forming the inner barrel surface (33). Recently, the three-dimensional structure of a hexamer of the Listeria monocytogenes EutB protein was determined (PDB accession no. 2qez), confirming these earlier predictions. In addition to the alpha/beta TIM barrel domain, however, a smaller alpha-helical N-terminal domain was found, consisting of residues 1 to 140, wrapped around the external surface of the TIM barrel and making contacts with the adjoining monomers. The homologous large subunits of other cobalamin-dependent enzymes also have the additional N-terminal sequence regions, which are missing from the available crystal structures.
Using sensitive comparison of probabilistic models of protein families with the HHsearch program (31), we found that the N-terminal alpha-helical domains of cobalamin-dependent lyase large subunits are homologous: for example, the first 140 aligned positions of the dehydratase large subunits specified by the Hidden Markov Model automatically built by the HHsearch program from the sequence of propanediol hydratase match the N-terminal region of the EutB family model with a probability (P) value of 1.7·10−4. Interestingly, the residues most conserved between different lyases are not the same as the ones involved in the interactions of EutB within the hexamer (see Fig. S1 in the supplemental material), suggesting either that these interactions in the crystallized form are not representative of the EutBC complex in vivo or that the N-terminal domains of the lyase large subunits play roles in addition to homooligomerization.
The three-dimensional organization of the small ethanolamine-ammonia lyase subunit (EutC) remains unknown. Prediction of the secondary and tertiary structures of EutC suggests an alpha/beta structure in this protein and a borderline structural similarity to NADP-dependent methylenetetrahydrofolate dehydrogenase from M. tuberculosis (PDB accession no. 2c2x), covering more than 60% of residues in both molecules (see Fig. S1 in the supplemental material). This is compatible with the Rossmann-like alpha/beta fold in EutC, which is also the fold adopted by the beta subunit of propanediol dehydratase (37). Interestingly, in an analogy with the large subunit, EutC is also predicted to have a small N-terminal alpha-helical domain, and this domain, or at least its longest helix, is clearly conserved in the propanediol dehydratase beta subunit. Moreover, specific sequence similarity to this region can also be detected in the N termini of three other proteins involved in the same pathways but having completely different structures, namely, in the all-helical gamma subunit of propanediol dehydratase (which actually gives better alignment to EutC than the apparently homologous beta subunit); in the beta-barrel protein EutQ, a member of the cupin superfamily; and in the phosphotransacylase PduL. Some of these similarities span relatively short numbers of residues, e.g., only 36 in the case of EutC-EutQ alignment, but nonetheless are specifically recovered with HHsearch (E value below 10−4) (see Fig. S1 in the supplemental material). Only the 17-residue EutC-PduL match is reported without statistical support.
EutB and EutC phylogeny.
To elucidate the evolutionary history of the core eut pathway, we aligned sequences of EutB and EutC and inferred the phylogenies of these proteins. The results were largely in agreement for both subunits and for all algorithms of phylogenetic inference. In particular, the subdivisions of Proteobacteria (Alphaproteobacteria, Betaproteobacteria, Deltaproteobacteria, and Gammaproteobacteria) form distinct clades that are clustered together in all trees. One exception is Enterobacteriaceae, which appear as a long branch distant from other Gammaproteobacteria. The other is actinobacteria branches, which are nested within Proteobacteria and, more specifically, within Alphaproteobacteria, whereas most of Firmicutes form a sister clade with the Enterobacteriaceae (Fig. 2). Sequences from Acidobacteria and Chlorophlexi are typically found within Proteobacteria, and the Fusobacteria sequences are clustered with Firmicutes. Inclusion of an outgroup (assuming that the subunits of 1,2-propanediol lyase are paralogous to EutBC) (see Discussion) suggests the root position on the long branch leading to Proteobacteria, though a relatively low level of statistical support on the deep branches makes this assignment tentative.
Two major subtrees, one including Firmicutes and the Enterobacteriaceae and the other including the rest of the Proteobacteria with the nested Actinomycetales, correspond to the main two types of eutBC contexts that were discussed in the previous section, i.e., the long and short versions, respectively (Fig. 2). Even in the species with two eut operons (K. pneumoniae, M. aquoeolei, and P. fluorescens), the two copies of EutB (and EutC) cluster in the trees not with each other but with the orthologs from the species that share long- or short-operon context.
Comparative genomics of the eutBC regulation.
The eut operons are controlled by at least two types of regulatory systems: in most Enterobacteriaceae and in some Betaproteobacteria (including Polaromonas naphthalenivorans CJ2, Methylibium petroleiphilum PM1, and all sequenced Burkholderiales), the operon is regulated by EutR, while Firmicutes and F. nucleatum have a two-component regulatory system adjacent to the eut genes (for example, EutV and EutW in Enterococcus faecalis [12]). Interestingly, this two-component system also appears to have a high rate of coinheritance with the cobalamin biosynthesis genes, again pointing in the same functional direction (18). Actinobacteria and a subset of the Proteobacteria lack orthologs of these genes, so these bacterial groups may possess yet other systems of the eut operon regulation.
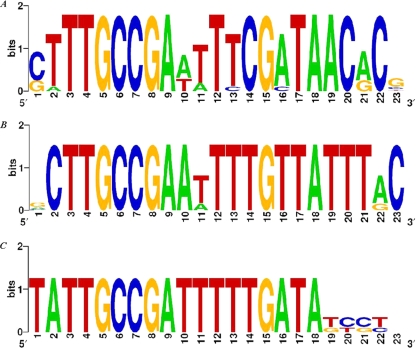
We analyzed sequence conservation in the upstream regions of the eut operons in two groups of EutR-containing genomes that include several diverse species, i.e., Enterobacteriaceae with 6 species and Betaproteobacteria with 17 species. Multiple sequence alignment of the putative regulatory regions in the first group revealed two conserved sequences (see Fig. S2 in the supplemental material). In S. Typhimurium, the global transcription factor Crp is known to control the paralogous pdu operon and may be involved in eut regulation as well (1), and the first region that we discovered, wwwTGTGAtyyrgwTCACTtWt, which is similar to the canonical Crp-binding motif (4), may indeed play a role in recognizing Crp. The other conserved region in the Enterobacteriaceae did not match any known regulatory sites. However, it was closely similar to the separately defined nucleotide motif in the regions located upstream of the eut operons in Betaproteobacteria, which was the only conserved element in the latter group of species (Fig. 3). We constructed positional weight matrices of this motif and scanned the intergenic regions of the various bacterial species with this model. In Betaproteobacteria, there were no significant similarities other than self-matches in the eut regulatory region, and we did not find this or any other conserved DNA motifs upstream of the eut operons in Firmicutes or any other bacterial groups. Interestingly, in Enterobacteriaceae, the next-best match after the self-matches was the intergenic region preceding cbiA, the 5′-proximal gene in the cob operon required for de novo cobalamin biosynthesis (see Table S3 in the supplemental material).
FIG. 3.
Conserved elements in proteobacteria that may bind EutR. (A) Conserved element preceding the eut operon in Betaproteobacteria (for site scores and locations, see Table S3 in the supplemental material); (B) conserved binding element preceding the eut operon in Enterobacteriaceae; (C) conserved element upstream of the cbiA gene in Enterobacteriaceae.
What causes food poisoning?
Ethanolamine or propanediol degradation pathways have been implicated in human food poisoning by the large-scale mining of genomic data (16). Propanediol utilization genes are paralogous to the eut genes and distributed relatively narrowly among the completely sequenced genomes. There is a considerable overlap between the lists of species that contain the pdu operon and those that contain the eut operon. The species that have both systems include a subset of enterobacteria (Salmonella spp., Escherichia coli E24377A, S. sonnei, Citrobacter koseri, and K. pneumoniae), clostridia (Clostridium perfringens ATCC 13124), and Listeria spp. We asked whether the pathogenicity phenotype can be specifically associated with a particular gene in either the ethanolamine utilization or the propanediol utilization pathway. We used enrichment statistics, given by the hypergeometric distribution (23), to assess the strength of the link between each eut and pdu gene and the pathogenic phenotype (Table 1). The calculated P value indicates how much the set of pathogenic bacteria is enriched with an analyzed gene compared to nonpathogenic species. There was no strong association between the enzymes, regulators, or transporters of either pathway and food poisoning, except for two enzymes, EutT and EutP, which are involved in cofactor biosynthesis and its reactivation, respectively. In contrast, the auxiliary components of the long ethanolamine utilization operon, notably metabolosome shell components, showed strong mutual enrichment with the food poisoning phenotype.
TABLE 1.
Strength of links between eut and pdu genes and food poisoning
| Gene(s)a | Pb | Protein function |
|---|---|---|
| eutS-pduU | 1,17 × 10−10 (14/11) | Metabolosome structural protein |
| eutL-pduB | 3.8 × 10−9 (14/16) | Metabolosome structural protein |
| eutP-pduV | 4.98 × 10−9 (13/11) | Putative GTPase |
| pduM | 1.1 × 10−8 (9/1) | Possible small-molecule kinase |
| eutMK-pduAJT | 9.99 × 10−8 (14/22) | Metabolosome structural protein |
| eutN | 2.5 × 10−6 (13/22) | Metabolosome structural protein |
| pduH | 4.012 × 10−6 (10/9) | Beta subunit of the reactivation enzyme of propanediol dehydratase; structural mimic of PduD that does not bind cobalamin |
| eutT | 3.48 × 10−5 (9/9) | Cobalamin adenosyltransferase |
| pduL | 0.00012 (11/20) | Propanediol utilization phosphotransacetylase |
| eutH | 0.00024 (8/9) | Transport protein |
| pduG | 0.00025 (10/17) | Propanediol utilization protein |
| eutQ | 0.00066 (9/15) | Ethanolamine utilization protein |
| pduS | 0.00066 (9/15) | Propanediol utilization protein |
| pduCDE | 0.0007 (10/20) | Propanediol utilization dehydratase |
| eutA | 0.001 (8/12) | Reactivating factor |
| eutR | 0.0069 (7/13) | Regulator |
| eutJ | 0.0096 (7/14) | Putative chaperone |
| eutBC | 0.07 (14/69) | Ethanolamine ammonia-lyase |
| pduO | 0.777 (10/65) | Corrinoid adenosyltransferase |
| eat | 0.43 (6/31) | Ethanolamine permease |
The metabolosome genes from the ethanolamine and the propanediol degradation pathways are closely similar and difficult to distinguish, particularly when they occur in mixed operons (for example, in L. monocytogenes).
The first number in parentheses is the number of food pathogens (out of a total of 14) where the gene was found, and the second number in parentheses is the number of nonpathogenic bacterial species (out of a total of 85) where the gene was found.
DISCUSSION
The relatively high degree of sequence conservation in the two distinct small alpha-helical regions at the N termini of large and small diol lyase subunits indicates that these regions may have a conserved, perhaps sensory or regulatory, role in the metabolism of propanediol and ethanolamine. The provenance of these extensions is unclear, but the evolutionary origin of the main catalytic domains in EutB and EutC is quite transparent: they must have been produced by recruitment of two of the most abundant “superfolds” (9), a TIM barrel and a Rossmann fold, respectively. To identify the evolutionary lineage in which this recruitment may have occurred, we analyzed the phylogeny and genomic context of the EutBC genes. The EutB and EutC phylogeny suggests that the evolutionary history of EutBC subunits included several unusual events. In particular, the most direct way of explaining the position of Actinobacteria in our trees appears to be a horizontal transfer of the eutBC gene pair from an ancient proteobacterium, perhaps an alphaproteobacterium, to Actinobacteria, followed by vertical inheritance and occasional loss of these genes in actinomycetes.
The EutB and EutC proteins of the Enterobacteriaceae cluster with the orthologs from Firmicutes and not with those from other Proteobacteria. Both Enterobacteriaceae and Firmicutes are the tips of long branches in our trees, and it is possible that their adjoining positions are due to the long branch attraction artifacts (5). We feel, however, that two other factors may contribute to this tree topology: first, the evolution of the EutBC enzymes in the long operons in Enterobacteriaceae and Firmicutes may be constrained in similar ways by the interaction with the metabolosome, and second, there may have been another act of horizontal gene transfer in the early evolution of these operons.
In order to understand the ancient evolutionary events better, we attempted to reconstruct the ancestral states of the eut operon using a simple parsimony model of gene gain and loss implemented in the Mesquite software package (19). Under this model, the ancestral proteobacterium is inferred to have contained eutBC and eat genes. Other components of the pathway appear in the branch leading to the Enterobacteriaceae, as does the transcriptional regulator eutR (also present in some Betaproteobacteria, to which it might have been transferred from the Enterobacteriaceae). The ancestral operon in Firmicutes likely included eutABC and the two-component regulatory system, but the ancestral state of other eut genes in this lineage cannot be determined unambiguously given the current data.
A conservative estimate of about five genes in an ancestral firmicute, together with an even smaller set of genes in the ancestral proteobacterium, may suggest the following tentative evolutionary scenario. The earliest version of the eut operon may have emerged by cooptation of a TIM barrel and a Rossmann fold, by adornment of them with additional N-terminal helical domains, and by recruitment of a permease gene for transportation of ethanolamine from the environment (perhaps of the eat type, which seems to be spread more widely in the extant species and more closely associated with the short operons than eutH). The small set of genes was supplemented by a recycling factor and a two-component regulatory system in Firmicutes, which may have replaced the Eat transporter with EutH (though the eat gene is retained in Clostridium acetobutylicum). More-recent evolution of the operon included acquisition of genes encoding the structural components of the metabolosome and accrual of other eut genes. Gains and losses of auxiliary ethanolamine degradation genes resulted in the diversity of gene contexts of eutBC.
Enterobacteriaceae may have acquired a partially formed eut operon with metabolosome shell genes from Firmicutes. Such direction of horizontal transfer is more plausible than the opposite one, given that the Enterobacteriaceae is a younger evolutionary lineage than Firmicutes and also that some deep-branching Proteobacteria, such as Photorhabdus luminescens, do not include eut genes.
The scenario outlined above assumes three horizontal gene/operon transfer events, i.e., a transfer of a long eut operon from an ancestral firmicute to the Enterobacteriaceae, a transfer of a short operon from an alphaproteobacterium to the Actinobacteria, and acquisition of the metabolosome shell genes by an ancestral firmicute, probably from a cyanobacterium that had a carboxysome. Evolutionary histories with fewer horizontal transfers can also be proposed, yet those typically include massive operon losses or unlikely evolutionary events, such as parallel accrual of similar sets of orthologous genes in long operons. On balance, we feel that our hypothesis of gradual buildup of the eut operon within Firmicutes and its transfer to the Enterobacteriaceae with another transfer from the Alphaproteobacteria to the Actinobacteria is best compatible with the available biochemical and genomic evidence.
The EutR regulator in S. Typhimurium responds to two effectors, cobalamin and ethanolamine. In the absence of one or both effectors, there is a weak basal constitutive expression of eutR from the PII promoter. Elevated concentration of the effectors induces eut operon activation by EutR through the PI promoter (see Fig. S3 in the supplemental material). EutR is hypothesized to sense cobalamin and ethanolamine directly (29), but the molecular basis for this recognition is not known. We found a conserved sequence in the upstream regions of the EutR gene-containing operons in Enterobacteriaceae and Betaproteobacteria, which is also present upstream of the cobalamin biosynthesis operon in Enterobacteriaceae. It is plausible that Enterobacteriaceae uses this control element to coordinate production of the EutBC apoenzyme and simultaneous synthesis or import of its cobalamin cofactor. Such coregulation may be achieved if EutR indeed serves as a sensor of both compounds and if its activated form upregulates the expression of both eut and cbi operons. On the other hand, cobalamin is required for the activity of several enzymes in addition to EutBC, and therefore, negative regulation of eut operon by depletion of EutR has to be decoupled from cbi operon regulation. This may be achieved via positive regulation of vitamin B12 production by multiple inputs (i.e., at least PocR in addition to EutR) and negative regulation by the B12-responsive riboswitch (22, 36).
Analysis of links between distribution of individual genes and the food poisoning phenotype suggests that the pathogenic phenotype may be related to the presence of some reaction intermediate, such as perhaps highly active cobalamin-derived radical species produced in the course of catalysis (3), or even some spurious compound, when it is driven to local high concentrations in the metabolosome. On the other hand, the core component of the ethanolamine utilization reaction, the EutBC enzyme, as well as ethanolamine transporters, appears to be relatively benign.
Supplementary Material
Acknowledgments
We are grateful to M. S. Gelfand and A. E. Kazakov for valuable discussions and to H. Li and D. Zhu for help with calculations.
This study was supported by the Stowers Institute and by grants from the Howard Hughes Medical Institute to M. S. Gelfand (55005610), the Russian Academy of Science (Molecular and Cellular Biology program), and the Russian Foundation for Basic Research (08-04-01000-a).
O.T. and A.M. conceived the study, and all authors analyzed the data, wrote the manuscript, and approved its final form.
Footnotes
Published ahead of print on 25 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Ailion, M., T. A Bobik, and J. R. Roth. 1993. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J. Bacteriol. 175:7200-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, G., R. R. Poyner, and G. H. Reed. 2008. Identification of the substrate radical intermediate derived from ethanolamine during catalysis by ethanolamine ammonia-lyase. Biochemistry 47:11360-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, O. G., and P. H. von Hippel. 1988. Selection of DNA binding sites by regulatory proteins. II. The bindingspecificity of cyclic AMP receptor protein to recognition sites. J. Mol. Biol. 200:709-723. [DOI] [PubMed] [Google Scholar]
- 5.Bergsten, J. 2005. A review of long-branch attraction. Cladistics 21:163-193. [DOI] [PubMed] [Google Scholar]
- 6.Bobik, T. A., M. Ailion, and J. R. Roth. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 174:2253-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinsmade, S. R., T. Paldon, and J. C. Escalante-Semerena. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187:8039-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, G. W., and J. T. Chang. 1975. Evidence for the B12-dependent enzyme ethanolamine deaminase in Salmonella. Nature 254:150-151. [DOI] [PubMed] [Google Scholar]
- 9.Coulson, A. F., and J. Moult. 2002. A unifold, mesofold, and superfold model of protein fold use. Proteins 46:61-71. [DOI] [PubMed] [Google Scholar]
- 10.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Fox, K. A., A. Ramesh, J. E. Stearns, A. Bourgogne, A. Reyes-Jara, W. C. Winkler, and D. A. Garsin. 2009. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 106:4435-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelfand, M. S., P. S. Novichkov, E. S. Novichkova, and A. A. Mironov. 2000. Comparative analysis of regulatory patterns in bacterial genomes. Brief. Bioinform. 1:357-371. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, N. L., S. Kotz, and A. W. Kemp. 1992. Univariate discrete distributions, 2nd ed., p. 266-269. Wiley, New York, NY.
- 15.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korbel, J. O., T. Doerks, L. J. Jensen, C. Perez-Iratxeta, S. Kaczanowski, S. D. Hooper, M. A. Andrade, and P. Bork. 2005. Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 3:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Letunic, I., and P. Bork. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127-128. [DOI] [PubMed] [Google Scholar]
- 18.Li, H., D. M. Kristensen, M. K. Coleman, and A. Mushegian. 2009. Detection of biochemical pathways by probabilistic matching of phyletic vectors. PLoS ONE 4:e5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddison, W. P., and D. R. Maddison. 1 June 2009, accession date. Mesquite: a modular system for evolutionary analysis, version 2.6. http://mesquiteproject.org/mesquite2.6/Mesquite_Folder/docs/mesquite/manual.html.
- 20.Mironov, A. A., N. P. Vinokurova, and M. S. Gelfand. 2000. GenomeExplorer: software for analysis of complete bacterial genomes. Mol. Biol. 34:222-231. [Google Scholar]
- 21.Orus, M. I., M. L. Rodriguez, F. Martinez, and E. Marco. 1995. Biogenesis and ultrastructure of carboxysomes from wild type and mutants of Synechococcus sp. strain PCC 7942. Plant Physiol. 107:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richter-Dahlfors, A. A., S. Ravnum, and D. I. Andersson. 1994. Vitamin B12 repression of the cob operon in Salmonella typhimurium: translational control of the cbiA gene. Mol. Microbiol. 13:541-553. [DOI] [PubMed] [Google Scholar]
- 23.Rivals, I., L. Personnaz, L. Taing, and M. C. Potier. 2007. Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23:401-407. [DOI] [PubMed] [Google Scholar]
- 24.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278:41148-41159. [DOI] [PubMed] [Google Scholar]
- 25.Rondon, M. R., and J. C. Escalante-Semerena. 1996. In vitro analysis of the interactions between the PocR regulatory protein and the promoter region of the cobalamin biosynthetic (cob) operon of Salmonella typhimurium LT2. J. Bacteriol. 178:2196-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronquist, F., and J. P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 27.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roof, D. M., and J. R. Roth. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171:3316-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roof, D. M., and J. R. Roth. 1992. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuler, G. D., S. F. Altschul, and D. J. Lipman. 1991. A workbench for multiple alignment construction and analysis. Proteins 9:180-190. [DOI] [PubMed] [Google Scholar]
- 31.Söding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951-960. [DOI] [PubMed] [Google Scholar]
- 32.Sun, L., and K. Warncke. 2006. Comparative model of EutB from coenzyme B12-dependent ethanolamine ammonia-lyase reveals a beta8alpha8, TIM-barrel fold and radical catalytic site structural features. Proteins 64:308-319. [DOI] [PubMed] [Google Scholar]
- 33.Sun, L., O. A. Groover, J. M. Canfield, and K. Warncke. 2008. Critical role of arginine 160 of the EutB protein subunit for active site structure and radical catalysis in coenzyme B12-dependent ethanolamine ammonia-lyase. Biochemistry 47:5523-5535. [DOI] [PubMed] [Google Scholar]
- 34.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2003. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9:1084-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamanishi, M., M. Yunoki, T. Tobimatsu, H. Sato, J. Matsui, A. Dokiya, Y. Iuchi, K. Oe, K. Suto, N. Shibata, Y. Morimoto, N. Yasuoka, and T. Toraya. 2002. The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol. Eur. J. Biochem. 269:4484-4494. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.