Abstract
Bacterial species can communicate by producing and sensing small autoinducer molecules by a process known as quorum sensing. Salmonella enterica produces autoinducer 2 (AI-2) via the luxS synthase gene, which is used by some bacterial pathogens to coordinate virulence gene expression with population density. We investigated whether the luxS gene might affect the ability of Salmonella enterica serovar Typhimurium to invade epithelial cells. No differences were found between the wild-type strain of S. Typhimurium, SL1344, and its isogenic luxS mutant with respect to the number and morphology of the membrane ruffles induced or their ability to invade epithelial cells. The dynamics of the ruffling process were also similar in the wild-type strain (SL1344) and the luxS mutant. Furthermore, comparing the Salmonella pathogenicity island 1 (SPI-1) type 3 secretion profiles of wild-type SL1344 and the luxS mutant by Western blotting and measuring the expression of a single-copy green fluorescent protein fusion to the prgH (an essential SPI-1 gene) promoter indicated that SPI-1 expression and activity are similar in the wild-type SL1344 and luxS mutant. Genetic deletion of luxS did not alter the virulence of S. Typhimurium in the mouse model, and therefore, it appears that luxS does not play a significant role in regulating invasion of Salmonella in vitro or in vivo.
Quorum sensing is an intercellular signaling mechanism used by bacteria to regulate gene expression in response to changes in cell population density (30). Quorum sensing relies on the production and secretion of a small signaling molecule during growth, so that as the size of a bacterial population grows, there is a corresponding increase in the extracellular concentration of the autoinducer. Once the population has reached a certain cell density, a critical threshold concentration of the autoinducer will be exceeded and a response will be triggered, typically leading to changes in gene transcription and ultimately, modulation of population behavior. This response is usually one that is productive only when it is carried out simultaneously by many cells and includes responses such as bioluminescence, antibiotic production, production of virulence factors, and biofilm formation (8, 11, 19, 29). By coordinating the gene expression of a bacterial community, quorum sensing enables bacteria to behave like multicellular organisms (24).
A diverse range of bacterial quorum-sensing signaling molecules have been identified. Many gram-negative species are known to produce N-acyl homoserine lactones. In many cases, these are generated by LuxI homologues and sensed by transcriptional regulators of the LuxR family (3). In contrast, most gram-positive bacteria use either linear or cyclic peptides which are generated from short precursor proteins (16). The only system currently known to exist in both gram-negative and gram-positive bacteria relies upon the production of a signal molecule termed autoinducer 2 (AI-2) by an AI synthase, encoded by the luxS gene (26). Since the luxS gene has been found in over 55 species of bacteria (32), it has been suggested that AI-2 may be used for interspecies signaling, rather than the intraspecies communication associated with other quorum-sensing signaling molecules. Hence, AI-2 may enable differential gene expression depending on whether bacteria exist in pure culture or in a consortium (2).
The luxS gene has been found in Salmonella enterica serovar Typhimurium (26), with the AI-2 signal being identified as 2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (20). This is a derivative of 4,5-dihydroxy-2,3-pentanedione (DPD), a by-product of the activated methyl cycle in which LuxS plays a role (31). An initial screen for genes regulated by luxS in Salmonella enterica serovar Typhimurium identified those of the luxS-regulated (lsr) operon (28). The lsr operon encodes a transporter for AI-2, homologous to the ribose ABC transporter, and enzymes that modify this signal upon its entrance into cells (27, 28, 33). The Lsr receptor/transporter complex has subsequently been found to predominate in pathogenic bacteria associated with endotherms (23), indicating that quorum sensing may play an important role in the interaction of such bacteria with their hosts.
The virulence of Salmonella enterica depends upon their abilities to enter and survive in host cells, and genes involved with these processes are tightly controlled, often in relation to environmental cues (10). Quorum sensing could potentially act as one such cue. The purpose of this study was to examine whether Salmonella enterica serovar Typhimurium may use quorum sensing to regulate virulence, specifically examining the effect of deletion of luxS on the ability of S. Typhimurium to induce membrane ruffles and invade epithelial cells. We found no evidence for a major role for LuxS in Salmonella invasion or in regulation of Salmonella pathogenicity island 1 (SPI-1) expression or secretion. Significantly, luxS had no significant impact on the virulence of the strain in vivo. These data contrast with a recent report that the luxS gene is required for expression of a subset of genes in SPI-1 and to optimize invasion (4).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study are listed in Table 1. A complete deletion of luxS from the start codon to the stop codon was constructed in Salmonella enterica serovar Typhimurium SL1344 using the lambda Red system (7) to generate strain SL1344LS. The deletion was confirmed by PCR, Western blotting with LuxS antibodies, and AI-2 detection using the well-established Vibrio bioluminescence reporter bioassay (25). Strain SL1344LS was complemented using the plasmid pBRluxS (15). SL1344 derivative strains JH3010 and JH3016 with chromosomal integration of single-copy fusions of prgH::gfp+ or rpsM::gfp+ (13) were kindly provided by I. Hautefort, together with plasmids pZEP10 and pZEP16 (13) that were used to make equivalent transcriptional gene fusions in strain SL1344LS using the lambda Red system. Strains SH008 and SH009 were constructed by P22 transduction of the prgH::gfp+ reporter construct into strains SL1344 and SR3306, respectively (both strains kindly provided by S. Ryu) (4). Colonies were screened by PCR for correct insertions, and the abilities of the colonies to induce ruffling and invade epithelial cells were checked and were comparable to the abilities of their isogenic parent. For standard invasion and green fluorescent protein (GFP) expression assays, strains were grown overnight at 37°C as a static culture in LB (Miller) broth supplemented with antibiotics as appropriate at the following concentrations: carbenicillin, 100 μg/ml; chloramphenicol, 15 μg/ml. Cultures were diluted 1:100 into LB (plus antibiotics) and grown for 3.5 h prior to infection at 37°C in an orbital shaker. To examine the effect of anaerobic growth conditions, an overnight static culture was diluted 1:100 into LB and grown statically in a sealed 2-liter gas jar made anaerobic with an AnaeroGen pack (Oxoid, Basingstoke, United Kingdom) at 37°C for 20 h.
TABLE 1.
S. enterica serovar Typhimurium strains used in this study
| Strain | Description | Reference |
|---|---|---|
| SL1344 | Wild type (our culture collection) | 14 |
| SL1344LS | SL1344 luxS | 15 |
| SL1344LS(pBRluxS) | SL1344 luxS with pBRluxS | 15 |
| JH3010 | SL1344 with transcriptional gene fusion prgH::gfp+ | 13 |
| SL1344LS prgH::gfp+ | SL1344 luxS with prgH::gfp+ | This study |
| SL1344LS prgH::gfp+(pBRluxS) | SL1344 luxS with prgH::gfp+ with pBRluxS | This study |
| JH3016 | SL1344 with transcriptional gene fusion rpsM::gfp+ | 13 |
| SL1344LS rpsM::gfp+ | SL1344 luxS with rpsM::gfp+ | This study |
| SL1344LS rpsM::gfp+(pBRluxS) | SL1344 luxS with rpsM::gfp+ with pBRluxS | This study |
| SL1344 (Choi et al.) | Wild type (culture collection of Ryu group) | 4 |
| SR3306 | SL1344 luxS | 4 |
| SH008 | SL1344 (Choi et al.) with transcriptional gene fusion prgH::gfp+ | This study |
| SH009 | SR3306 (luxS mutant) with transcriptional gene fusion prgH::gfp+ | This study |
Infection of cultured cells.
For experiments in which epithelial cells were infected with S. Typhimurium and subsequently fixed and stained, Madin-Darby canine kidney (MDCK) strain I and HeLa cells were seeded on 13-mm coverslips (2.5 × 104 cells per coverslip) and maintained at 37°C in a humidified atmosphere of 5% CO2 for 3 days. Prior to infection, the medium was replaced with a modified Krebs' buffer (137 mM NaCl, 5.4 mM KCl, 1 mM MgSO4, 0.3 mM KH2PO4, 0.3 mM NaH2PO4, 2.4 mM CaCl2, 10 mM glucose, and 10 mM Tris, adjusted to pH 7.4 at 37°C with HCl). After equilibration of the cells in this medium at 37°C in air, S. Typhimurium was added at a multiplicity of infection (MOI) of approximately 50 to 100 for 15 min for standard invasion assays. Increased infection times and/or decreased MOI were also examined to confirm data obtained under standard infection assay conditions.
Quantification of invasion.
S. Typhimurium invasion was quantified using differential immunofluorescence staining by the method of Perrett and Jepson (21). Coverslips were examined using a Leica DM LB2 upright microscope (Leica Microsystems, Mannheim, Germany). Quantification of MDCK cells (labeled with 4′,6′-diamidino-2-phenylindole [DAPI]), adherent bacteria (labeled with fluorescein isothiocyanate), and total bacteria (labeled with tetramethyl rhodamine isothiocyanate [TRITC]) in 10 randomly selected areas of each coverslip was performed to calculate the number of invading bacteria per cell. The identity of the strains was hidden from the person assessing invasion to eliminate any possible bias. The total number of cells counted per coverslip was 250 or greater.
Analysis of membrane ruffling by cytochemical staining of F-actin.
After the cells were infected with S. Typhimurium for the required time, the coverslips were washed thoroughly in phosphate-buffered saline (PBS), fixed in 2% paraformaldehyde, permeabilized with 0.1% Triton X-100, and sequentially incubated with goat anti-Salmonella CSA-1 antibody and fluorescein isothiocyanate-conjugated rabbit anti-goat immunoglobulin alongside TRITC-phalloidin, as described previously (21). The coverslips were then washed thoroughly in PBS and mounted in Vectashield containing DAPI and examined using a Leica DM LB2 upright microscope to assess the frequency and morphology of Salmonella-induced membrane ruffles in 10 randomly selected fields per coverslip. The identity of the strains was hidden from the person quantifying ruffling and assessing morphology to eliminate any possible bias. The total number of cells counted per coverslip was 250 or greater.
Phase-contrast time-lapse microscopy.
MDCK strain I cells were seeded on 22-mm coverslips (2.5 × 104 cells per coverslip) and maintained at 37°C in a humidified atmosphere of 5% CO2. Three to 4 days after seeding, the coverslip was washed twice with warm (37°C) modified Krebs' buffer and mounted in a coverslip holder with 1 ml of modified Krebs' buffer. The coverslip holder was then placed on the stage of a Leica DM IRB inverted epifluorescence microscope enclosed within a stage incubator (Solent Scientific) at 37°C. S. Typhimurium grown for 3.5 h in LB Miller was added to the chamber of the coverslip holder at an MOI of approximately 50. Phase-contrast images were obtained with a 40× oil immersion lens (numerical aperture of 1.0). Improvision Openlab 4 software was used to automate the acquisition of images at each of three focal depths (1.5-μm steps) at 10-s intervals over a 20-min time course with a Hamamatsu ORCA ER cooled charge-coupled-device camera. Data on the time interval between bacterial binding and induction of membrane ruffling were pooled from at least four experiments, which represents the study of 100 individual ruffles for each strain. All data are expressed as medians, with the range of values indicated. Significance of differences between median values was assessed using a Kruskal-Wallis test followed by Dunn's multiple-comparison test.
Analysis of T3S profiles.
Salmonella bacteria were grown in LB broth overnight before being diluted 1:50 in fresh LB broth and grown under conditions that stimulate expression of the SPI-1 type III secretion system (9). The culture supernatants were collected and filter sterilized (pore size, 0.22 μm), and proteins were precipitated with ammonium sulfate (4 g/10 ml of supernatant) overnight at 4°C. Precipitated secreted proteins were pelleted by centrifugation at 42,000 × g and 4°C for 1 h. The secreted protein pellet was resuspended in 500 μl of sterile distilled H2O and stored at −20°C until required. Samples were electrophoresed on 12% sodium dodecyl sulfate-polyacrylamide gels and transferred to Protran nitrocellulose transfer membranes (Schleicher and Schuell) by using a wet transfer apparatus (Bio-Rad). Western blot analysis on type 3 secretion system (T3SS) components and effectors was performed using rabbit antibodies at a dilution of 1:2,000 coupled with a goat anti-rabbit horseradish peroxidase-labeled secondary antibody (Dako Cytomation). Detection was carried out by using 4-chloro-1-naphthol (Sigma) according to the manufacturer's instructions.
Flow cytometry.
For measurement of GFP in S. Typhimurium, samples were taken after 3.5 h of growth in LB at 37°C and fixed in 2% paraformaldehyde for 15 min at 4°C. Fixed bacteria were subsequently washed and diluted in PBS to obtain a maximum of approximately 107 particles per ml. Samples were kept in the dark at 4°C until analysis.
Flow cytometric analysis was performed on a FACSVantage SE flow cytometer (Becton Dickinson, Franklin Lakes, NJ) equipped with a 15-mW air-cooled argon ion laser as the excitation light source (488 nm). S. Typhimurium SL1344 without the single chromosomal gfp+ gene fusions was used as a negative control, and rpsM::gfp+ mutants, which constitutively express GFP, were used as a positive control. Approximately 50,000 events identified as Salmonella cells were collected per sample. GFP fluorescence intensity values are presented as means for the populations.
Mouse infection.
Bacterial strains were grown in 10 ml of LB at 37°C for 16 h with shaking (200 rpm). Female BALB/c mice (six per group) were challenged orally for each strain with 105, 106, or 107 CFU of S. Typhimurium SL1344, the isogenic luxS strain (SL1344LS), or sterile PBS as a control. For each strain, the number of mice reaching the clinical end point after 12 days was noted. The experiment was repeated twice with similar results.
Statistical analysis.
Data were analyzed using PRISM 3.0 software. Data are expressed as means ± standard errors, with statistical significance set at a P of ≤0.05. Data were compared using a one-way analysis of variance, with a Tukey test applied post hoc.
RESULTS
LuxS does not affect membrane ruffling or invasion of epithelial cell lines.
A complete deletion of luxS was constructed in Salmonella enterica serovar Typhimurium SL1344 to generate SL1344LS. AI-2 secretion assays confirmed that the luxS mutant did not produce AI-2, while the wild-type strain (SL1344) and plasmid-complemented SL1344LS [SL1344LS(pBRluxS)] exhibited the previously reported AI-2 peak during log-phase growth (4, 26; data not shown). To elucidate whether LuxS has an effect on the virulence of S. Typhimurium, we compared the abilities of the wild-type SL1344 strain and its isogenic luxS mutant (SL1344LS) to invade and induce ruffling in two cell lines, MDCK and HeLa. The ability to produce membrane ruffles was assessed using TRITC-phalloidin staining of F-actin, while invasion was quantified using differential immunocytochemical staining of adhered and total cell-associated S. Typhimurium. Infection assays examined cultures in late log phase (3.5 h), when the AI-2 concentration in wild-type S. Typhimurium culture medium was maximal (data not shown).
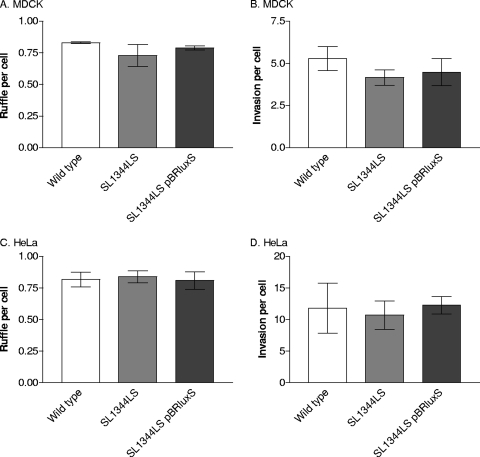
The numbers of ruffles propagated by the wild-type strain (SL1344) and strain SL1344LS were similar after a 15-min infection in both cell types (Fig. 1A and C). Similarly, the mean number of SL1344LS bacteria invading cells during a 15-min infection was not significantly different from those of the wild type and the complemented strain, SL1344LS(pBRluxS) (Fig. 1B and D). Increasing the infection time (to 60 min) or decreasing the MOI (to 2:1 or 20:1) confirmed that the numbers of invading bacteria of the wild-type and SL1344LS strains were comparable (data not shown). Following anaerobic growth, Salmonella invasion of MDCK cells was also unaffected by luxS deletion (data not shown). Measurements of invasion using polarized Caco-2 cells also showed no difference between the wild-type and SL1344LS strains (data not shown). Thus, LuxS does not affect entry of S. Typhimurium into epithelial cells under a range of growth conditions and infection models.
FIG. 1.
Comparison of invasion and ruffle induction by S. enterica serovar Typhimurium strains SL1344 (wild type), SL1344LS, and SL1344LS(pBRluxS) in MDCK and HeLa cells infected for 15 min with each strain. Ruffle formation was measured using TRITC-phalloidin staining of F-actin (A and C), and the ability to enter cells was measured using differential staining of adhered and invaded bacteria (B and D). Values are the means of three independent experiments ± standard errors of the means (error bars).
LuxS does not affect ruffle morphology or propagation dynamics.
Despite the fact that there was no measured difference in the number of ruffles induced or invading bacteria, it remained possible that subtle differences in the induction and/or propagation of ruffles may exist between the wild-type and SL1344LS strains, as has been shown with other Salmonella mutants (22). Images of the ruffles propagated by strains SL1344, SL1344LS, and SL1344LS(pBRluxS), generated using confocal microscopy, were compared in each cell line, but no obvious differences between the morphology of the ruffles and the position of the bacteria relative to the ruffles were identified (data not shown). This observation was confirmed with time-lapse phase-contrast microscopy where the kinetics of ruffle induction were also measured (Table 2). Strain SL1344LS had a median time interval that was 10 s greater than those of both the wild type and SL1344LS(pBRluxS), a difference that was not statistically significant. Therefore, ruffle induction and propagation appear to occur in the same manner in strains SL1344 and SL1344LS.
TABLE 2.
Comparison of the time taken for membrane ruffle initiation after bacterial adherence by three S. enterica serovar Typhimurium strainsa
| S. Typhimurium strain | nb | Ruffle induction time after adherence (s)c |
|
|---|---|---|---|
| Median | Range | ||
| SL1344 | 111 | 60 | 20-630 |
| SL1344LS | 119 | 70 | 20-490 |
| SL1344LS(pBRluxS) | 127 | 60 | 20-460 |
S. enterica serovar Typhimurium strains SL1344 (wild type), SL1344LS, and SL1344LS(pBRluxS) were grown for 3.5 h in LB.
The number of productive bacterium-cell interactions analyzed (n) is shown.
The time interval between bacterial binding to MDCK cells and induction of membrane ruffling was obtained from phase-contrast video microscopy. Data are expressed as median values, and the range in time intervals (minimum to maximum) was measured.
Contribution of LuxS to SPI-1 type 3 secretion.
Although no difference was observed in the invasion behavior of the S. Typhimurium luxS mutant, we decided to assess whether the expression of Salmonella pathogenicity island 1 may be different, which may indicate that LuxS does play a role in S. Typhimurium virulence. The secretion profiles of wild-type SL1344 and SL1344LS strains were compared by Western blotting using antibodies directed against both the SPI-1 type 3 secretion system apparatus proteins InvG, InvJ, PrgH, and also the effector proteins SipA, SipB, SipC, and SipD (12) (Fig. 2). The levels of SPI-1 apparatus and secreted effector proteins were comparable in strains SL1344 and SL1344LS, indicating that SPI-1 T3S protein expression and secretion are independent of luxS.
FIG. 2.
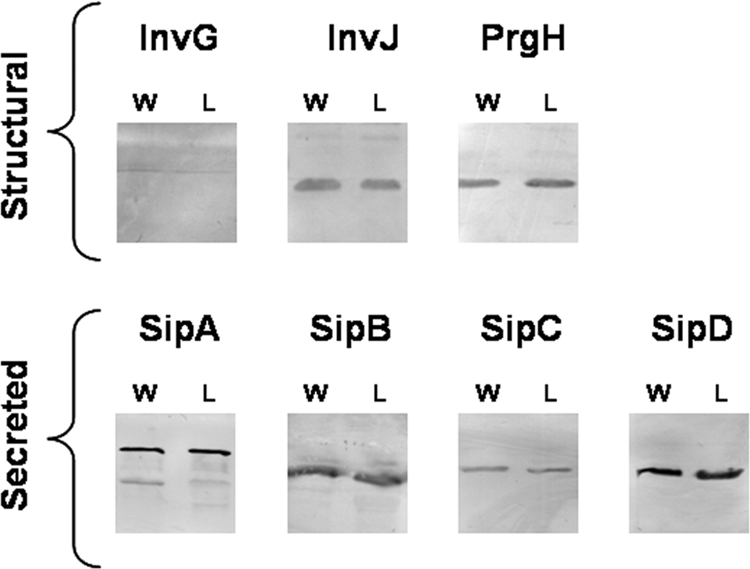
SPI-1 secretion is not dependent upon luxS. Western blot analysis of whole-cell T3S structural proteins (InvG, InvJ, and PrgH) and extracellular secreted proteins (SipA to SipD) from the wild-type strain, SL1344 (W), and the isogenic luxS mutant, SL1344LS (L), shows no difference in the quantity of SPI-1 effectors secreted.
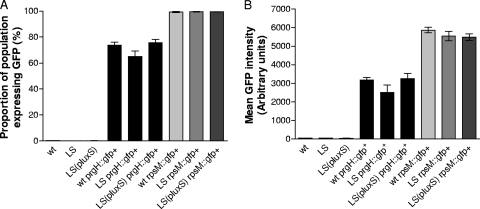
SPI-1 T3SS gene expression was also examined using single-copy prgH::gfp+ fusions inserted into the chromosomes of strains SL1344 (wild type), SL1344LS, and SL1344LS(pBRluxS). prgH encodes a structural component of the SPI-1 T3SS-1 apparatus (17) and has previously been used as a reliable measure of SPI-1 expression (5, 13). Strains without gfp+ fusions and with single-copy rpsM::gfp+ fusions were used as negative and positive controls, respectively. When the wild-type SL1344 strain and luxS mutant carrying a single copy of the prgH::gfp+ fusion in their chromosome were compared by flow cytometry at 3.5 and 6 h, a slight reduction was found in the proportion of the population expressing GFP, and therefore PrgH, in the luxS mutant, but this difference was not statistically significant (Fig. 3). The amount of PrgH expression per cell was also slightly lower in strain SL1344LS than in the wild type but again was not statistically different. The possibility that LuxS might influence the kinetics of prgH induction was explored further by quantifying the proportion of S. Typhimurium SL1344 prgH::gfp+ and SL1344LS prgH::gfp+ expressing GFP at hourly intervals over a 6-h time course from overnight static culture through log to stationary growth phases. No difference was evident between the wild type and luxS mutant with respect to prgH expression at any point (data not shown), suggesting that LuxS does not affect prgH expression kinetics. The similar levels of SPI-1 expression and protein secretion in the wild type and SL1344LS provide further support to our conclusion that luxS does not contribute to regulation of SPI-1 T3SS expression.
FIG. 3.
Expression of SPI-1 in S. enterica serovar Typhimurium strains SL1344 (wild type), SL1344LS, and SL1344LS(pBRluxS) grown for 3.5 h in LB. Flow cytometry was used to measure the expression of GFP in the wild type (wt), luxS mutant (LS), and complemented mutant [LS(pluxS)] bacteria with no transcriptional gfp+ gene fusion, transcriptional gene fusion rpsM::gfp+ or transcriptional gene fusion prgH::gfp+. (A) Proportion of each bacterial population with detectable fluorescence in the GFP channel; (B) mean fluorescence intensity. Data represent means ± standard errors of the means (error bars) from three independent experiments. Of the population with no transcriptional gfp+ gene fusion, ≤0.3% have detectable fluorescence in the GFP channel (negative control), while approximately 99% of the S. Typhimurium cells containing the constitutive fusion (rpsM::gfp+) express GFP above the threshold intensity (positive control).
LuxS does not attenuate virulence in mice.
To confirm our in vitro results, the contribution of luxS to virulence was tested in a mouse model. Female BALB/c mice were challenged orally with wild-type strain SL1344, SL1344LS, or PBS as a control. Table 3 shows no significant differences in the number of mice reaching the clinical end point 12 days after oral challenge with either the wild-type strain or luxS mutant. These results suggest that luxS has no significant impact on the pathogenicity of Salmonella in vivo.
TABLE 3.
Virulence of S. enterica serovar Typhimurium in the mouse infection modela
| Strain or control | No. of mice (n = 6) that reached the clinical end point challenged with the following doseb: |
||
|---|---|---|---|
| 105 CFU | 106 CFU | 107 CFU | |
| SL1344 | 5 | 6 | 6 |
| SL1344LS | 6 | 5 | 6 |
| PBS (control) | 0 | 0 | 0 |
Female BALB/c mice were challenged orally with 105, 106, or 107 CFU. of S. Typhimurium SL1344 (wild type), the isogenic luxS mutant SL1344LS, or sterile PBS as a control.
The number of mice that reached the clinical end point at 12 days postchallenge is shown. Mice that reached the clinical end point, i.e., the point of infection where severe symptoms appeared to be due to high bacterial burdens, were removed from the group. In the control group, all mice remained well when inoculated with PBS.
DISCUSSION
The possession of the luxS gene, coding for autoinducer 2 synthase, by Salmonella enterica has led to the proposal that Salmonella may regulate gene expression in response to population size, i.e., quorum sense (25, 26). Initial screening of genes regulated by luxS and AI-2 in S. enterica serovar Typhimurium identified only those genes involved in the synthesis and detection of AI-2. These include metE, which encodes an enzyme with a role in the activated methyl cycle, from which AI-2 is a by-product, and those of the luxS-regulated (lsr) operon, which encode a transporter for AI-2 and enzymes which modify this signal upon its entrance into cells (28). Thus, it is not clear whether luxS-based quorum sensing per se regulates any biological activities in Salmonella, especially with the recent report that LuxS has been found to affect flagellar phase variation in S. Typhimurium independently of the AI-2 signal (15). However, one report has implicated luxS in the regulation of S. Typhimurium pathogenesis, with LuxS being necessary for optimal expression of a subset of type 3 secretion genes in SPI-1, under the control of InvF (4). We have used alternative methods to examine whether S. Typhimurium uses luxS-based quorum sensing to regulate its ability to invade host cells.
No differences were found between the wild-type SL1344 strain and its isogenic luxS mutant in the number of membrane ruffles propagated or the number of bacteria invading MDCK, HeLa, and Caco-2 cell lines. Visualization of the infection process in MDCK cells using phase-contrast time-lapse microscopy confirmed the results of the immunocytochemical staining assays, since no clear difference was found between the wild type (SL1344) and SL1344LS in the kinetics or morphology of membrane ruffles or in the positions of invading bacteria. Analysis of multiple host cell lines indicates that the similar behavior of the wild type and mutant in these assays is not cell line dependent. Similar to our in vitro observations, the luxS mutant was found not to differ from the wild type when comparing virulence via the oral route in the mouse model of infection. Thus, it appears that luxS is not essential for S. Typhimurium pathogenicity.
To complement the work examining the contribution of luxS to pathogenicity, the expression of SPI-1 T3SS in the wild type and the luxS deletion mutant were compared. SPI-1 encodes a type 3 secretion system (T3SS-1) and several effector proteins, secreted through this macromolecular needle-complex, which are essential for invasion (6). Therefore, if luxS-based quorum sensing were to play a role in regulating S. Typhimurium invasion, a change in SPI-1 expression would be the expected mechanistic basis. In complete agreement with the results of the ruffling and invasion assays, the expression of the SPI-1 T3S apparatus proteins and the secreted effectors were found to be very similar in the wild type and SL1344LS. Choi et al. used real-time PCR to compare expression of SPI-1 genes in the wild type (SL1344) and luxS mutant grown under anaerobic conditions to stationary phase (4). Decreased expression of invF and several genes, sicA, sigD, and sopE, which are regulated by InvF, in the luxS mutant was reversed by introduction of the luxS gene in trans or by addition of 4,5-dihydroxy-2,3-pentanedione. DPD, a precursor of AI-2, or synthetic AI-2 is required to chemically complement the luxS gene in order to indicate that a phenotype is indeed regulated by quorum sensing and not related to the metabolic role of luxS (31). Choi et al. (4) also reported that, in contrast to our findings, prgH expression was approximately twofold lower in the luxS mutant than in the wild type. A smaller, and not statistically confirmed, reduction in hilA expression was also observed, suggesting that LuxS might regulate prgH via HilA, the predominant transcriptional activator of prgH expression (1). In contrast to their findings with invF and associated genes, expression of prgH and hilA in the luxS mutant was restored by pLuxS but not by DPD, indicating that any downregulation of hilA or prgH in the luxS mutant is independent of quorum sensing. Taken together with our findings that LuxS does not significantly affect prgH expression, SPI-1 type 3 secretion, invasion of a range of epithelial cell lines, and virulence in a mouse model, all these results support our conclusion that quorum sensing does not have a major role in either regulation of SPI-1 T3S or virulence.
We examined whether differences in the mutant strains tested and/or experimental conditions used might explain the disparity between our findings and those of Choi et al. with respect to our inability to detect any effect of LuxS on Salmonella virulence or SPI-1 gene expression as previously reported (4). We eliminated the possibility that the differences in invasion and SPI-1 regulation noted by Choi et al. (4) were related to some difference in the strains used by repeating our invasion, gene expression, and protein secretion assays with their luxS mutant and wild-type strains. These experiments revealed no difference in invasion under two alternative MOIs at either 15 or 60 min postinfection (data not shown). Similarly, we detected no difference in prgH promoter activity, expression of the SPI-1 proteins PrgH and InvJ in cellular fractions or in secretion of the SPI-1 effector proteins SipA, SipB, and SipC (regulated by InvF) between the wild type and luxS mutants examined by Choi et al. (4). It remained possible that differences in the bacterial growth phase examined are important, since our initial assays were performed at late log phase, conditions selected because extracellular AI-2 concentrations and SPI-1 expression are maximal (4, 18, 27), while Choi et al. (4) measured those genes regulated by HilA and InvF at stationary phase when SPI-1 expression is suboptimal and when AI-2 has been internalized and thus extracellular AI-2 concentrations are low. In addition, Choi et al. (4) grew Salmonella strains under anaerobic conditions, while most studies of invasion and SPI-1 expression use aerobic growth conditions. The differences reported by Choi et al. with respect to invasion and prgH gene expression in luxS mutant could not be replicated by growing either luxS mutant and their respective parent strains under anaerobic conditions to stationary phase (data not shown). Thus, we conclude that the previously reported role of luxS in these aspects of Salmonella biology are most likely related to the precise experimental conditions employed by Choi et al. (4) and are of negligible importance under the much more extensive range of growth conditions and infection protocols we have examined.
In summary, it was found that in S. Typhimurium SL1344, luxS has no measured effect on the expression of SPI-1 T3SS apparatus proteins or on its ability to secrete SPI-1 T3S effector proteins required for invasion. As a consequence, the epithelial cell invasion phenotype of the luxS mutant was similar to that of the wild type, with no difference being exhibited in either the time it takes to induce membrane ruffling after adherence to a host cell, the number of ruffles propagated, or on the rate of invasion. Additionally, the virulence of the luxS mutant was similar to that of the wild type in a mouse model of infection.
Acknowledgments
We are grateful to Jay Hinton, Isabelle Hautefort, Sangryeol Ryu, and Jeongjoon Choi for providing bacterial strains. We thank Alan Leard for assistance with microscopy and Mick Bailey for help with the flow cytometry.
The Medical Research Council supported this work by providing a Ph.D. studentship to C.A.P. (G74/77) and funding microscopes in Bristol University's Cell Imaging Facility. Research in the laboratories of M.A.J., C.M.A.K., and P.W. was also supported by MRC and BBSRC research grants. S.H. is supported by BBSRC and Unilever under a CASE Ph.D. studentship, and H.S. was a recipient of a BBSRC DTA Ph.D. studentship.
Footnotes
Published ahead of print on 25 September 2009.
REFERENCES
- 1.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L. 1999. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol. 2:582-587. [DOI] [PubMed] [Google Scholar]
- 3.Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi--sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. [DOI] [PubMed] [Google Scholar]
- 4.Choi, J., D. Shin, and S. Ryu. 2007. Implication of quorum sensing in Salmonella virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect. Immun. 75:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, L., I. Martinez-Argudo, T. J. Humphrey, and M. A. Jepson. 2009. GFP plasmid-induced defects in Salmonella invasion depend on plasmid architecture, not protein expression. Microbiology 155:461-467. [DOI] [PubMed] [Google Scholar]
- 6.Collazo, C. M., and J. E. Galán. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.Eichelberg, K., and J. E. Galán. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24-29. [DOI] [PubMed] [Google Scholar]
- 11.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene-products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galán, J. E., and H. Wolf-Watz. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444:567-573. [DOI] [PubMed] [Google Scholar]
- 13.Hautefort, I., M. J. Proenca, and J. C. D. Hinton. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 15.Karavolos, M. H., D. M. Bulmer, K. Winzer, M. Wilson, P. Mastroeni, P. Williams, and C. M. Khan. 2008. LuxS affects flagellar phase variation independently of quorum sensing in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190:769-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleerebezem, M., L. E. N. Quadri, O. P. Kuipers, and W. M. deVos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 17.Kubori, T., Y. Matsushima, D. Nakamura, J. Uralil, M. Lara-Tejero, A. Sukhan, J. E. Galan, and S. Aizawa. 1998. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280:602-605. [DOI] [PubMed] [Google Scholar]
- 18.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian-cells is affected by bacterial-growth state. Proc. Natl. Acad. Sci. USA 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 20.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 21.Perrett, C. A., and M. A. Jepson. 2007. Applications of cell imaging in Salmonella research. Methods Mol. Biol. 394:235-273. [DOI] [PubMed] [Google Scholar]
- 22.Perrett, C. A., and M. A. Jepson. 2009. Regulation of Salmonella-induced membrane ruffling by SipA differs in strains lacking other effectors. Cell. Microbiol. 11:475-487. [DOI] [PubMed] [Google Scholar]
- 23.Rezzonico, F., and B. Duffy. 2008. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxS in most bacteria. BMC Microbiol. 8:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 25.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of Al-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 28.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer Al-2 controls the expression of an ABC transporter that functions in Al-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 29.Williams, P., N. J. Bainton, S. Swift, S. R. Chhabra, M. K. Winson, G. S. A. B. Stewart, G. P. C. Salmond, and B. W. Bycroft. 1992. Small molecule-mediated density-dependent control of gene expression in prokaryotes: bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol. Lett. 79:161-167. [DOI] [PubMed] [Google Scholar]
- 30.Williams, P., K. Winzer, W. C. Chan, and M. Cámara. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B 362:1119-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 32.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 33.Xavier, K. B., S. T. Miller, W. Y. Lu, J. H. Kim, J. Rabinowitz, I. Pelczer, M. F. Semmelhack, and B. L. Bassler. 2007. Phosphorylation and processing of the quorum-sensing molecule autoinducer-2 in enteric bacteria. ACS Chem. Biol. 2:128-136. [DOI] [PubMed] [Google Scholar]