Abstract
The σE-directed envelope stress response maintains outer membrane homeostasis and is an important virulence determinant upon host infection in Escherichia coli and related bacteria. σE is activated by at least two distinct mechanisms: accumulation of outer membrane porin precursors and an increase in the alarmone ppGpp upon transition to stationary phase. Expression of the σE regulon is driven from a suite of approximately 60 σE-dependent promoters. Using green fluorescent protein fusions to each of these promoters, we dissected promoter contributions to the output of the regulon under a variety of in vivo conditions. We found that the σE promoters exhibit a large dynamic range, with a few strong and many weak promoters. Interestingly, the strongest promoters control either transcriptional regulators or functions related to porin homeostasis, the very functions conserved among E. coli and its close relatives. We found that (i) the strength of most promoters is significantly affected by the presence of the upstream (−35 to −65) region of the promoter, which encompasses the UP element, a binding site for the C-terminal domain of the α-subunit of RNA polymerase; (ii) ppGpp generally activates σE promoters, and (iii) σE promoters are responsive to changing σE holoenzyme levels under physiological conditions, reinforcing the idea that the σE regulon is extremely dynamic, enabling cellular adaptation to a constantly changing environment.
In bacteria, a repertoire of σs directs RNA polymerase to discrete sets of promoters. The housekeeping σ directs transcription from promoters of the thousands of genes encoding functions required generally for growth, whereas the alternative σs orchestrate specialized responses to cellular, developmental, and environmental signals (19, 23). Most alternative σs transcribe on the order of 50 to 100 genes and use few auxiliary regulatory factors. These characteristics make it feasible to examine the behaviors of an entire set of promoters transcribed by a particular σ, thereby determining how the promoters themselves contribute to the transcriptional output of the response. In this report, we analyze the behaviors of promoters recognized by σE, an alternative σ factor in Escherichia coli.
The σE-directed envelope stress response maintains outer membrane homeostasis and is an important virulence determinant upon host infection in E. coli and related bacteria (22, 41). Studies across several organisms have revealed that a small core group of promoters is conserved across these organisms, whereas a larger group varies between organisms (38). The two most conserved promoters drive regulatory circuits: for the transcription of σE itself and for σ32, responsible for the cytoplasmic heat shock response. A second core conserved group of promoters drives genes with a coherent function primarily directed to the synthesis and/or assembly primarily of porins and to a lesser extent lipopolysaccharide (LPS), and together these constitute the two unique constituents of the outer membrane (32). This group includes the two small RNAs (sRNAs) that downregulate the expression of several porins (5, 25, 34, 48, 49), all four members of the recently discovered machine that inserts β-barrel proteins into the outer membrane (44, 53), and the chaperones that deliver porin precursors to the β-barrel insertion machine. Indeed, upon inactivation of σE in E. coli, cells lyse and bleb, indicating its key role in envelope integrity (22). The more variable members of the regulon are enriched for functions important in pathogenesis, suggesting that this response may be an early warning system to bacteria entering a host environment (38).
Three different signals are believed to alter the activity of σE. First, σE activity is sensitive to the assembly status of porins (1, 29). This fairly well-characterized signal transduction pathway involves destruction of RseA, the membrane-spanning anti-sigma factor that inactivates σE (1-2). RseA binding to σE holds it in an inactive state during normal growth (13, 17, 30). Upon accumulation of unfolded porins, DegS cleavage of RseA initiates a protease cascade that degrades RseA, thereby increasing the pool of σE able to form holoenzyme and initiate transcription. σE holoenzyme transcribes both itself and its negative regulator. Therefore, as long as the signal persists, σE increases but its negative regulator is rapidly degraded (3, 9, 16, 26).The DegS protease itself is the sensor of the porin signal (51). The protease domain of DegS is held in an inactive state by binding to its PDZ domain (45, 46). Inhibition is relieved when the DegS PDZ domain binds a YXF motif located in the C terminals of most porins and several other outer membrane proteins. As porin C termini are buried in the trimer interface of assembled porins, accumulation of exposed YXF residues signals a block in porin assembly. Thus, this signal ties activation of σE to porin status in the cell. Second, it has been proposed that σE activity is sensitive to the status of LPS, although a molecular mechanism has not been elucidated (47). A plausible scenario is that this pathway is the second arm of the stress pathway just described. LPS or some derivative of it could bind to the N-terminal domain of RseB, which is structurally very similar to the lipoprotein binding domain of LolB (28, 52). RseB is a periplasmic protein that binds tightly to RseA and inhibits cleavage by DegS (8). Binding of an LPS-related substrate to RseB could weaken its interaction with RseA so that DegS cleavage could ensue. These two signals together could constitute a “stress” arm of the response, enabling the response to stress-induced imbalances in the outer membrane of the cell.
A third mode of regulation, independent of the RseA and RseB pathway, activates σE in response to intracellular signals related to growth phase and nutrient availability (10, 11). σE activity is significantly upregulated as cells enter stationary phase. ppGpp and its coactivator DksA mediate this response in two different ways: (i) directly, by facilitating transcription by σE holoenzyme, as demonstrated in in vitro transcription studies (11); (ii) indirectly, by redistributing the σ70 holoenzyme away from rRNA operons (responsible for >50% of the transcription during exponential phase [6]), thereby increasing the availability of RNA polymerase for σE-dependent transcription. Indirect effects are possible because it is believed that there are more σs than core RNA polymerase in the cell, resulting in competition among σs for binding to core RNA polymerase (18). When RNA polymerase is relieved from rRNA transcription, more RNA polymerase is either free or nonspecifically bound to DNA and able to freely exchange its σs, thereby increasing the ability of σs to bind to core RNA polymerase and mediate transcription. Additionally, ppGpp may increase the ability of alternative σs to complete for RNA polymerase (reviewed in reference 33). Taken together, it is clear that the σE holoenzyme is under complex control in the bacterial cell.
Thus far, there have been only a few quantitative in vivo dissections of promoter contributions to a regulon, and most of these examined variations in transcription factor binding sites. For example, different configurations of PhoP binding sites have been correlated with promoter strength (31, 35, 36), and differential responses of Cpx-regulated promoters have been examined (37). Only a single study has focused on the strength of promoters driven by alternative σs. Expression driven by the regions upstream of putative σS genes has been examined, but this analysis used 500-bp regions and did not dissect contributions of particular promoters (43).
In the present work, we explore the contributions of all previously validated σE-dependent promoters to the complex σE holoenzyme response. This includes unique σE-dependent promoters from two E. coli strains (K-12 MG1655 and CFT073) and Salmonella typhimurium (38). We show that the strongest promoters control either transcriptional regulators or functions related to porin homeostasis, both core functions of the regulon, that the strength of most promoters is significantly affected by the presence of the upstream (−35 to −65) region of the promoter, the region encompassing the UP element, a binding site for the C-terminal domain of the α-subunit of RNA polymerase, that ppGpp generally activates promoters, an inference that had thus far been derived from only a few promoters, and that promoter output is responsive to physiologically relevant variations in the amount of σE.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Strains and plasmids used in this study are listed in Table 1. All strains were grown in Luria-Bertani (LB) broth or M9 complete minimal medium supplemented with appropriate antibiotics at 30°C with shaking. M9 complete minimal medium was prepared as described previously (42) and supplemented with 0.2% glucose, 1 mM MgSO4, vitamins, and all amino acids (40 μg/ml). Medium was supplemented with 30 μg/ml kanamycin, 100 μg/ml ampicillin, 10 μg/ml tetracycline, and/or 20 μg/ml chloramphenicol as required.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype | Source, reference, or P1 donor |
|---|---|---|
| E. coli strains | ||
| MG1655 (CAG45113) | K-12 (MG1655) rph-1 | E. coli Genetic Stock Center (20, 24) |
| MC1061 | K-12 araD Δ(ara-leu)7697 Δ(codB-lacI) galK16 galE15 mcrA0 relA1 rpsL150 spoT1 mcrB9999 hsdR2 | E. coli Genetic Stock Center (7) |
| CAG 22216 | MC1061 ΔlacX74 [φλrpoH P3::lacZ] rpoE::ΩCm | 14 |
| CAG45114 (CAG25196) | MG1655 ΔlacX74 [φλrpoH P3::lacZ] | 38 |
| CAG58200 (CAG25197) | CAG45114 pLC245 | 38 |
| CAG55701 to -55760 | CAG58200 pUA E1 to E60 (long σE promoter library; 60 strains)a | This work |
| CAG58301 to -58360 | CAG58200 pUA Et1 to Et60 (short σE promoter library; 60 strains)a | This work |
| CAG55601 to -55660 | CAG22216 pUA E1 to E60 (long σE promoter library; 60 strains)a | This work |
| CAG58140 to -58197, -58138 and -9 | CAG22216 pUA Et1 to Et60 (short σE promoter library; 60 strains)a | This work |
| CAG55801 to -55860 | CAG45113 pUA E1 to E60 (long σE promoter library; 60 strains)a | This work |
| CAG58201 to -58260 | CAG45113 pUA Et1 to Et60 (short σE promoter library; 60 strains)a | This work |
| CAG55761 | CAG58200 pUA66 | This work |
| CAG55661 | CAG22216 pUA66 | This work |
| CAG55861 | CAG45113 pUA66 | This work |
| CAG55906 | CAG45114 ΔrelA | This work, P1 ΔrelA::kan (Keio collection [4]) |
| CAG55907 | CAG55906 ΔspoT::cam | This work, P1 SEA2010 (11) |
| CAG55912 to -55921 | CAG55907 pUA E1 to E60 (select long σE promoters; 10 strains)a | This work |
| CAG55934 to -55943 | CAG55907 pUA Et1 to Et60 (select short σE promoters; 10 strains)a | This work |
| CAG55922 | CAG55907 pUA66 | This work |
| ΔrelA::kan (Keio collection) | BW25113 (K-12 rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1) ΔrelA::kan | Keio collection (4) |
| SEA2010 | MG1655 ΔlacX74 [φλrpoH P3::lacZ] ΔrelA251::kan ΔspoT207::cam | 11 |
| Plasmids | ||
| pUA66 | Vector, SC101 ori, Kanr GFP reporter plasmid carrying gfpmut2 | 55 |
| pUA E1-E60 | Long σE promoter library, used to measure activity of σE-dependent long promoter fragments (−65 to +20) cloned in XhoI-BamHI sites upstream of gfpmut2 in pUA66 (60 constructs)a | This work |
| pUA Et1-Et60 | Short σE promoter library, used to measure activity of σE-dependent short promoter fragments (−35 to +20) cloned in XhoI-BamHI sites upstream of gfpmut2 in pUA66 (60 constructs)a | This work |
| pTrc99A | Vector, pBR322 ori, Apr, expression vector containing an IPTG-inducible trc promoter | 38 |
| pLC245 | rpoE cloned in pTrc99A downstream of the IPTG-inducible trc promoter, Apr | 38 |
See Table S1 in the supplemental material.
Construction of assay strains.
Assay strains and promoters are listed in Table S1 of the supplemental material. All assay strains are derivatives of E. coli K-12 strains MG1655 and MC1061. Transformations were performed using standard methods; P1 transductions were performed using P1 vir, and transductants were selected on LB plates with appropriate antibiotics. Assay strains are derivatives of strains CAG22216, CAG45113, CAG55907, and CAG58200, transformed with derivatives of the green fluorescent protein (GFP) expression vector, pUA66, carrying the long and short σE promoter libraries (see Table S1 in the supplemental material). Transformants were grown overnight in LB medium supplemented with appropriate antibiotics in deep 96-well plates and stored as glycerol stocks. Assays in the absence of σE, or with basal levels of σE, were performed with derivatives of CAG22216 (MC1061 ΔlacX74 [φλrpoHP3::lacZ] rpoE::ΩCm) or CAG45113 (MG1655), respectively. Assays with overexpression of σE were performed with derivatives of CAG58200 (MG1655 ΔlacX74 [φλrpoHP3::lacZ]) carrying the plasmid pLC245 expressing rpoE from the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Ptrc promoter. Assays with basal levels of σE in ppGpp0 strains were performed with derivatives of CAG55907 (MG1655 lacX74 [φλrpoHP3::lacZ] ΔrelA ΔspoT::cam).
The ppGpp0 strain CAG55907 was made as follows. First, strain CAG55906 was constructed by transducing the ΔrelA::kan allele from the Keio collection (4) into CAG45114 (MG1655 lacX74 [φλrpoHP3::lacZ]) and removing the kan marker by transforming with pCP20, screening for Kan sensitivity, and subsequently screening for loss of pCP20 (12). CAG55907 was then constructed by transducing CAG55906 with SEA2010, selecting for the ΔspoT207::cam allele. Alleles were confirmed by PCR, and the ppGpp0 phenotype was confirmed as an amino acid auxotroph (inability to grow on M9 minimal medium lacking amino acids [54]). All assays were repeated using at least two independent transductants.
Promoter library plasmid construction.
All σE promoter constructs were carried on the low-copy-number vector pUA66, driving the expression of GFP from the reporter gene gfpmut2. Promoter sequences are displayed in Table S1 of the supplemental material. The long σE promoter library (plasmids pUA E1 to E60) was constructed by PCR amplifying genomic sequences from −65 to +20 with respect to the transcription start point from E. coli K-12 MG1655, E. coli CFT073, and Salmonella enterica subsp. enterica serovar Typhimurium LT2 for each promoter, using forward primers containing an XhoI site and reverse primers containing a BamHI site. The PCR fragments were cloned into the XhoI-BamHI sites of pUA66, and final constructs were verified by sequencing. The short σE promoter library (plasmids pUA Et1 to pUA Et60) was constructed by PCR amplifying sequences from the −35 motif to +20 downstream of the transcription start point for each promoter, using the long promoter plasmid library as templates and forward primers containing an XhoI site and reverse primers containing a BamHI site. The −35 motif was identified by searching for the overrepresented GGAACTT motif in aligned promoter sequences as described by Rhodius et al. (38). As before, the PCR fragments were cloned into the XhoI-BamHI sites of pUA66, and final constructs were verified by sequencing.
In vivo promoter assays.
Assay strains were stored as glycerol stocks in deep 96-well plates (1 ml). Cultures were grown in 96-well plates containing 150 μl of M9 complete or LB medium with appropriate antibiotics, inoculating from glycerol stocks using a sterilized 96-pin metal pinner. Cultures were grown overnight (∼16 h) in 96-well U-bottom tissue culture plates covered with sterile breathable sealing film (sterile AeraSeal; E&K Scientific) at 30°C with shaking at 850 rpm on Elmi shakers. The following day the overnight cultures were diluted 1:200 into a final volume of 150 μl of fresh medium with appropriate antibiotics in clear-bottom black plates, covered with a clear lid, and incubated in a multimode microplate reader-incubator-shaker (Varioskan; Thermo Fisher Scientific). For strains overexpressing σE (derivatives of CAG58200) the fresh medium also contained 100 μM IPTG to induce rpoE expression. Cultures were grown for 8 h with orbital shaking at 480 rpm at 6-mm diameter. Repeated measurements of the optical density at 450 nm (OD450) for M9 complete (or OD600 for LB) and fluorescence (relative fluorescence units [RFU]; excitation at 481 nm and emission at 507 nm) were performed every 15 min. All experiments were repeated at least three times, with less than 5% standard error between replicate experiments. Differences in growth and expression profiles of randomly chosen strains between shake flask batch cultures and microplate reader plates were found to be negligible. This was also confirmed by changing the well positions of strains in the microplates to avoid any local effects.
Data analysis.
Culture background fluorescence was determined using a promoterless pUA66 vector transformed into the parent strains of each promoter library. This pUA66 vector strain was grown and assayed in triplicate with each promoter library. The triplicate data points were averaged and used to generate a standard curve for OD against fluorescence (RFU). This standard curve was then used to subtract background fluorescence from the reporter strain fluorescence value at the same OD (not necessarily at the same time point), generating a background-subtracted OD versus RFU differential rate plot for each reporter strain. The slope of the linear portion of each differential rate plot corresponds to the promoter activity of the specific promoter-GFP fusion in that reporter strain. Background fluorescence slopes (OD versus RFU) were between −2 and −0.8 for OD600 values between 0.7 and 1.4 and were ∼0 for an OD600 values between 1.5 and 2.7.
A standard curve was generated to compare OD values for cultures measured either as 150-μl samples in 96-well plates by the Varioskan or in a 1-ml path length cuvette by a regular spectrophotometer, in either M9 complete medium or LB broth. The conversion factors were 3.8 at OD450 for cultures grown on M9 complete medium and 3.4 at OD600 for cultures grown in LB broth. Using this conversion factor, the growth profiles of reporter strains were divided into exponential and stationary phases to compare with reported growth phase regulation of σE activity (11). The slopes of linear portions of the OD versus RFU differential plots at exponential and stationary phases of growth were then used to obtain the promoter activities at respective growth phases. The reported promoter activities are averages from at least three independent experiments.
Preliminary data analysis was performed using the SkanIT software for the Varioskan. Subsequent data analysis was performed automatically using Matlab software (Mathworks) with in-house-developed scripts (available upon request).
RESULTS AND DISCUSSION
Methodology of the experiments.
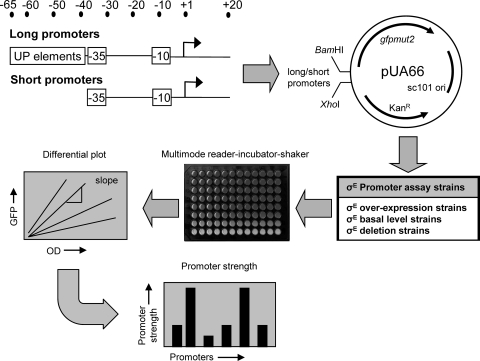
To quantitatively determine expression of all σE-dependent promoters in vivo, we created a series of GFP reporter strains, each carrying a derivative of the low-copy-number plasmid pUA66 with a unique σE promoter driving expression of a rapidly folding GFP. This promoter library contains two versions of each of the 60 validated σE promoters: a “short” promoter, extending from −35 to +20 relative to the start site of transcription, and a “long” promoter, extending from −65 to +20. Short promoters contain only the core promoter (−10 and −35 motifs), whereas long promoters include the upstream UP element sequences (39). Cultures were grown in 96-well plates in the multimode Varioskan, which allows aerated growth under controlled temperature conditions and automatically monitors both cell growth (OD) and GFP fluorescence at timed intervals. We measured the change in GFP fluorescence as a function of cell growth (differential plot). Promoter strength is taken as the slope of this line, and this parameter is plotted as a bar graph in all subsequent figures (Fig. 1). Control experiments by us (data not shown) and the Alon laboratory (27) indicate that plasmid copy number does not change under any of the conditions tested. The entire methodology is described in detail in Materials and Methods. We note that the σE-dependent rpoH promoter has a closely overlapping σ70 promoter that is present in our constructs (15). The commonly utilized σE-dependent rpoHP3::lacZ reporter does not contain this additional promoter (29).
FIG. 1.
Methodology for measuring the strength of σE-dependent promoters. For 60 σE-dependent promoters, short fragments containing the core −35/−10 elements (−35 to +20) and long fragments that also included the UP element (−65 to +20) were cloned into the GFP expression vector pUA66. These promoter plasmid libraries were transformed into assay strains that either overexpressed σE (derivatives of CAG58200), contained basal levels of σE (derivatives of CAG45113 and CAG55907), or with σE deleted (derivatives of CAG22216 [contains a suppressor of σE essentiality]). Strains were grown in 96-well plates at 32°C with aeration (shaking) in a multimode Varioskan. Cell growth (optical density) and GFP fluorescence (RFU) were measured every 15 min. For each strain, differential plots were constructed of GFP fluorescence versus cell growth. Promoter strength was calculated from the slope of the linear portions of the plot, corresponding to the exponential or stationary growth phase. All calculated promoter strengths were subtracted for background culture and medium fluorescence by using control assay strains carrying the promoterless GFP vector pUA66. See Materials and Methods for further details.
Activities of σE promoters under “stress activation” conditions.
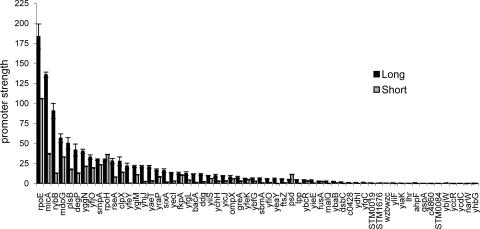
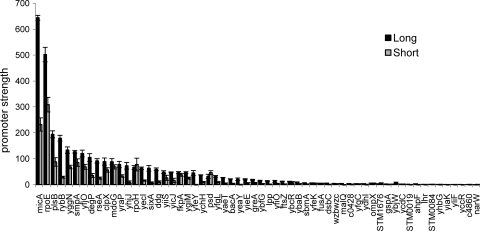
During stress, the anti-sigma RseA is degraded rapidly, freeing σE to carry out transcription (1); additionally, as σE transcribes its own gene (rpoE) (40), a positive feedback loop increases the amount of σE until stress subsides and degradation of RseA slows. To mimic these conditions, we overexpressed σE in cells growing in LB medium and examined transcription from the “long” and “short” derivatives of each of the validated σE promoters both during exponential-phase growth (OD600, 0.7 to 1.4) (Fig. 2) and during the transition to stationary phase (OD600, 1.5 to 2.7) (Fig. 3). In both figures, promoters are listed in ranked order from most to least active “long” promoter variant. We observed that in almost all cases, the “long” promoter variants were more active than the “short” constructs, suggesting that “UP element” binding by the α-C-terminal domain contributes to promoter strength. However, the magnitude of the contribution varied, with most promoters exhibiting ∼1.5- to 3.0-fold increases in activity (e.g., rpoE and clpX) but with some exhibiting as much as a 5- to 7-fold increase in activity (e.g., rybB and micA). Similar results were obtained with cells grown in M9 minimal medium supplemented with amino acids (data not shown). We note that our cloned promoter fragments end at +20. We chose this endpoint because we wanted to include the initial transcribed sequences, which can affect promoter escape. As a result, the stability and translation of GFP mRNA could differ in vivo. However, we have found a similar hierarchy of promoter activities using in vitro transcription assays (V. A. Rhodius, data not shown), indicating that the promoter-gfp reporter system provides a reasonable measure of promoter strength and that any influences on stability and translation of gfp mRNA are minimal.
FIG. 2.
Exponential-phase strengths of long and short σE-dependent promoters after σE overexpression. Promoter strengths are shown for the long and short σE promoter plasmid libraries in derivatives of the σE overexpression strain CAG58200, carrying the rpoE overexpression plasmid pLC245 during exponential-phase growth (OD600 of 0.7 to 1.4) at 32°C in LB. σE overexpression was induced by growth in 100 μm IPTG. The bars represent the averages of three independent experiments, and error bars represent 1 standard deviation.
FIG. 3.
Stationary-phase strengths of long and short σE-dependent promoters after σE overexpression. Promoter strengths are shown for the long and short σE promoter plasmid libraries in derivatives of the σE overexpression strain CAG58200, carrying the rpoE overexpression plasmid pLC245, during stationary-phase growth (OD600 of 1.5 to 2.7) at 32°C in LB. σE overexpression was induced by growth in 100 μm IPTG. The bars represent the averages of three independent experiments, and error bars represent 1 standard deviation.
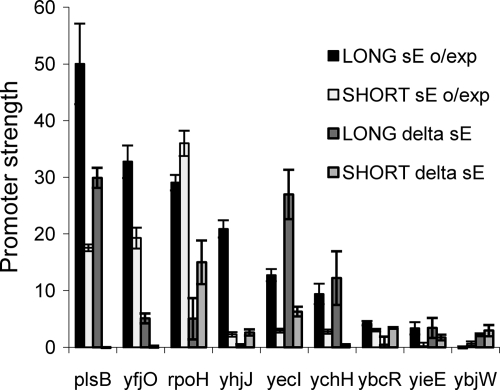
We tested whether any of the promoters had significant σE-independent activity by assaying them in a strain lacking σE but with a mutation that suppresses σE essentiality (14) (Fig. 4). σE-independent activity was detected from only nine promoters. For three of these (plsB, yfjO, and rpoH), both the “long” and “short” forms of the promoters were more active in the presence of σE, suggesting that they are predominantly driven by σE. The six remaining promoters (yhjJ, yieE, ybcR, ybjW, yecI, and ychH) had approximately similar activities with and without σE, for either or both the long and short forms. However, in vitro transcriptions assays identified σE-dependent transcripts from four of these promoters (yhjJ, yieE, ybcR, and ychH) (data not shown), suggesting that these sequences are still recognized by σE and, in addition, are also regulated by other σs. We have previously mapped the transcript start sites of four of the nine promoter regions (plsB, rpoH, yhjJ, and yieE) using 5′ random amplification of cDNA ends (38). All four regions show σE-dependent starts as well as adjacent σE-independent starts that are absent or much weaker in the presence of σE, indicating nearby promoters that are outcompeted in the presence of σE. In toto, these data show that most σE promoters are completely σE dependent with little direct overlap of promoters regulated by other σs.
FIG. 4.
σE-independent strengths of the long and short promoters. The graph compares long and short promoter strengths in σE overexpressing (o/exp; derivatives of CAG58200 carrying the rpoE overexpression plasmid pLC245) and no-σE (delta sE; derivatives of CAG22216) strains. Only nine promoters gave significant σE-independent activities. All promoter strengths were determined from exponential-phase cultures (OD600 of 0.7 to 1.4) grown at 32°C in LB. σE overexpression was induced by growth in 100 μm IPTG. The bars represent the averages of three independent experiments, and error bars represent 1 standard deviation.
Examination of promoter strength data from the exponential phase of growth revealed that about 40 promoters have significant activity, and their promoter strength varies widely (∼100-fold). Interestingly, the most highly active “long” promoters are predominantly comprised of those carrying out conserved functions of σE: regulatory transcription of σE and its anti-sigma factor (rpoE and rseA), as well as σ32 (rpoH), and functions related to porin homeostasis, the two sRNAs responsible for downregulating porins (rybB and micA) and portions of the machinery required for insertion of proteins into the outer membrane (yaeT, smpA, and yfgL) and the degP chaperone, implicated in porin transport. Importantly, almost all of the 10 most active “long” promoters are also the most active “short” promoters, indicating that core promoter strength contributes heavily to expression. The promoter driving rybB is an interesting exception, as its “UP” element boosts strength by about sevenfold, making the strength of this promoter heavily dependent on its UP element.
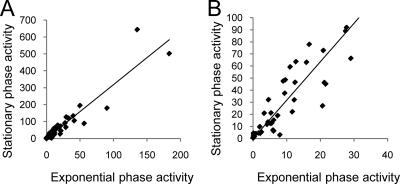
Comparison of promoter strength data between exponential and stationary phases of growth revealed that most promoters increase in strength around threefold with remarkably little deviation from this, apart from promoters that remain inactive under both conditions (Fig. 5). Consequently, the hierarchy of promoter strength remains relatively constant (compare Fig. 2 and 3). For example, 8 of the 10 most active promoters in exponential phase are among the 10 most active in stationary phase, and those promoters that do switch positions in the hierarchy generally show only small changes (e.g., rpoE is the most active promoter in exponential phase and the second most active promoter in stationary phase). Increased activity of σE promoters during stationary phase has been carefully studied for the rpoH promoter, and it has been concluded that this effect derives from direct and indirect effects of increased ppGpp during this growth phase (10, 11). The results presented here (see Fig. 7 and other findings, below) indicate that most σE-controlled promoters are subject to this effect and that promoters both with and without an UP element are activated during this growth phase. These data suggest that under these conditions most promoters increase their activity by similar amounts upon entry into stationary phase.
FIG. 5.
Comparison of stationary-phase and exponential-phase promoter strengths. Scatter plots comparing long promoter strengths (A) and short promoter strengths (B) during stationary and exponential phase after σE overexpression. Experimental data are from Fig. 2 and 3.
FIG. 7.
The requirement for ppGpp in the strength of σE-dependent promoters. The graphs show promoter strengths of the top 10 active long (A) and short (B) promoters under basal levels of σE expression during exponential (expo) or stationary (stat) phases of growth in either wild-type (derivatives of CAG45113) or ppGpp0 (derivatives of CAG55907; ΔrelA ΔspoT::cam) strains. The bars represent the averages of three independent experiments, and error bars represent 1 standard deviation.
The activity of σE promoters under normal growth conditions.
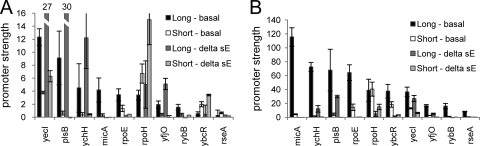
The experiments performed thus far used σE overexpression conditions to examine behavior of promoters during stress. However, during normal cell growth, the amount of active σE in the cell is much lower. It is of great interest to compare promoter behavior under σE-limiting (basal) and σE excess (overexpression) conditions. The high autofluorescence of E. coli cells limits reliable detection (promoter activity is ≥2-fold autofluorescence) to only the 10 most active promoters (Fig. 6). Six of these promoters have some σE-independent activity (Fig. 4), making it important to consider only the σE-dependent contribution to activity. Indeed, four promoters exhibited greater activity in exponential phase in the absence of σE (e.g., yecI, plsB, ychH, and rpoH) than in its presence, even though in vitro transcription data indicate that they are σE dependent (data not shown). A plausible explanation is that these promoter fragments have overlapping σ70 promoters (50), a scenario known to be true for PrpoH (15). The activity of these additional competing promoters may be decreased in the presence of σE, due to occupancy and increased activity of the σE promoter by EσE. As it is not possible to quantify this accurately, we made the assumption here that the σE-independent activity of these promoters remains unchanged whether or not σE is present. Therefore, the quantitative arguments presented below are based on the 4 promoters that have no σE-independent promoter activity (micA, rpoE, rybB, and rseA), although all 10 promoters exhibit qualitatively similar behaviors. These results reveal two very significant differences in promoter behavior during σE-limiting and excess conditions: behavior upon entry to stationary phase and the magnitude of the UP element effect (Fig. 6A and B; Table 2).
FIG. 6.
Strengths of long and short σE-dependent promoters under basal levels of σE expression. The graphs show promoter strengths of the top 10 most active long and short σE-dependent promoters in strains expressing basal levels of σE (basal; derivatives of CAG45113) compared with promoter strengths in strains without σE (delta sE; derivatives of CAG22216). Promoter strengths were measured during the exponential phase (A) or stationary phase (B) of growth at 32°C in LB. The bars represent the averages of three independent experiments, and error bars represent 1 standard deviation. Note the delta σE activities of the long yecI and plsB promoters in exponential phase are represented numerically.
TABLE 2.
Comparison of promoter properties between basal and high (overexpression) levels of σE
| Promoterc | Fold change in promoter strength |
|||||||
|---|---|---|---|---|---|---|---|---|
| Growth phasea |
UP effectb |
|||||||
| Long (stat/exp) |
Short (stat/exp) |
Basal (long/short) |
o/Exp (long/short) |
|||||
| Basal | o/exp | Basal | o/exp | Exp | Stat | Exp | Stat | |
| micA | 27.6 | 4.8 | 14.3 | 6.3 | 13.7 | 26.4 | 3.7 | 2.8 |
| rpoE | 18.7 | 2.7 | 10.6 | 2.9 | 2.5 | 4.5 | 1.7 | 1.6 |
| rseA | 29.8 | 3.3 | 0.9 | 3.2 | 0.4 | 13.7 | 3.5 | 3.7 |
| rybB | 10.5 | 2.0 | 2.3 | 2.3 | 3.9 | 17.9 | 7.2 | 6.3 |
| plsB* | 7.4 | 3.9 | 6.9 | 5.0 | 14.3 | 15.4 | 2.9 | 2.2 |
| rpoH* | 11.4 | 2.3 | 6.0 | 2.2 | 0.5 | 1.0 | 0.8 | 0.9 |
| ybcR* | 68.4 | 2.7 | 9.3 | 2.3 | 0.3 | 2.0 | 1.4 | 1.7 |
| ychH* | 16.1 | 4.0 | 5.3 | 3.4 | 13.6 | 41.4 | 3.4 | 4.0 |
| yecI* | 3.0 | 5.0 | 3.4 | 5.3 | 3.2 | 2.8 | 4.3 | 4.1 |
| yfjO* | 8.6 | 3.7 | 6.8 | 3.6 | 4.9 | 6.2 | 1.7 | 1.8 |
| Avg | 20.1 | 3.4 | 6.6 | 3.6 | 5.7 | 13.1 | 3.1 | 2.9 |
Comparing change in promoter activity over growth phase (stationary/exponential) for long and short promoters under either basal or overexpressed (o/exp) levels of σE. Values are ratios of stationary/exponential promoter activities.
Comparing the UP effect of long and short promoter activities for basal or overexpressed (o/exp) levels of σE under either the exponential (exp) or stationary (stat) growth phase. Values are ratios of long and short promoter activities.
*, promoters exhibiting significant σE-independent activity (Fig. 6).
There was a much greater increase in promoter activity upon entry into stationary phase when σE was limiting than when it was in excess. Whereas long promoters increased an average of 20-fold and short promoters increased an average of about 7-fold under σE-limiting conditions, these same promoters increased only about 3- to 4-fold under excess σE conditions (Table 2). Importantly, these same average values are obtained whether one considers only the four completely σE-dependent promoters or whether the increase is computed over the entire data set, indicating that this is a σE-dependent property. We showed that ppGpp is responsible for these large increases in promoter activity. Upon transition into stationary phase under σE-limiting conditions, cells lacking ppGpp had little or no increase in activity of these promoters (Fig. 7), as was shown previously for rpoH and rybB (10, 11, 48).
Under σE-limiting conditions, the UP element also has a proportionately greater effect on promoter strength than under σE excess conditions. This parameter can be reliably quantified only in stationary-phase cultures, where activities of both the long promoters (with UP element) and the short promoters are relatively high (Fig. 6; see also Table S2 in the supplemental material). The presence of an UP element increased expression 15-fold under σE-limiting conditions but only 3.6-fold under σE excess conditions for the four promoters completely dependent on σE, indicating that this is a σE-dependent property. Similar effects were seen for the whole data set (Table 2). Thus, the UP element plays a major role in determining σE promoter strength under limiting σE conditions.
Our data suggest that when σE is limiting, the strength of σE-dependent promoters is very sensitive to conditions, i.e., exponential versus stationary phase and the presence/absence of UP sequences. Interestingly, UP elements at σ70-dependent promoters predominantly enhance promoter binding (KB) (21). If a similar mechanism is true at σE-dependent promoters, then this explains why UP elements have little effect under σE excess conditions, where promoters are mostly saturated with σE holoenzyme, but contribute so greatly to promoter strength under σE-limiting conditions where σE holoenzyme binding proficiency determines strength. In this regard, it is interesting that during exponential phase under basal σE levels, UP elements have little effect (Table 2), although these data were close to background and may not be reliable. Nonetheless, they suggest the interesting hypothesis that under basal conditions, there is too little available σE holoenzyme for UP elements to enhance promoter activity. Consequently, this line of reasoning suggests that the role of UP elements in σE promoters is to enable the promoter to dramatically change its strength in response to physiologically relevant changes in σE holoenzyme levels from exponential to stationary phase (Table 2; compare long and short promoter activities under basal σE levels during growth phase).
Conclusions and future prospects.
This is the first study that analyzes the complete target promoter set of a σ factor. We found that the σE promoters exhibit a large dynamic range and that promoter strength is asymmetrically distributed. There are just a few very strong promoters, somewhat more of the moderate-strength promoters, and many weak promoters. Very interestingly, promoter strength is strongly correlated with the function of the genes. The three strongest promoters all carry out regulatory roles in the σE response: the strongest promoter transcribes σE itself and its negative regulators, and the next two strongest transcribe the two sRNAs that downregulate porin production, thereby reducing the signal that generates the stress response in the first place. These promoter regions are highly conserved across the various E. coli strains and their close relatives, suggesting that their promoter strength is likely to be conserved across these organisms. This finding is somewhat reminiscent of a recent study of the PhoP regulon across multiple genomes, which found that the only targets directly regulated by PhoP in all species were the phoPQ operon itself and its negative regulator slyB (36). The moderately active promoters all carry out functions of the regulon that are conserved across many bacteria, and most of the weak promoters carry out less-conserved functions. That less-conserved functions of the regulon are generally driven by weak σE promoters is interesting and may reflect the fact that they have recently been incorporated into the regulon, either following horizontal transfer from a different organism or mutational alteration in the upstream region to permit some σE-mediated expression. The large number of low-activity promoters driving nonconserved functions underlines the tremendous plasticity of bacterial regulons.
Most of the promoters are completely σE dependent and are regulated during growth phases by ppGpp. In addition, most σE promoters require sequences upstream of their core motifs for full activity. In our previous work we identified AT-rich upstream sequences within the −65 to −35 regions of σE promoters that are candidates for α-binding UP elements. This work supports the idea that these are functional UP elements. Our ongoing in vitro transcription analysis of these promoters indicates that these sequences enhance promoter strength in the absence of additional transcription factors, further arguing that they encode UP element sequences (data not shown). This work also indicates that the σE promoters are extremely responsive to changing σE holoenzyme levels under physiological conditions, reinforcing the idea that the σE regulon is extremely dynamic, enabling cellular adaptation to a constantly changing environment.
The range of promoter strengths and their dynamics exhibited in vivo illustrate that in order to fully understand a response, promoter identification and knowledge of promoter strength is crucial. However, successful prediction of promoters and their strength from just sequence data remains a challenging problem. We are using these libraries of promoters to examine the correlation between promoter sequence and promoter strength to further improve predictive promoter models (V. A. Rhodius and V. Mutalik, unpublished data). Our findings thus far suggest that the degree of conservation of select motifs is moderately predictive of promoter strength and that higher-order relationships between motifs may be important for promoter function.
Supplementary Material
Acknowledgments
We thank Athanasios Typas for many helpful discussions and Monica Guo for careful reading of the manuscript.
This work was supported by National Institutes of Health grant GM57755 (to C.A.G.) and National Science Foundation grant MCB-0347302 (to S.E.A.).
Footnotes
Published ahead of print on 25 September 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E., L. E. Connolly, B. M. Alba, and C. A. Gross. 1999. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes Dev. 13:2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 3.Alba, B. M., J. A. Leeds, C. Onufryk, C. Z. Lu, and C. A. Gross. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes Dev. 16:2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bossi, L., and N. Figueroa-Bossi. 2007. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 65:799-810. [DOI] [PubMed] [Google Scholar]
- 6.Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other paramters of the cell by growth rate, p. 1553-1569. In R. Curtiss III, J. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, DC. [Google Scholar]
- 7.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 8.Cezairliyan, B. O., and R. T. Sauer. 2007. Inhibition of regulated proteolysis by RseB. Proc. Natl. Acad. Sci. USA 104:3771-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaba, R., I. L. Grigorova, J. M. Flynn, T. A. Baker, and C. A. Gross. 2007. Design principles of the proteolytic cascade governing the σE-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 21:124-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costanzo, A., and S. E. Ades. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costanzo, A., H. Nicoloff, S. E. Barchinger, A. B. Banta, R. L. Gourse, and S. E. Ades. 2008. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor σE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 67:619-632. [DOI] [PubMed] [Google Scholar]
- 12.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 24:373-385. [DOI] [PubMed] [Google Scholar]
- 14.De Las Penas, A., L. Connolly, and C. A. Gross. 1997. σE is an essential sigma factor in Escherichia coli. J. Bacteriol. 179:6862-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson, J. W., and C. A. Gross. 1989. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 16.Flynn, J. M., I. Levchenko, R. T. Sauer, and T. A. Baker. 2004. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 18:2292-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grigorova, I. L., R. Chaba, H. J. Zhong, B. M. Alba, V. Rhodius, C. Herman, and C. A. Gross. 2004. Fine-tuning of the Escherichia coli σE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 18:2686-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grigorova, I. L., N. J. Phleger, V. K. Mutalik, and C. A. Gross. 2006. Insights into transcriptional regulation and sigma competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl. Acad. Sci. USA 103:5332-5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruber, T. M., and C. A. Gross. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57:441-466. [DOI] [PubMed] [Google Scholar]
- 20.Guyer, M. S., R. R. Reed, J. A. Steitz, and K. B. Low. 1981. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp. Quant. Biol. 45:135-140. [DOI] [PubMed] [Google Scholar]
- 21.Haugen, S. P., W. Ross, and R. L. Gourse. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6:507-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden, J. D., and S. E. Ades. 2008. The extracytoplasmic stress factor, σE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3:e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, K. F. 1993. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 175:3401-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen, J., A. A. Rasmussen, M. Overgaard, and P. Valentin-Hansen. 2006. Conserved small non-coding RNAs that belong to the σE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 364:416-424. [DOI] [PubMed] [Google Scholar]
- 26.Kanehara, K., K. Ito, and Y. Akiyama. 2002. YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σE, RseA. Genes Dev. 16:2147-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan, S., A. Bren, A. Zaslaver, E. Dekel, and U. Alon. 2008. Diverse two-dimensional input functions control bacterial sugar genes. Mol. Cell 29:786-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, D. Y., K. S. Jin, E. Kwon, M. Ree, and K. K. Kim. 2007. Crystal structure of RseB and a model of its binding mode to RseA. Proc. Natl. Acad. Sci. USA 104:8779-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 30.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355-371. [DOI] [PubMed] [Google Scholar]
- 31.Miyashiro, T., and M. Goulian. 2007. Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc. Natl. Acad. Sci. USA 104:16305-16310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nystrom, T. 2004. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol. Microbiol. 54:855-862. [DOI] [PubMed] [Google Scholar]
- 34.Papenfort, K., V. Pfieffer, F. MIka, S. Lucchini, J. Hinton, and J. Vogel. 2006. Sigma E-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62:1674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez, J. C., and E. A. Groisman. 2009. Transcription factor function and promoter architecture govern the evolution of bacterial regulons. Proc. Natl. Acad. Sci. USA 106:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez, J. C., D. Shin, I. Zwir, T. Latifi, T. J. Hadley, and E. A. Groisman. 2009. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 5:e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, N. L., and T. L. Raivio. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191:1798-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhodius, V. A., W. C. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross, W., K. K. Gosink, J. Salomon, K. Igarashi, C. Zou, A. Ishihama, K. Severinov, and R. L. Gourse. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407-1413. [DOI] [PubMed] [Google Scholar]
- 40.Rouviere, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowley, G., M. Spector, J. Kormanec, and M. Roberts. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4:383-394. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY.
- 43.Shimada, T., H. Makinoshima, Y. Ogawa, T. Miki, M. Maeda, and A. Ishihama. 2004. Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J. Bacteriol. 186:7112-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sklar, J. G., T. Wu, L. S. Gronenberg, J. C. Malinverni, D. Kahne, and T. J. Silhavy. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 104:6400-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sohn, J., R. A. Grant, and R. T. Sauer. 2007. Allosteric activation of DegS, a stress sensor PDZ protease. Cell 131:572-583. [DOI] [PubMed] [Google Scholar]
- 46.Sohn, J., and R. T. Sauer. 2009. OMP peptides modulate the activity of DegS protease by differential binding to active and inactive conformations. Mol. Cell 33:64-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tam, C., and D. Missiakas. 2005. Changes in lipopolysaccharide structure induce the σE-dependent response of Escherichia coli. Mol. Microbiol. 55:1403-1412. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, K. M., V. A. Rhodius, and S. Gottesman. 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 189:4243-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Udekwu, K. I., and E. G. Wagner. 2007. Sigma E controls biogenesis of the antisense RNA MicA. Nucleic Acids Res. 35:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wade, J. T., D. C. Roa, D. C. Grainger, D. Hurd, S. J. Busby, K. Struhl, and E. Nudler. 2006. Extensive functional overlap between sigma factors in Escherichia coli. Nat. Struct. Mol. Biol. 13:806-814. [DOI] [PubMed] [Google Scholar]
- 51.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 52.Wollmann, P., and K. Zeth. 2007. The structure of RseB: a sensor in periplasmic stress response of E. coli. J. Mol. Biol. 372:927-941. [DOI] [PubMed] [Google Scholar]
- 53.Wu, T., J. Malinverni, N. Ruiz, S. Kim, T. J. Silhavy, and D. Kahne. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121:235-245. [DOI] [PubMed] [Google Scholar]
- 54.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 55.Zaslaver, A., A. E. Mayo, R. Rosenberg, P. Bashkin, H. Sberro, M. Tsalyuk, M. G. Surette, and U. Alon. 2004. Just-in-time transcription program in metabolic pathways. Nat. Genet. 36:486-491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.