Abstract
Intracellular polysaccharide (IPS) is accumulated by Streptococcus mutans when the bacteria are grown in excess sugar and can contribute toward the cariogenicity of S. mutans. Here we show that inactivation of the glgA gene (SMU1536), encoding a putative glycogen synthase, prevented accumulation of IPS. IPS is important for the persistence of S. mutans grown in batch culture with excess glucose and then starved of glucose. The IPS was largely used up within 1 day of glucose starvation, and yet survival of the parental strain was extended by at least 15 days beyond that of a glgA mutant; potentially, some feature of IPS metabolism distinct from providing nutrients is important for persistence. IPS was not needed for persistence when sucrose was the carbon source or when mucin was present.
Streptococcus mutans is a facultative colonizer of the human dental plaque, the microbial pellicle that covers the surface of the teeth. It is the major etiological agent of dental caries (17). Sugar metabolism is central to the behavior of S. mutans (4, 7). It can use a variety of sugars. The sugars are fermented by glycolysis with production of organic acids, particularly lactic acid (4, 7). In addition to providing energy, sucrose is used to produce extracellular polysaccharides to form the biofilm matrix that aids in the association of S. mutans with the dental plaque. Once the S. mutans biofilm becomes part of the dental plaque, the acidic by-products of sugar fermentation dissolve tooth enamel, eventually resulting in dental caries (17). The presence of sugars in the dental plaque is periodic and reflects the intake of dietary sugars. If there is excess sugar available, in addition to producing organic acids and matrix, intracellular (iodophilic) polysaccharide (IPS; glycogen) is formed.
The IPS of S. mutans is a polymer of the glycogen-amylopectin type, with α-(1, 4)- and α-(1, 6)-linked glucose, and is stored as intracellular granules (10). Intracellular glycogen storage reserves in various bacterial species are synthesized from glucose-1-P via ADP-glucose (1). The synthesis involves at least three enzymes: glycogen synthase, glucose-1-phosphate pyrophosphorylase, and branching enzyme. The genes encoding these enzymes are commonly found in a glg operon, although the order of genes differs between species. In two gram-positive species, Bacillus subtilis and Bacillus stearothermophilus, the gene order is glgB-glgC-glgD-glgA-glgP (15, 29): glgA encodes glycogen synthase, glgB encodes glucan branching enzyme, and glgC and glgD encode subunits of glucose-1-phosphate pyrophosphorylase. The glgP gene encodes glycogen phosphorylase, which is unlikely to be involved in glycogen synthesis (29). Genes putatively encoding similar enzymes are present in the same order in the genome of S. mutans (29); they are thought also to form an operon.
The IPS can be used as a source of carbohydrate for fermentation upon nutrient depletion (11, 13). In planktonic cultures, IPS reserves are largely consumed within 12 h of the imposition of sugar starvation (11, 13, 32). In S. mutans, IPS utilization may prolong acid production and hence the period of lowered pH of the resting (between meals) plaque, a factor that contributes to the incidence of caries. Indeed, IPS is implicated in dental caries: a mutant that synthesized elevated levels of IPS was hypercariogenic in germfree rats (27). Strains isolated from human carious lesions were nearly all stable IPS producers, whereas most strains from caries-inactive persons were variable IPS producers (13, 33).
Since S. mutans deep in the dental plaque may not have access to nutrients because of competition with the bacteria at the surface of the plaque, the bacteria may need to survive longer periods of nutrient starvation. Previous studies in our laboratory showed that S. mutans can survive under sugar starvation conditions, provided that the pH remains above ∼5.5 (22). The presence of spent medium and mucin significantly prolonged survival of sugar-starved biofilms and batch cultures (22; also unpublished observations). Here we examine the role of IPS.
The role of IPS (glycogen) in bacterial survival has been tested for several other bacterial species. It was found to extend survival of Aerobacter aerogenes (8) and Escherichia coli (28). Intracellular glycogen was also shown to support the survival of Streptococcus mitis during stationary-phase starvation (32). In contrast, glycogen-rich Sarcina lutea died at a higher rate during starvation than did bacteria without glycogen (2).
In order to test the role of IPS in S. mutans survival, we constructed an IPS-deficient mutant by inactivating glgA (GenBank SMU.1536) (http://www.oralgen.lanl.gov/), putatively encoding the glycogen synthase. We also constructed a mutant potentially altered in IPS metabolism by inactivating the putative pullulanase structural gene, pul (SMU.1541). Pullulanases are responsible for hydrolyzing α-(1,6) linkages (and in some cases 1,4 linkages) in pullulan and in other polysaccharides (35) and may be important in determining the branching in IPS and/or affecting the catabolism of IPS. We studied the persistence of bacteria under conditions of sugar limitation and of sugar excess in both batch cultures and biofilms. We found that IPS can play a role in supporting S. mutans persistence in batch cultures but found no role for IPS in survival in biofilms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The parental strain was S. mutans UA159. Other strains are listed in Table 1. Strains were stored in 15% glycerol at −76°C and revived for experiments by growth overnight at 37°C in a 5% CO2 incubator in either Todd-Hewitt (TH) broth (Difco, Detroit, MI) or chemically defined medium (FMC) (30) supplemented with 24 mM glucose or on TH agar. The FMC (30) was supplemented with 100 mM glucose or 50 mM sucrose to achieve sugar excess conditions. It was supplemented with 6 mM glucose or 3 mM sucrose to achieve sugar-limited conditions (22). The flow cell biofilm starvation medium was fresh FMC with no sugar. TH agar was used; when appropriate, it was supplemented with 2% glucose. A 5% stock solution of type III pig gastric mucin (Sigma, St. Louis, MO) was prepared by dissolving the mucin powder in 0.01 M potassium phosphate buffer (K2HPO4-KH2PO4, 21:4). When indicated, it was added to the culture medium to a final concentration of 0.5% and filter sterilized. When appropriate, S. mutans was grown in the presence of antibiotics at the concentrations indicated: kanamycin, 300 μg/ml; chloramphenicol, 10 μg/ml; erythromycin, 25 μg/ml. All the cloning procedures were carried out with E. coli DH5α, which was grown in Luria-Bertani lysogeny broth (LB). The antibiotic concentrations used for E. coli were as indicated: kanamycin, 50 μg/ml; chloramphenicol, 50 μg/ml; erythromycin, 300 μg/ml.
TABLE 1.
S. mutans strains used
| Straina | Relevant genotype | Source |
|---|---|---|
| SL13178 | Δpul (pul::kan) | This study |
| SL13180 | ΔglgA (glgA::kan) | This study |
| SL14602 | ΔfruAB (fruAB::cat) | This study |
| SL14642 | ΔglgA ΔfruAB (glgA::kanfruAB::cat) | This study |
| SL14646 | Δpul ΔfruAB (pul::kanfruAB::cat) | This study |
| SL15068 | UA159 with pMC50 [Pspac(Hy)] | This study |
| SL15070 | UA159 with pMC51 [Pspac(Hy)-glgA] | This study |
| SL15072 | SL13180 with pMC50 [glgA::kan Pspac(Hy)] | This study |
| SL15074 | SL13180 with pMC51 [glgA::kan Pspac(Hy)-glgA] | This study |
All strains have the genetic background of S. mutans UA159.
Batch culture growth and survival.
Overnight cultures grown in FMC with 24 mM glucose were diluted 25-fold into fresh FMC containing limiting or excess sugar. Bacteria were grown in stationary culture tubes in a 5% CO2 incubator at 37°C. Growth was monitored with a BioMate 3 spectrophotometer (Thermo Electron Scientific Instrument Corporation) by measuring the optical density at 675 nm (OD675). For determinations of survival, 25-ml cultures were grown in 50-ml culture tubes at 37°C and 5% CO2. At different times, samples were removed and serial dilutions of each culture were made in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.4 mM KH2PO4) and plated onto TH agar. Survival was documented as CFU per ml of culture. The limit of detection was 10 CFU/ml.
Transformation of S. mutans.
S. mutans was transformed using the method of Lindler and Macrina (16). Briefly, a 5-ml culture of S. mutans UA159 was grown overnight in TH broth, at 37°C. The overnight culture was diluted 25-fold into fresh TH broth containing 10% glucose and 10% heat-inactivated horse serum (MP Biomedicals, Inc.), which has been shown to induce competence in S. mutans (19). The bacteria were allowed to grow at 37°C for 3.5 h, a time reported to give optimal transformation of S. mutans (16). A 0.5-ml portion of this culture was transferred to a 13-ml culture tube containing various concentrations (0.5 to 10 μg/ml) of plasmid or chromosomal DNA and incubated at 37°C for 2 h. Transformants were selected on TH agar containing the appropriate antibiotic; transformant colonies were generally obtained after 2 days of incubation at 37°C in a 5% CO2 incubator.
Construction of S. mutans deletion mutants.
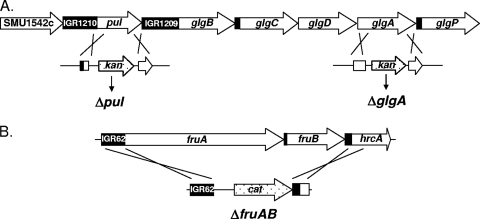
The glgA and pul genes were disrupted by double crossover recombination with plasmid donors. The plasmids were constructed using a vector, pVK262, that lacks an origin of replication for gram-positive organisms and contains a kanamycin resistance cassette. The resulting plasmids contained fragments of about 600 bp of S. mutans DNA flanking the kan cassette. These fragments were regions immediately 5′ and 3′ and extending into glgA (pKM6) or pul (pKM4) and were PCR amplified from the parental strain chromosomal DNA by using specific primers (Table 2). The pKM4 and pKM6 plasmids was designed to replace a DNA fragment between codon 117 and codon 500 of the pul open reading frame (ORF) and from codon 176 to codon 330 of the glgA ORF, respectively, with the kan gene; there was no transcription terminator downstream of the kan gene. The plasmids were used to transform S. mutans UA159, selecting for kanamycin resistance. Transformants were isolated and tested for disruption of glgA or pul by double crossover recombination (Fig. 1). Replacement of pul and glgA with kan by double crossover in strains SL13778 and SL13780, respectively (Fig. 1), was confirmed by appropriate PCR.
TABLE 2.
Primers used in this study
| Region, fragment, or gene | Primer | Sequence (5′-3′) |
|---|---|---|
| glgA 3′ region | glgA Fw.5 | TACATGAATTCCATCAGGCTATGCTGATTT |
| glgA Rev.6 | ATGTATACGTAAGCTAGTTCTTTATATAGA | |
| glgA 5′ region | glgA Fw.7 | ATACAGCCGGAAGGGCTAGCAGCGGAAGG |
| glgA Rev.8 | TTCAATCCTTCTATGTTAGGGCCCTTCCGGCGTAT | |
| pul 3′ region | 3′ pul Fw.7 | ATCGAGAATTCGGTGCTGAAGTTTACGGCGGGCTCCT |
| 3′ pul Rev.8 | TGCTAGTACGTAGCGTTTTATATTTGT | |
| pul 5′ region | pul Fw.5 | TCATGAAGGCACCATTAGAATTCTTTGCTA |
| pul Rev.6 | TAGTAGATTAGGTGCCAGTTTTATCT | |
| Overlapping PCR fragment 1 (IGR62-cat) | OL PCR Fw.1 | GATATCTGACTGATGAAACAGCATCG |
| OL PCR Rev.2 cat-1 | AACAAATTTTCATCAATTCCTTGCTCTCCTTT | |
| Overlapping PCR fragment 2 (cat-IGR63-hrcA) | OL PCR Fw.3 cat-2 | ATGACTGGCTTTTATAACAGCAATAAAACT |
| OL PCR Rev.4 | CGAAAAGCTTCAAAATCAAATGCC | |
| cata gene for overlapping PCR | cat gene Fw.1 | TTGATGAAAATTTGTTTGATTTT |
| cat gene Rev.2 | TTATAAAAGCCAGTCATTAGGCC | |
| glgA gene for complementation studies | glgA gene Complem. Fw.1 SmaI | CCAGCCCGGGAAAGCAAGTGAAGTTG |
| glgA gene Complem. Rev.2 BglI | CGGCGCCATTCAGGCTAAGCTAGTTCTTTATATAG |
The cat gene was amplified from pR326 (6).
FIG. 1.
Schematic representation of genetic regions studied. (A) The pul and glg region of the S. mutans UA159 genome. The glg and pul genes are indicated by arrows. IGR indicates an intergenic region. The sites of gene replacement of pul and glgA by a kan cassette are shown below. The pul gene was inactivated by replacing an internal fragment of the gene (positions 1469911 to 1468763 of the S. mutans genome [http://www.oralgen.lanl.gov/], the deleted 1,148 bp corresponding to 383 codons of the ORF) with a kan cassette. Similarly, the glgA gene was inactivated by replacing an internal fragment (positions 1462665 to 1463113, the deleted 448 bp corresponding to 149 codons of the ORF) with a kan cassette. (B) The fru region. The fruA and fruB genes are indicated by arrows. The region of gene replacement by a cat cassette is shown below; the region replaced was between positions 77576 and 83477 and encompassed both ORFs (http://www.oralgen.lanl.gov/).
A strain with the adjacent fruA and fruB genes deleted was constructed by long flanking PCR (34). Specific primers were designed to replace the entire fruA and fruB ORFs with a chloramphenicol resistance (cat) cassette (Table 2). Replacement of fruA-fruB with cat by double crossover recombination in strain SL14602 was confirmed by appropriate PCR. The ΔfruAB::cat construct was introduced into strains SL13778 and SL13780 to produce strains SL14646 and SL14642, respectively. Details of constructions are available on request.
Complementation of a ΔglgA mutant.
Plasmids pMC50 and pMC51 were constructed using the pJAR2 plasmid, which has an origin of replication for S. mutans and carries an erythromycin resistance gene, erm (5). pJAR2 was digested with PvuI, the ends were made blunt and then digested with EcoRV, and the 6.3-kb vector fragment was purified by electrophoresis. The Pspac(Hy) promoter (21), lacking the associated repressor sequence and thus being constitutively expressed, was extracted from pVK55 as a 321-bp EcoRI-BglII fragment, and the ends were filled in with Klenow polymerase. This promoter fragment was ligated to the 6.3-kb fragment from pJAR2 to yield pMC50, which is the control empty vector plasmid. The glgA gene was PCR amplified using UA159 chromosomal DNA as template and primers containing SmaI and BglI restriction enzyme sites (Table 2). These sites were also present downstream of the Pspac(Hy) promoter in pMC50, allowing for directional cloning of the glgA gene. The resulting plasmid, pMC51, was introduced in UA159 (parental strain) and SL13180 (ΔglgA), giving rise to SL15070 and SL15074, respectively. For vector control, we introduced pMC50 into the parental strain and the ΔglgA mutant, giving strains SL15068 and SL15072, respectively.
IPS determination.
Bacteria were streaked on TH agar containing 2% glucose and incubated for 2 days either in a 5% CO2 incubator or in a candle jar at 37°C. Then the agar plates were flooded with 3 ml of 0.2% (wt/vol) iodine in 2.0% (wt/vol) potassium iodide solution. IPS was detected by the rapid formation of a brown pigment.
The IPS content of bacterial cultures was determined by the chemical method of DiPersio et al. (9, 10). Ten-milliliter samples from cultures were heated for 5 min at 100°C. Bacteria were collected by centrifugation at 4,000 × g for 10 min and washed twice with ice-cold water. Bacterial suspensions were adjusted to 5.0 nominal OD675 units in 1 ml ice-cold water for the IPS determinations. One-milliliter amounts were placed in 15-ml conical tubes, and 0.3 ml of 5.3 M KOH was added to each tube. Tubes were covered and placed in a boiling water bath for 90 min. After cooling, the now-clear solutions were neutralized by the addition of 0.3 ml of HCl (5.3 M). The tubes were vortexed, and 1.0 ml of 1.0 M potassium phosphate, pH 7.0, was added. After mixing, 0.6 ml of freshly prepared 0.2% (wt/vol) iodine in 2.0% (wt/vol) potassium iodide solution was added with mixing. The OD520 was determined using a BioMate 3 spectrophotometer. A 1.0-ml amount of water was treated in a manner similar to that for the bacterial samples and used as a blank. IPS content was expressed in adjusted OD units (9, 10). For determination of IPS production, cultures were assayed approximately 8 h after the end of exponential growth in FMC containing different concentrations of glucose or sucrose. The amount of IPS present did not change substantially during the first 16 h after the end of exponential growth, provided that sugar remained present (9, 10; unpublished observations).
Flow cell biofilms.
Biofilms were grown in flow cell chambers as described previously (18, 22). S. mutans was grown overnight in batch cultures in FMC containing 24 mM glucose in a 5% CO2 incubator at 37°C. The bacteria were diluted 25-fold into fresh FMC containing 24 mM glucose and were allowed to grow for 4 to 5 h. The bacteria were washed twice with PBS or sterile water and then diluted to an OD675 of 0.1. Five hundred microliters of the diluted culture was injected directly into the flow chamber tubing with a syringe. Immediately after all chambers were inoculated, a flow (200 μl min−1) of FMC containing 50 mM sucrose and 15 mM NaHCO3 was started using an Ismatec (Glattbrugg, Switzerland) digital pump. The chambers were initially inverted for 20 min to allow the bacteria to adhere to the glass coverslip, and then the chambers were returned to an upright position and incubated for 16 to 18 h at 37°C before examination. When the role of IPS in biofilm survival was being tested, chambers were incubated with FMC with 50 mM sucrose and 15 mM NaHCO3 for 10 h before the imposition of sugar starvation; chambers established for longer times were prone to clogging.
Static biofilms.
Static biofilms of S. mutans were established in 24-well plates containing 12-mm-diameter sterile coverslips (Fisherbrand; Fisher Scientific, Pittsburgh, PA). S. mutans was inoculated in 5 ml FMC containing 24 mM glucose and incubated overnight at 37°C in a 5% CO2 incubator. These cultures were diluted 25-fold into fresh FMC containing 24 mM glucose and incubated for 4 to 6 h. The bacteria were harvested by centrifugation, washed twice with 5 ml PBS, and then diluted to a nominal OD675 of 0.001 in FMC containing the indicated concentration of sugar. Each well was inoculated with 1 ml of culture, and the plates were incubated at 37°C in a 5% CO2 incubator for 14 h. When appropriate, the spent medium was removed from biofilms developed with 3 mM sucrose, filter sterilized, and kept on ice. The supernatant of the biofilms grown in excess sugar was also removed; the biofilms were washed by gently pipetting 1 ml of PBS in each chamber, and then 1 ml of either spent medium from the biofilms developed in limiting sugar concentrations or fresh FMC was added to the chambers to induce starvation.
To monitor biofilm survival, supernatant was removed and the well was washed twice with 1 ml PBS to remove planktonic bacteria. The glass coverslip was extracted from the chamber with sterile forceps and placed in 5 ml PBS in a 15-ml conical tube, which was kept on ice. To disperse bacteria from the biofilm, the biofilm-covered coverslip was sonicated using a cell disrupter (Sonic Dismembrator, model 500; Fisher Scientific, Pittsburgh, PA) with a microtip for 20 s at a voltage amplitude of approximately 60%. The suspension was serially diluted in PBS and plated on TH agar. The results were recorded in CFU per well.
Biofilm imaging.
Flow cell chambers were disconnected from the tubing and inverted for microscopy. Static biofilms were visualized by removing the glass coverslip from the microtiter plate well and carefully inverting it over a 10-well multitest slide (ICN Biomedicals, Inc.). Biofilms were stained using the BacLight stain (Invitrogen; Molecular Probes) as described previously (19). Images were captured using a Leica DM IRE2 microscope with a TCS SL system, using a 100× oil-immersion objective and Leica imaging software.
RESULTS
Inactivation of putative genes for IPS metabolism.
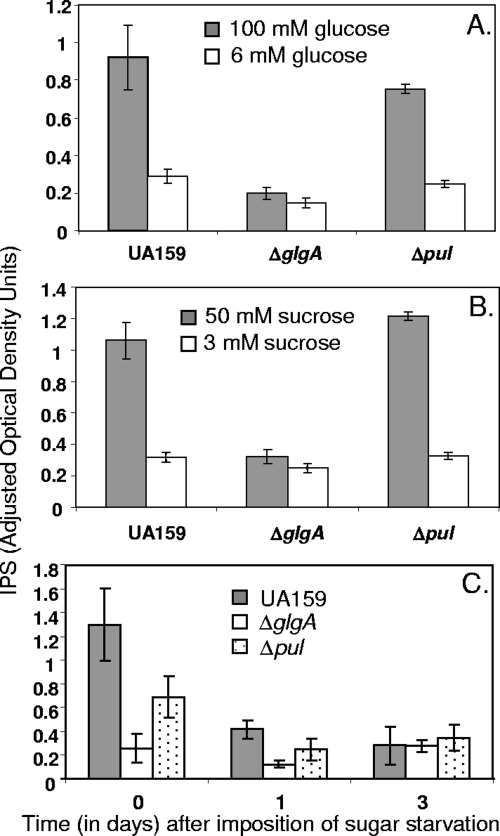
Analysis of the annotated S. mutans genome sequence (http://www.oralgen.lanl.gov/) suggested that the putative genes for glycogen synthesis form an operon, with the order being glgB-glgC-glgD-glgA-glgP (Fig. 1A). The pul gene lies immediately upstream of this glg operon and is separated from it by an intergenic space of 219 nucleotides, which contains a likely transcription terminator (http://www.oralgen.lanl.gov/). The putative glycogen synthase for IPS synthesis is encoded by glgA (SMU1536). We inactivated glgA by replacement of a 448-bp internal fragment of the gene with a kanamycin resistance cassette via double crossover recombination (Fig. 1). We separately inactivated pul, thought to encode a pullulanase for modification and/or metabolism of IPS, by a similar method. The starch-iodide method was used to test for IPS synthesis in bacterial colonies that had been grown on TH agar containing 2% glucose for 48 h at 37°C. Colonies of the parental strain UA159 gave a dark brown color, whereas those of the glgA mutant gave a yellow color, suggesting that the mutant lacked IPS. Colonies of the pul mutant gave a brown color, similar to colonies of the parental strain. The chemical method of DiPersio et al. (9) was used to quantify IPS production. Bacteria were grown in the chemically defined medium FMC (30) containing 100 mM glucose or 50 mM sucrose. The parental strain and the pul mutant, SL13178, formed substantial amounts of iodine-reactive material (IPS), whereas the glgA mutant, SL13180, exhibited only background levels of iodine-reactive material (Fig. 2). Production of IPS was restored by the introduction into SL13180 of pMC51, an autonomously replicating plasmid containing an intact copy of the glgA gene expressed from the Pspac(Hy) promoter (as determined by plate assay). No IPS accumulation was detected when SL13180 was transformed with the vector control plasmid pMC50. Thus, the loss of IPS production in strain SL13180 was because of glgA inactivation and not because of any polar effect on a downstream gene. When bacteria were grown in FMC with only 6 mM glucose or 3 mM sucrose, none of the strains formed a significant amount of IPS (Fig. 2). We were unable to assay IPS accurately for bacteria grown in biofilms, probably because of inefficient removal of the extracellular polysaccharide matrix of the biofilms.
FIG. 2.
Comparison of IPSs formed by the parental strain S. mutans UA159 and glgA and pul mutants (SL13180 and SL13178, respectively) grown at different sugar concentrations. (A and B) Bacteria were grown in FMC containing glucose (A) or sucrose (B), and IPS was determined 8 h after the end of exponential growth. (C) Change in IPS content after transfer to spent medium of bacteria grown in FMC containing 100 mM glucose. Numbers are the averages of three experiments ± standard deviations.
Role of IPS in the persistence of S. mutans in batch cultures.
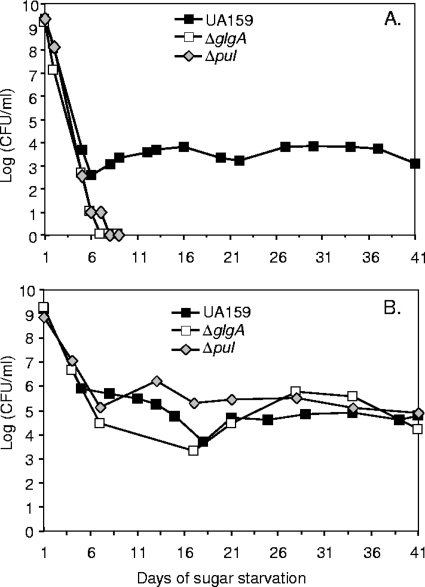
We tested the persistence of the glgA and pul mutants under conditions that were modified from those described by Renye et al. (22). Bacteria were grown in FMC containing 100 mM glucose for 14 h. At this time, the bacteria had accumulated IPS. Bacteria were then harvested by centrifugation, washed, and resuspended in an equal volume of filter-sterilized spent medium from a 14-h culture of the corresponding strain grown in 6 mM glucose. The transfer to the spent medium had previously been shown to prolong survival, presumably because it provided some important product that had been secreted into the medium and also avoided the damaging effects of low pH (spent medium from cultures grown with 100 mM glucose had a pH below the critical threshold for survival [22]). Under these conditions, the glgA and pul mutants survived for 5 to 8 days (Table 3), with a representative survival curve shown in Fig. 3A. In contrast a subpopulation of the parental strain UA159 persisted for much longer (Fig. 3A). The same results were obtained whether the spent medium was derived from strain UA159, the glgA mutant, or the pul mutant. When a plasmid-borne copy of the intact glgA gene was introduced into the glgA mutant, survival (>35 days; strain SL15074, Table 3) was comparable to that of the parental strain, UA159. In contrast, introduction of the empty plasmid vector did not affect survival of the glgA mutant. Neither plasmid affected the survival of the parental strain, UA159. Thus, the effect on survival of glgA inactivation was direct and not because of a polar effect on a downstream gene. The extent of the persistence of the parent varied considerably between experiments, but the persistence was always at least 15 days longer than that of the mutants and always at least 21 days (Table 3).
TABLE 3.
Persistence of S. mutans strains in batch culturesb
| Strain | Mucin | Latest time at which viable bacteria were recovered (days in stationary phase) in different experiments |
|---|---|---|
| SL13180 (glgA::kan) | − | 6, 6, 6, 6, 7, 8 |
| SL13178 (pul::kan) | − | 5, 5, 5, 6, 6, 6 |
| UA159 | − | 21, 35, 35, 97, 104 |
| SL13180 (glgA::kan) | + | 51, 65, 66, 75 |
| SL13178 (pul::kan) | + | 70, 102, 126 |
| UA159 | + | 71, 75, 87, 95 |
| SL15072 [glgA::kan Pspac(Hy)] | − | 3, 3a |
| SL15074 [glgA::kan Pspac(Hy)-glgA] | − | 59, 59 |
These cultures were alive at day 3 and dead at day 10.
Bacteria were cultured in FMC containing 100 mM glucose and then transferred to FMC containing spent medium from cultures of the same strain grown in FMC with 6 mM glucose. Cultures did (+) or did not (−) contain 0.5% mucin.
FIG. 3.
Effect of glgA and pul mutations on the persistence of S. mutans cultures grown in FMC containing 100 mM glucose. After overnight growth, bacteria were transferred to spent medium without (A) or with (B) 0.5% mucin. Survival was determined as CFU obtained from 1 ml of culture. Filled squares, S. mutans UA159; open squares, glgA mutant SL13180; diamonds, pul mutant SL13178. In each case, the results of an experiment representative of at least three experiments are shown.
There was no difference between the mutants and the parent strain in the pH of cultures: the pH after 16 h of growth in FMC containing 100 mM glucose or 50 mM sucrose was ∼4.5; the corresponding cultures with 6 mM glucose or 3 mM sucrose were at pH ∼6.0, which was maintained throughout survival experiments. It is formally possible that the mutants were more sensitive to stress even though they were not subject to lower pH than was the parent. However, we detected no difference between the mutants and the parent strain in their sensitivity to the oxidative stress caused by 0.2% hydrogen peroxide, suggesting that the mutants did not display a general sensitivity to stress.
The survival of the parent was longer than that obtained by Renye et al. (22) because cultures were incubated in an atmosphere containing 5% CO2 as opposed to ambient atmosphere in the presence of bicarbonate buffer. We think it likely that the bicarbonate buffer failed to maintain an adequate CO2 environment, and so survival was impaired.
The IPS content of the parental strain UA159 and the pul mutant had declined substantially within 1 day of transfer to the starved medium and by day 3 approached background level, comparable to the level in the glgA mutant (Fig. 2C). This result is in good agreement with previous studies of oral bacteria, which showed that the intracellular glycogen reserves are largely metabolized within 12 h of the removal of sugar (11, 13, 32). Thus, the effect of IPS accumulation on survival extends well beyond its utilization.
We had previously found that inclusion of 0.5% mucin in FMC could extend the survival of batch cultures of the parental S. mutans strain UA159 (22). Addition of 0.5% mucin also extended the survival of the glgA and pul mutants grown in FMC containing 100 mM glucose and transferred to spent medium (Fig. 3B). Indeed their survival reached values comparable to those of the parental strain UA159 (Table 3). Addition of 0.5% starch had an effect very similar to that of the addition of mucin. The mucin had little if any effect on the accumulation or metabolism of IPS by strain UA159 or the pul mutant (data not shown); the presence of exogenous starch interfered with the assay for IPS.
IPS deficiency and the persistence of S. mutans in biofilms.
To test the importance of IPS in biofilm survival, we established flow cell biofilms using FMC with 50 mM sucrose and 15 mM NaHCO3 for 10 h. The biofilms were then perfused with fresh FMC containing NaHCO3 but lacking sucrose. Survival of bacteria in the biofilms was monitored by dispersing the bacteria and testing their viability on TH agar. Under these conditions, UA159 survived for about 8 days. There was no significant difference in survival between the parental strain and the glgA and pul mutants. Nor was there any difference in biofilm structure. It is plausible to think that strain UA159 would have accumulated IPS, although the presence of the extracellular matrix prevented direct determination of IPS. If this inference is correct, then IPS did not support longer survival of the parental strain, in contrast to the effect observed with batch cultures.
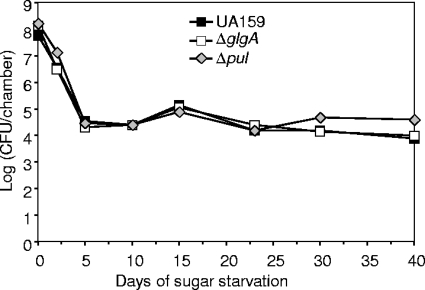
A significant difference between the studies of batch cultures and those of flow cell biofilms was the way in which sugar starvation was imposed. With batch cultures, starvation was imposed by transferring bacteria from sugar-rich growth medium into spent medium from a sugar-limited culture. In contrast, sugar starvation was induced in flow cell biofilms by sudden removal of sucrose from the fresh medium being supplied to the biofilm. Our studies with batch cultures indicate that the difference in sugar starvation conditions between spent medium and fresh medium can be important for survival (J. A. Renye, Jr., P. J. Piggot, and B. A. Buttaro, unpublished data); it may be that the spent medium contains metabolites that prolong survival. To more closely reproduce the starvation conditions of the batch cultures, we developed static biofilms in 24-well microtiter plates. Biofilms were established for 14 h by using FMC containing 50 mM sucrose. They were then placed in spent medium from 3 mM sucrose biofilms (which mimics the batch culture setup). The biofilms for all strains were viable for more than 40 days. There was no difference in persistence between the mutants and the parental strain under the conditions tested (Fig. 4).
FIG. 4.
Persistence of S. mutans strains in static biofilms. Wells of microtiter plates were loaded with cultures in FMC containing 50 mM sucrose. After 14 h the supernatants and associated planktonic bacteria were removed and replaced with spent medium. Filled squares, S. mutans UA159; open squares, glgA mutant SL13180; diamonds, pul mutant SL13178. Results of an experiment representative of three experiments are shown.
We also tested the possibility that the difference between batch cultures and biofilms was a consequence of the sugar used (glucose for batch cultures versus sucrose for biofilms) by examining the survival of bacteria grown in batch cultures with sucrose instead of glucose. Bacteria were grown in FMC containing 50 mM sucrose and then transferred to spent medium from bacteria grown in FMC containing 3 mM sucrose. Under these conditions, the bacteria grew embedded in extracellular polysaccharides, forming large aggregates, which sedimented to the bottom of the culture tubes. The glgA and pul mutants persisted for times comparable to that for the parental strain UA159; experiments were terminated after approximately 40, 80, and 240 days, and in each case all the strains were still viable.
Inactivation of fruAB did not influence persistence of S. mutans.
The results reported above indicate that IPS extended survival when glucose was a carbon source but not when sucrose was a carbon source. During growth in excess sucrose, S. mutans synthesizes two types of extracellular polysaccharides, fructan and glucan. Fructan is thought to be a reserve carbohydrate, ready to be used when the environment is depleted of nutrients. In contrast, glucan is primarily used for the assembly of the extracellular matrix, which is an essential part of a biofilm (24, 36). We hypothesized that fructan accumulated during growth in excess sucrose could be used as a source of carbohydrate during starvation, through the hydrolytic activity of fructanase, and thus could help in extending survival regardless of the presence of IPS. The fru operon in S. mutans UA159 consists of fruA and fruB genes (Fig. 1B). fruA encodes a fructanase (FruA), which is thought to be the main enzyme responsible for the degradation of fructan. fruB encodes a similar enzyme, but its role in the degradation of fructan is minor (12, 14, 24).
To test for the role of fructan, we inactivated the fruA and fruB genes in the parental strain, UA159, and in the glgA and pul mutants. Accumulation of IPS was not impaired by inactivation of the fruAB genes (data not shown). To test the survival, we cultured the fruAB mutant and the glgA fruAB and pul fruAB double mutants under conditions that permitted IPS accumulation (batch cultures in FMC containing 50 mM sucrose). Then the bacteria were washed and starved by switching to sterile spent medium from cultures of the same strains grown in 3 mM sucrose. The fruAB mutation did not impair the survival of the parental strain or of the glgA and pul mutants; experiments were terminated after approximately 40 and 80 days, and in each case all the strains were viable.
DISCUSSION
In this study, we report that inactivation of the glgA gene prevents formation of IPS by S. mutans (Fig. 2). The homologous glgA gene of other species encodes glycogen synthase, responsible for synthesis of α-1,4-glucan chains using ADP-glucose as a glucose donor (15, 29). Insertional inactivation of glgA blocked IPS synthesis. Introduction of an intact plasmid-borne copy of glgA restored IPS synthesis to the glgA mutant. Thus, we consider that loss of IPS synthesis was not a polar effect on glgP. Rather, we infer that glgA is the structural gene for IPS synthase in S. mutans; further, that glgB encodes glucan branching enzyme; and that glgC and glgD encode subunits of glucose 1-phosphate pyrophosphorylase. Previous studies by Spatafora and coworkers have shown that inactivation of dltA also impairs IPS synthesis; the dltA gene encodes a putative d-alanine-activating enzyme, so that its involvement in IPS synthesis is indirect (26).
IPS accumulates toward the end of exponential growth when S. mutans is grown in the chemically defined medium FMC containing excess glucose or sucrose (9, 10) (Fig. 2). We have found that inactivation of glgA severely impaired the persistence of bacteria grown in batch culture with excess glucose and then transferred to spent medium lacking glucose. The mutant survived for only about 6 days in stationary phase, whereas the parental strain persisted for at least 21 days and generally for substantially longer, as did the glgA mutant when a plasmid-borne intact copy of glgA had been introduced into the mutant. Yet the IPS was largely used up within 24 h of transfer of the bacteria to FMC lacking glucose (Fig. 2C), which is consistent with previous observations (11, 13, 32). Thus, the advantage of the IPS to the parent strain is probably not as an energy reserve that is parsimoniously used up over an extended period. Rather, the advantage may consist of the priming by IPS, within the first day of starvation, of some metabolic pathway and/or trait that leads to the long-term persistence. The effect is unlikely to result from a distinctive secreted nutrient or signal, because survival depended on the strains themselves and not on any difference between their culture supernatants. Intriguingly, the survival of the pul mutant was as severely impaired as that of the glgA mutant (Fig. 3A; Table 3). This result lends support to the idea that some feature of IPS metabolism, distinct from providing nutrients, is important for persistence.
The protein encoded by the pul gene has putative N-terminal pullulanase and C-terminal alpha-amylase domains, potentially able to act on pullulan, amylopectin, or glycogen (20, 35). The pul gene lies directly upstream from the glg operon, suggesting that the encoded protein might be involved in determining IPS branch structure and/or IPS metabolism. Inactivation of pul did not impair the accumulation of IPS or, within the limits of our assay, its degradation (Fig. 2). This result indicates that the mutation did not have a polar effect on expression of the glg operon, although we were unable to clone the intact pul gene and did not directly test by complementation for possible polar effects. However, the effects on survival of pul inactivation were very similar to those of glgA inactivation under the conditions tested (Fig. 3 and 4). Thus, we think it plausible that the putative pullulanase is indeed involved in determining IPS properties. The difference in survival between the pul mutant and the parental strain UA159 suggests that the strains differ in the structure of IPS and/or the path of its degradation. Plausibly, the degradation products of IPS in the pul mutant cannot prime the metabolic trait that confers an advantage in survival on the parental strain. However, we cannot exclude an alternative explanation, that pul inactivation affects survival in some way that is unrelated to IPS metabolism.
IPS conferred a clear advantage for persistence in batch cultures with glucose as carbon source. In contrast, no advantage was observed when mucin or starch was added to the medium or when sucrose was used as a carbon source. Thus, under conditions that may be closer to those of the dental plaque than batch culture with glucose, IPS had at most a redundant role in promoting survival. Sucrose enables S. mutans to synthesize extracellular glucan from the glucosyl moiety and fructan from the fructosyl moiety of the sucrose. The glucan functions as an extracellular matrix to facilitate adherence of the bacteria to the dental plaque (7). However, the fructan is thought to function primarily as a reserve of carbohydrate, ready to be used when the environment is depleted of nutrients (36); it seemed a plausible candidate to serve as an alternative to IPS in enhancing survival. Fructan utilization by S. mutans is through the action of the fructanase encoded by fruA (23, 36). The fruA gene is cotranscribed with fruB, which encodes a similar protein, although no fructanase activity has been found for FruB (3). To curtail possible use of fructan as a source of carbohydrate during survival, we deleted both genes of the fruAB operon. The deletion did not affect the survival of strain UA159 or of the glgA mutant when the strains were grown in the presence of sucrose (with no mucin) either in batch cultures or in biofilms. Thus, within the limits of this test, fructan utilization did not explain the persistence of the glgA mutant when sucrose was used as a carbon source. It is possible that some other (still-unknown) enzyme can degrade fructan or that the extracellular glucan might be utilized (7, 25). Indeed, we think it possible that various carbon sources are able to support survival, although they are not efficiently utilized and are not able to support growth.
This possibility is supported by several observations. First, S. mutans survives in spent medium after exhaustion of glucose where the only carbon and energy sources available are either lactic acid, which could potentially be utilized via pyruvate metabolism, or amino acids, which could enter pyruvate metabolism or be directly used to produce ATP. In our studies, IPS may influence those pathways. Second, mucin is insufficient to promote growth of S. mutans (31) but is capable of prolonging survival (22) (Fig. 3). In addition, S. mutans cannot grow with starch as a sole carbon source (25), but we found that starch also prolonged survival. The biofilm matrix of S. mutans contains both fructans and glucans that potentially could be metabolized. While these disparate carbon sources are not used as a primary carbon and energy source, they can potentially be used to prolong survival. Further experimentation is necessary to determine if these various compounds are being used as carbon and energy sources or if they are somehow playing a protective role, preventing the cells from being damaged during survival.
Acknowledgments
This work was supported by Public Health Service grant DE14640 to P.J.P. from the National Institutes of Health.
Footnotes
Published ahead of print on 2 October 2009.
REFERENCES
- 1.Ballicora, M. A., A. A. Iglesias, and J. Preiss. 2003. ADP-glucose pyrophosphorylase, a regulatory enzyme for bacterial glycogen synthesis. Microbiol. Mol. Biol. Rev. 67:213-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burleigh, I. G., and E. A. Dawes. 1967. Studies on the endogenous metabolism and senescence of starved Sarcina lutea. Biochem. J. 102:236-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burne, R. A., K. Schilling, W. H. Bowen, and R. E. Yasbin. 1987. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J. Bacteriol. 169:4507-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlsson, J. 1997. Bacterial metabolism in dental biofilms. Adv. Dent. Res. 11:75-80. [DOI] [PubMed] [Google Scholar]
- 5.Chary, V. K., M. Busuioc, J. A. Renye, Jr., and P. J. Piggot. 2005. Vectors that facilitate the replacement of transcriptional lacZ fusions in Streptococcus mutans and Bacillus subtilis with fusions to gfp or gusA. FEMS Microbiol. Lett. 247:171-176. [DOI] [PubMed] [Google Scholar]
- 6.Claverys, J. P., A. Dintilhac, E. V. Pestova, B. Martin, and D. A. Morrison. 1995. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene 164:123-128. [DOI] [PubMed] [Google Scholar]
- 7.Colby, S. M., and R. R. Russell. 1997. Sugar metabolism by mutans streptococci. Soc. Appl. Bacteriol. Symp. Ser. 26:80S-88S. [PubMed] [Google Scholar]
- 8.Dawes, E. A., and D. W. Ribbons. 1964. Some aspects of the endogenous metabolism of bacteria. Bacteriol. Rev. 28:126-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiPersio, J. R., S. J. Mattingly, M. L. Higgins, and G. D. Shockman. 1974. Measurement of intracellular iodophilic polysaccharide in two cariogenic strains of Streptococcus mutans by cytochemical and chemical methods. Infect. Immun. 10:597-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiPersio, J. R., S. J. Mattingly, M. L. Higgins, and G. D. Shockman. 1978. A quantitative ultrastructural and chemical investigation of the accumulation of iodophilic polysaccharide in two cariogenic strains of Streptococcus mutans. Microbios 21:109-126. [PubMed] [Google Scholar]
- 11.Freedman, M. L., and A. L. Coykendall. 1975. Variation in internal polysaccharide synthesis among Streptococcus mutans strains. Infect. Immun. 12:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Germaine, G. R., S. K. Harlander, W. L. Leung, and C. F. Schachtele. 1977. Streptococcus mutans dextransucrase: functioning of primer dextran and endogenous dextranase in water-soluble and water-insoluble glucan synthesis. Infect. Immun. 16:637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibbons, R. J., and S. S. Socransky. 1962. Intracellular polysaccharide storage by organisms in dental plaques. Its relation to dental caries and microbial ecology of the oral cavity. Arch. Oral Biol. 7:73-79. [DOI] [PubMed] [Google Scholar]
- 14.Igarashi, T., A. Yamamoto, and N. Goto. 1992. Characterization of the dextranase purified from Streptococcus mutans Ingbritt. Microbiol. Immunol. 36:969-976. [DOI] [PubMed] [Google Scholar]
- 15.Kiel, J. A., J. M. Boels, G. Beldman, and G. Venema. 1994. Glycogen in Bacillus subtilis: molecular characterization of an operon encoding enzymes involved in glycogen biosynthesis and degradation. Mol. Microbiol. 11:203-218. [DOI] [PubMed] [Google Scholar]
- 16.Lindler, L. E., and F. L. Macrina. 1986. Characterization of genetic transformation in Streptococcus mutans by using a novel high-efficiency plasmid marker rescue system. J. Bacteriol. 166:658-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer, R. J., Jr. 1999. Microscopy flowcells: perfusion chambers for real-time study of biofilms. Methods Enzymol. 310:160-166. [DOI] [PubMed] [Google Scholar]
- 19.Perry, D., and H. K. Kuramitsu. 1981. Genetic transformation of Streptococcus mutans. Infect. Immun. 32:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plant, A. R., R. M. Clemens, H. W. Morgan, and R. M. Daniel. 1987. Active-site- and substrate-specificity of Thermoanaerobium Tok6-B1 pullulanase. Biochem. J. 246:537-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quisel, J. D., W. F. Burkholder, and A. D. Grossman. 2001. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J. Bacteriol. 183:6573-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renye, J. A., Jr., P. J. Piggot, L. Daneo-Moore, and B. A. Buttaro. 2004. Persistence of Streptococcus mutans in stationary-phase batch cultures and biofilms. Appl. Environ. Microbiol. 70:6181-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schachtele, C. F., A. E. Loken, and M. K. Schmitt. 1972. Use of specifically labeled sucrose for comparison of extracellular glucan and fructan metabolism by oral streptococci. Infect. Immun. 5:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schachtele, C. F., R. H. Staat, and S. K. Harlander. 1975. Dextranases from oral bacteria: inhibition of water-insoluble glucan production and adherence to smooth surfaces by Streptococcus mutans. Infect. Immun. 12:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson, C. L., and R. R. Russell. 1998. Intracellular alpha-amylase of Streptococcus mutans. J. Bacteriol. 180:4711-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spatafora, G., K. Rohrer, D. Barnard, and S. Michalek. 1995. A Streptococcus mutans mutant that synthesizes elevated levels of intracellular polysaccharide is hypercariogenic in vivo. Infect. Immun. 63:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spatafora, G. A., M. Sheets, R. June, D. Luyimbazi, K. Howard, R. Hulbert, D. Barnard, M. el Janne, and M. C. Hudson. 1999. Regulated expression of the Streptococcus mutans dlt genes correlates with intracellular polysaccharide accumulation. J. Bacteriol. 181:2363-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strange, R. E. 1968. Bacterial “glycogen” and survival. Nature 220:606-607. [DOI] [PubMed] [Google Scholar]
- 29.Takata, H., T. Takaha, S. Okada, M. Takagi, and T. Imanaka. 1997. Characterization of a gene cluster for glycogen biosynthesis and a heterotetrameric ADP-glucose pyrophosphorylase from Bacillus stearothermophilus. J. Bacteriol. 179:4689-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Hoeven, J. S., C. W. van den Kieboom, and P. J. Camp. 1990. Utilization of mucin by oral Streptococcus species. Antonie van Leeuwenhoek 57:165-172. [DOI] [PubMed] [Google Scholar]
- 32.van Houte, J., C. E. de Moor, and H. M. Jansen. 1970. Synthesis of iodophilic polysaccharide by human oral streptococci. Arch. Oral Biol. 15:263-266. [DOI] [PubMed] [Google Scholar]
- 33.van Houte, J., and H. M. Jansen. 1970. Role of glycogen in survival of Streptococcus mitis. J. Bacteriol. 101:1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 35.Walker, G. J. 1968. Metabolism of the reserve polysaccharide of Streptococcus mitis. Some properties of a pullulanase. Biochem. J. 108:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wexler, D. L., J. E. Penders, W. H. Bowen, and R. A. Burne. 1992. Characteristics and cariogenicity of a fructanase-defective Streptococcus mutant strain. Infect. Immun. 60:3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]